Abstract
Post-transcriptional regulatory processes may change transcript levels and affect cell-to-cell variability or noise. We study small-RNA downregulation to elucidate its effects on noise in the iron homeostasis network of Escherichia coli. In this network, the small-RNA RyhB undergoes stoichiometric degradation with the transcripts of target genes in response to iron stress. Using single-molecule fluorescence in situ hybridization, we measured transcript numbers of the RyhB-regulated genes sodB and fumA in individual cells as a function of iron deprivation. We observed a monotonic increase of noise with iron stress but no evidence of theoretically predicted, enhanced stoichiometric fluctuations in transcript numbers, nor of bistable behavior in transcript distributions. Direct detection of RyhB in individual cells shows that its noise is much smaller than that of these two targets, when RyhB production is significant. A generalized two-state model of bursty transcription that neglects RyhB fluctuations describes quantitatively the dependence of noise and transcript distributions on iron deprivation, enabling extraction of in vivo RyhB-mediated transcript degradation rates. The transcripts’ threshold-linear behavior indicates that the effective in vivo interaction strength between RyhB and its two target transcripts is comparable. Strikingly, the bacterial cell response exhibits Fur-dependent, switch-like activation instead of a graded response to iron deprivation.
INTRODUCTION
The remarkable capability of bacteria to adapt to variations in the external environment is achieved in large part through changes in gene expression mediated by small RNAs (sRNAs) (1–3). sRNAs play major roles in diverse cellular responses to environmental cues and stresses including iron deprivation, quorum sensing, oxidative stress, carbon metabolism and more (2,4). Most sRNAs downregulate the expression of their target genes by forming imperfect duplexes with target transcripts, through mediation by the Hfq chaperone protein. sRNA–mRNA complexes are then degraded by the RNAse E degradosome, or transcript translation is inhibited by either the occlusion of the ribosome binding site or of a translational enhancer (5). In both cases, regulation is stoichiometric in character, in contrast with the catalytic regulation of gene expression by transcription factors (6).
Stoichiometric degradation gives rise to unique features such as a threshold-linear switching behavior between an efficiently silenced regime and an active regime. Silencing occurs when the rate of production of the sRNA exceeds that of its target mRNA, generating low levels of mRNA and protein. The active regime occurs when the production rate of the sRNA is smaller than that of the mRNA, leading to a linear increase of target mRNA levels (7,8). Stoichiometric degradation has also been predicted by theoretical models to give rise to an enhancement of intrinsic noise of transcripts and proteins of sRNA target genes, at the crossover between the silenced and active regimes, i.e. when the production rates of the sRNA and an mRNA target are comparable (7,9,10). Intrinsic noise is the cell-to-cell variation in gene expression of genetically identical cells (11–16), stemming from the stochastic nature of biochemical processes such as transcription and translation (17). Enhanced stoichiometric fluctuations are different from those due to small numbers, which are expected to dominate noise as sRNA production increases deeper into the silenced regime.
The iron homeostasis network of E. coli, which includes the sRNA RyhB (18), is an attractive model system to characterize sRNA regulation and test theoretical predictions. This network is controlled by the master ferric uptake regulator (Fur), a transcription factor that senses the intracellular concentration of free iron. In its iron-bound form, Fur represses the expression of many genes involved in iron acquisition, storage, oxidative stress and more. When iron is limiting, Fur becomes iron-free, relieving the repression of iron homeostasis genes. A specific target of Fur repression is ryhB, encoding an sRNA that coordinately inhibits the production of some twenty iron-using proteins (4), by targeting the degradation of their mRNAs (19,20). RyhB was reported to inhibit Fur production in a mixed double-negative feedback loop configuration (21), suggesting the possibility of bistable behavior similar to that found in double-negative feedback motifs involving two transcription factors (22,23). Bistability in a mixed double-negative feedback loop involving a sRNA and a transcription factor has been observed in a stochastic numerical analysis, but only in a limited region of parameter space (24).
Recently, the effects of sRNA regulation on protein noise have been studied within the framework of the iron homeostasis network of E. coli (25). It was found that the total protein noise from two RyhB targets is robust over a wide range of RyhB production rates, despite large reductions in the mean concentrations of both proteins. Furthermore, it was shown that this robustness is due to the dominance of extrinsic noise sources, e.g. fluctuations in the number of global factors such as RNA polymerases, ribosomes and metabolites, which affect the expression of all genes in a cell equally (14,15,26). It was suggested that extrinsic noise dominated protein fluctuations, and prevented the observation of intrinsic enhanced fluctuations in the expression of target gene proteins. No evidence for bistable behavior was observed in the distributions of protein numbers of the two RyhB target genes studied (25).
Here, we report the results of a comprehensive study of the effects of regulation by an sRNA on mRNA transcript noise, using the iron homeostasis network of E. coli as a model system. The transcript level is appropriate to measure this noise because it responds directly to sRNA regulation, and unlike the protein level, is less prone to be masked by global extrinsic noise (27). We focused on the coupled degradation of RyhB sRNA and its target mRNAs sodB, which codes for iron-superoxide dismutase, and fumA, which codes for fumarase A, one of three fumarase isozymes participating in the TCA cycle. We measured numbers of individual mRNA molecules in individual cells using single-molecule fluorescence in situ hybridization (smFISH) (26–28), and looked for evidence of enhanced stoichiometric fluctuations in the noise of target transcripts. By measuring the fluorescence of RyhB tiled with a small amount of labeled oligomers in individual cells, we characterized sRNA noise as well and its dependence on iron levels. Furthermore, we tested whether a two-state model of transcription that has been highly successful in describing the statistics of transcript copy number (27,29–31), can be generalized by adding RyhB-mediated degradation, to account for the dependence of transcript noise and distributions on iron deprivation. We also tested whether the network exhibits bistability as iron deprivation is varied, and the extent to which threshold-linear behavior is observed in the wild-type. Previous demonstrations of threshold-linear behavior were carried out by controlling RyhB production from an inducible promoter on plasmids (8). As we will show below, our measurements also reveal that far from exhibiting a graded response to changes in iron deprivation by external iron availability, RyhB regulation becomes activated in a switch-like fashion within a very narrow range of iron concentrations.
MATERIALS AND METHODS
Growth media and conditions
Bacterial strains were propagated in LB medium at 37°C overnight from a single colony inoculum. For experiments, cultures were diluted 1:100 into fresh LB medium and grown to OD600 of 0.2–0.4 before the indicated treatment. Cells were then diluted 1:100 into LB medium with different concentrations of DTPA or arabinose as indicated, and allowed to grow for 3 h to OD600 of 0.4-0.6.
Bacterial strains and plasmids
Strains carrying mutations ΔsodB, ΔfumA, ΔsufC, ΔshiA and ΔfepB are part of the Keio Collection, Keio University, Japan, (32) and are in the background of the derivative of MG1655 called BW25113, All other strains used here are derived directly from MG1655. EM1238 carries ΔryhB (33); TM338 is rne-Flag-cat and TM528 is its mutated derivative rne701-Flag-cat (34); IRE109 carries the arabinose-inducible RyhB and IRE111 the arabinose-inducible YFP; NC499 is an RNAase III mutant strain; HL1188 is an Hfq mutant from S. Gottesman (see Supplemetary Materials and Methods). More details of all strains are presented in Supplementary Table S1.
Measurements of promoter activity
MG1655 (wild-type) bacterial cells bearing plasmids with a promoter fusion pryhB–YFP, psodB–YFP and pfumA–YFP (25,35) were grown as indicated above. Cell suspensions were then transferred to a plate reader, in order to measure their fluorescence intensity Ft every  min, for a period of 1.5 h. The fluorescence intensity was corrected by subtracting the background of samples of wild-type bacterial cells, also measured throughout the same period. For each time point the promoter activity was calculated as
min, for a period of 1.5 h. The fluorescence intensity was corrected by subtracting the background of samples of wild-type bacterial cells, also measured throughout the same period. For each time point the promoter activity was calculated as 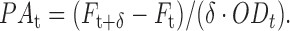 The final promoter activity PA is an average over all the values of the time series PAt, taking into account only points within the log phase of growth.
The final promoter activity PA is an average over all the values of the time series PAt, taking into account only points within the log phase of growth.
Half-life of sodB mRNA using quantitative real time PCR (qRT-PCR)
Bacterial cells were grown as indicated above. RNA lifetime measurements were carried out following previously published methods (27). Details about modifications and the subsequent qRT-PCR are given in the Supplementary Methods.
Single-molecule fluorescence in situ hybridization (smFISH)
Bacterial cells were grown and treated as indicated above. Single-molecule fluorescence in situ hybridization (smFISH) experiments were performed following previously published procedures with the indicated modifications provided here and in the Supplementary Methods (27). Images were taken on a Nikon Eclipse –Ti-E microscope controlled by the NIS-Elements software using a 100 × N.A 1.45 oil immersion phase contrast objective lens (Nikon plan-apochromat 100 × 1.45 λ) and an Andor iXon X3 EMCCD camera. All the filters are from Chroma. The filters used were ET-534/30× for excitation, 560lp as dichroic mirror and ET-585/40M for the emission. A phase contrast image was acquired followed by a z-stack of 13 slices and 250 nm spacing of fluorescent images with 2 s integration time of each slice. Each sample was imaged at multiple locations to get a total of at least 500 cells.
Direct detection of RyhB
RyhB was measured directly in individual cells by fluorescence in situ hybridization (FISH). Experiments were performed as described for smFISH through the fluorescence emitted by the labeled oligomer probes when hybridized to the complimentary RyhB transcripts. The short length of RyhB (90 nt long) allows hybridization to only three labeled oligomer probes, preventing counting of fluorescent loci as representing individual molecules as in smFISH. In spite of this, hybridization of RyhB to labeled oligomer probes can be exploited to detect RyhB directly by the mean fluorescence density in cells. Analysis of cell fluorescence was carried out following the same procedures as when analyzing fluorescently-labeled proteins (25).
Image and smFISH data analysis
Spot recognition was performed as described in the Supplementary Methods. Cell recognition is based on the MicrobeTracker suite (36), which was integrated into our custom Matlab program. To quantify the fluorescence intensity of each mRNA spot and correct for variation in cell labeling the following procedure was used: the average intensity from a diffraction-limited spot was measured. We then subtracted from the intensity of each spot in a bacterium the median of the intensity values of the pixels in this bacterium which do not contain a fluorescent spot. The variation of the mRNA numbers between experiments is ∼20% which is comparable to the errors in So et al (27). The discrete nature of the intensity of spatially separable fluorescence spots in our experiments is illustrated in Supplementary Figure S1. To estimate the amount of false positive transcript counts, we analyzed images of sodB and fumA transcripts in ΔsodB and ΔfumA strains respectively, using the same parameter values as in the analysis of images in the wild type strain. We obtained 0.04 ± 0.02 sodB transcript molecules and 0.05 ± 0.03 fumA transcript molecules, values which are ∼25 and ∼10 times smaller than the respective smallest mean transcript numbers obtained in the wild type strain at large DTPA concentrations. A similar procedure was carried out in the case of shiA and fepB transcripts. While it is customary to normalize smFISH data to a single gene copy (27,37), we have refrained from doing so with our data, since both sodB and fumA, at 37.36 and 36.31 centisomes, respectively, are quite close to the terminus region at 34 centisomes, and therefore their copy number is not expected to vary much with growth rate (38). In contrast, the ryhB gene, at 77.14 centisomes, is much closer to the oriC region (84.57 centisomes), and therefore variations in the number of copies with growth rate contribute significantly to the levels of RyhB.
Generalized two-state model of transcription regulation
A two-state model has been shown to give a satisfactory description of the stochastic properties of transcription, in particular bursting (27,29,37). According to this model, promoters can be either in an on state from which mRNA molecules are produced with a rate ktx, or in an off state, in which transcription does not occur. The rates of transition from the on to the off states and vice versa are koff and kon, respectively. Additionally, mRNA degradation can occur at a rate kdeg. When the half-life of mRNA molecules is larger than the average time in which the promoter is active 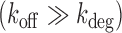 , the stationary solution for the distribution of mRNA molecules as function of the kinetic parameters is Equation (1), the negative binomial distribution (29,31):
, the stationary solution for the distribution of mRNA molecules as function of the kinetic parameters is Equation (1), the negative binomial distribution (29,31):
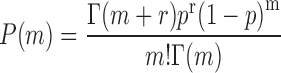 |
(1) |
where m is the number of mRNA molecules, 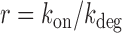 ,
,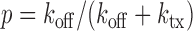 and
and 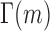 is the Gamma function. When
is the Gamma function. When 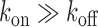 , i.e. when the gene is primarily in the on state, the negative binomial distribution reduces to a Poisson distribution. The mean μ and variance
, i.e. when the gene is primarily in the on state, the negative binomial distribution reduces to a Poisson distribution. The mean μ and variance 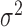 of P(m) are given respectively by
of P(m) are given respectively by 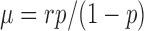 and
and 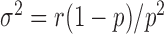 , from which the noise can be evaluated:
, from which the noise can be evaluated:
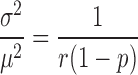 |
(2-a) |
or alternatively as:
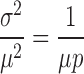 |
(2-b) |
The difference between Poissonian noise and this expression is the additional factor of 1/p in Equation (2-b).
Fitting a threshold-linear dependence to mean transcript numbers data
To carry out fits of Equation (4) for the mean transcript number per cell 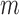 as function of the promoter activity of RyhB
as function of the promoter activity of RyhB 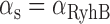 , with λ as the only fitting parameter, to the experimental data of
, with λ as the only fitting parameter, to the experimental data of 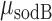 and
and 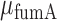 , we regarded the transcript production rates
, we regarded the transcript production rates 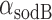 ,
, 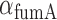 of both genes as fixed, and estimated their values from their respective degradation rates
of both genes as fixed, and estimated their values from their respective degradation rates 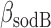 and
and 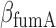 , under iron-rich conditions in which the production of RyhB can be neglected. The measured half-lives of both transcripts, 6.8 min for sodB (see Results) and 4 min for fumA (39), yield
, under iron-rich conditions in which the production of RyhB can be neglected. The measured half-lives of both transcripts, 6.8 min for sodB (see Results) and 4 min for fumA (39), yield 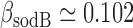 min−1 and
min−1 and 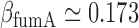 min−1. The lifetimes of both transcripts lie in the range of long values measured under iron-rich conditions in a genome-wide study of mRNA degradation in this E. coli strain (40). From Equation (4), we obtain
min−1. The lifetimes of both transcripts lie in the range of long values measured under iron-rich conditions in a genome-wide study of mRNA degradation in this E. coli strain (40). From Equation (4), we obtain 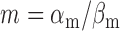 for
for 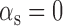 , and therefore
, and therefore 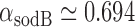 min−1 and
min−1 and 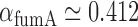 min−1. We note that while
min−1. We note that while 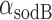 and
and 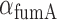 have been observed to decrease and increase respectively with the extent of iron deprivation (25), these variations are small over the regime in which fitting the present data with Equation (4) is relevant. The values of
have been observed to decrease and increase respectively with the extent of iron deprivation (25), these variations are small over the regime in which fitting the present data with Equation (4) is relevant. The values of 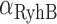 for different concentrations of DTPA were obtained from a fit of a Hill function to the measured promoter activity of RyhB shown in Figure 2A. Finally, the value of
for different concentrations of DTPA were obtained from a fit of a Hill function to the measured promoter activity of RyhB shown in Figure 2A. Finally, the value of 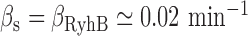 was deduced from previous measurements (19,41).
was deduced from previous measurements (19,41).
Figure 2.
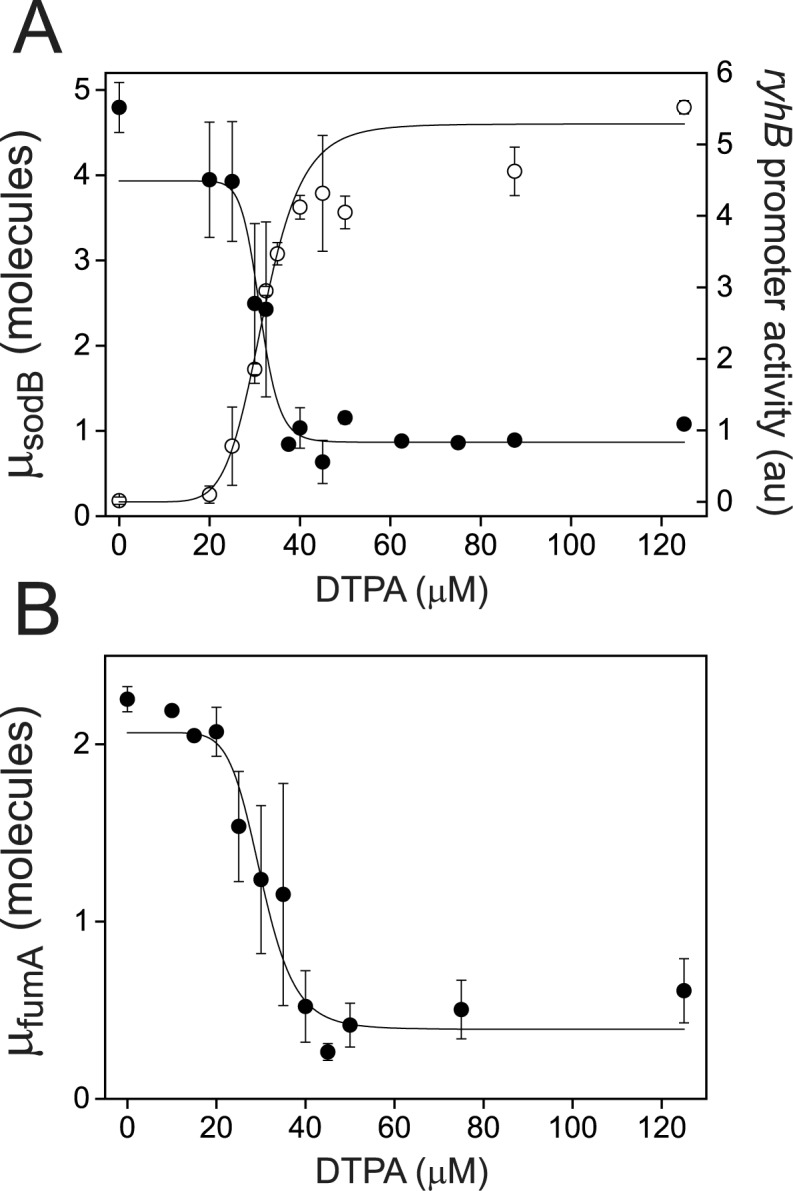
Switch-like behavior of the mean sodB and fumA transcript numbers upon iron deprivation. (A) Mean sodB transcript number μsodB in individual cells as function of the concentration of the cell-impermeable iron-chelator DTPA (black dots). Also plotted is the promoter activity of RyhB (empty circles), with reporter plasmids encoding a pryhB-yfp fusion. The black lines through both sets of data, are fits with Hill functions (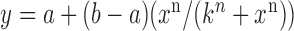 with
with 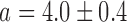 ,
, 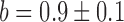 ,
, 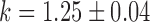 ,
, 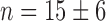 for sodB transcripts and
for sodB transcripts and 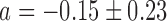 ,
, 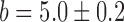 ,
, 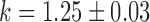 and
and 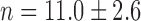 for pryhB). (B) Mean fumA transcript number μfumA in individual cells as function of the concentration of the cell-impermeable iron-chelator DTPA (black dots). Data were obtained from four independent experimental runs. Error bars represent standard errors. The black line through the data, represents a fit with a Hill function (
for pryhB). (B) Mean fumA transcript number μfumA in individual cells as function of the concentration of the cell-impermeable iron-chelator DTPA (black dots). Data were obtained from four independent experimental runs. Error bars represent standard errors. The black line through the data, represents a fit with a Hill function (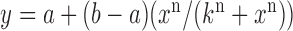 with
with 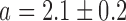 ,
, 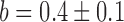 ,
, 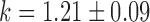 ,
, 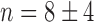 ).
).
RESULTS
Effects of iron deprivation on RyhB target transcript distributions
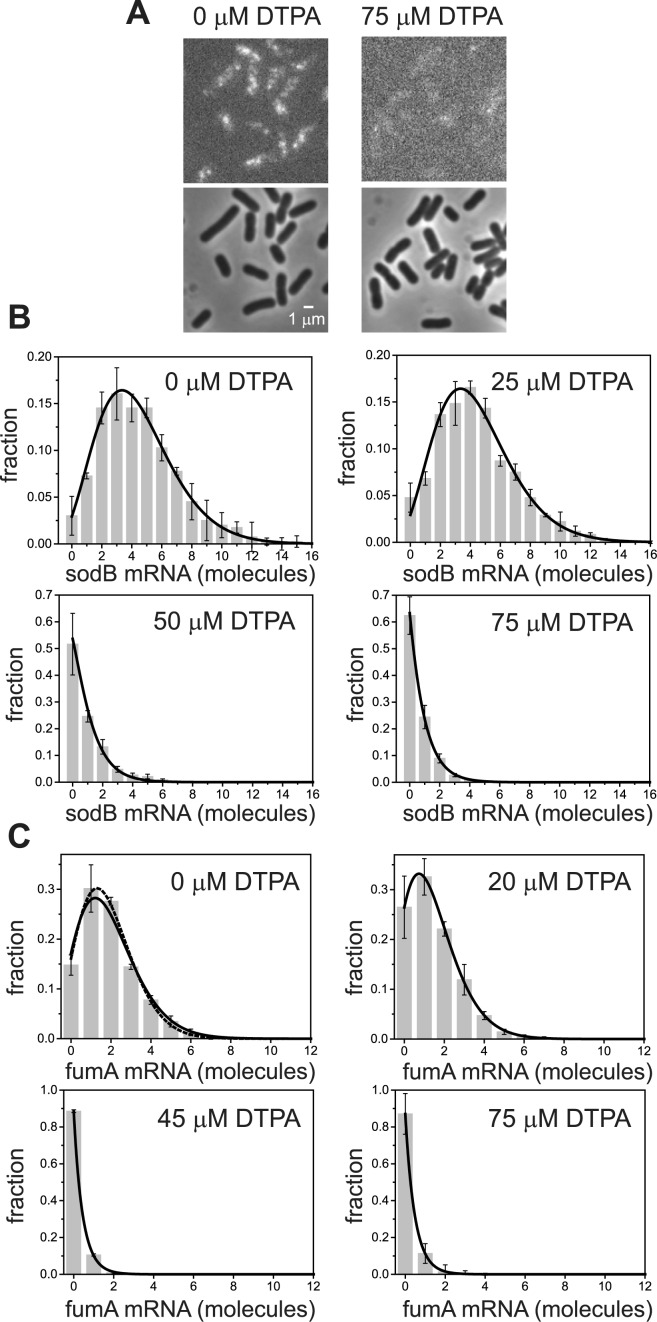
To characterize the effect of post-transcriptional regulation by RyhB on the cell–cell transcript variability of one of its main targets, sodB, we carried out smFISH measurements in individual cells, for different levels of iron deprivation. Fluorescence and phase contrast images of wild-type E. coli cells exposed to different concentrations of the cell-impermeable iron chelator diethylene triamine penta-acetic acid (DTPA) are shown in Figure 1A. Cells were fixed at least 3 h after exposure to DTPA, in order to allow adaptation to iron-poor conditions and avoid gene expression oscillations that result upon sudden exposure to DTPA (35). Spots within cells in the fluorescence images correspond to either single or few sodB transcripts, and the determination of the transcript number in each cell is carried out by quantifying localized fluorescence (see Supplementary Methods and Supplementary Figure S1), following well established procedures (42). Distributions of sodB transcript number per cell for four representative concentrations of DTPA that lead to different levels of RyhB production (43) are shown in Figure 1B. We carried out similar measurements of transcripts of fumA, another gene whose transcripts undergo stoichiometric degradation with RyhB. Distributions of fumA transcripts for four representative values of DTPA concentration are shown Figure 1C. Two features of these distributions are noteworthy. First, their respective means shift toward smaller values as the extent of iron deprivation increases. Second, each distribution corresponding to a given value of DTPA concentration is unimodal and similar to those characterizing mRNAs that are not sRNA targets (26), in spite of reports of a mixed double-negative feedback loop configuration involving RyhB and Fur (21). Such motifs may lead to bistability (3).
Figure 1.
Effects of iron deprivation on sodB and fumA transcripts visualized using single-molecule FISH (smFISH). (A) Top: typical images showing labeled sodB transcripts under iron-rich and iron-poor conditions. Bottom: phase contrast images of the same fields of view. (B) Distributions of sodB transcript number in individual cells, for the indicated concentrations of the iron chelator DTPA. The distributions represent an average over four experimental repeats while error bars represent standard errors. The solid lines are fits to the experimental data using the negative binomial distribution (Equation 1). (C) Distributions of fumA transcript number in individual cells, for the indicated concentrations of the iron chelator DTPA. The distributions represent an average over three experimental repeats while error bars represent standard errors. The solid and dashed lines are fits to the experimental data using the negative binomial Equation (1) and the Poisson distributions, respectively.
To confirm that the behavior of RyhB target transcripts we observed is indeed due to RyhB-induced degradation and components of the degradosome, we studied the effects of iron deprivation in various mutant strains (Supplementary Figure S2). As expected, we found that the mean transcript numbers of sodB and fumA are rather insensitive to iron deprivation when experiments were conducted in a ΔryhB mutant relative to the wild type strain (Supplementary Figure S2A). Similar measurements with a Δhfq mutant also display a weaker decay in mean transcript number with iron deprivation, when compared to the wild type strain (Supplementary Figure S2B). Control experiments with a strain in which RNase E function is impaired by truncation of the last 360 amino acids of the C-terminal domain (34) show that the degradation of two RyhB targets is visibly reduced with iron deprivation (Supplementary Figure S2C). Note that the three mutants in which the function of components of the degradosome was impaired, display larger values of the mean transcript number relative to the wild type strain. Finally, we also conducted similar experiments with a mutant in which the RNase III function was impaired and found that under our experimental conditions, the behavior of RyhB target transcripts was similar to the wild type (Supplementary Figure S2D), in agreement with previous findings (19).
Switch-like activation of the network with increasing iron deprivation
Our initial analysis of the effect of iron levels on the distributions of sodB and fumA mRNAs per cell revealed two types of behavior: one type characterized by a peak at a nonzero number of transcripts (at about 3–4 sodB mRNAs, as illustrated by the top histograms in Figure 1B; 1–2 fumA mRNAs, as illustrated by the top histograms in Figure 1C), and the other type having a peak at zero transcripts per cell (bottom histograms in Figure 1B and C). To monitor how the behavior of transcript distributions changes between these two types in more detail, we carried out measurements of transcript distributions with a finer resolution in DTPA concentration. Surprisingly, we found that the transition between the two types of sodB mRNA distributions occurred sharply, as illustrated by a plot of the mean sodB transcript number μ as a function of DTPA concentration (Figure 2A): μ decreases sharply from 4.9 ± 0.6 transcripts per cell to 0.9 ± 0.2, within a very narrow window of DTPA concentrations. A fit of a Hill function to the data, also shown in Figure 2A, yields an effective Hill coefficient of 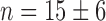 , a value that underscores the sharpness of the transition. This switch-like behavior of the mean sodB transcript number as a function of iron deprivation by addition of DTPA was confirmed using an qRT-PCR technique on total extracted sodB mRNA (Supplementary Figure S3). Switch-like behavior is also displayed by fumA transcripts as function of DTPA concentration (Figure 2B). To find out whether the switch-like change in μ is also observed upstream of sodB, we measured the promoter activity of ryhB in cell ensembles bearing plasmids with a promoter fusion, pryhB–YFP (Materials and Methods section). The promoter activity of ryhB, fitted with a Hill function is also shown in Figure 2A. Clearly, RyhB production also displays switch-like activation that mirrors the trend in the mean sodB transcript number, indicating that the origin for the switch lies upstream of ryhB transcription, i.e. in its regulation. Addition of iron to cells grown in M9 medium also resulted in a sharp reduction in the promoter activity of ryhB, and switch-like behavior of the mean sodB transcript number, with a Hill coefficient
, a value that underscores the sharpness of the transition. This switch-like behavior of the mean sodB transcript number as a function of iron deprivation by addition of DTPA was confirmed using an qRT-PCR technique on total extracted sodB mRNA (Supplementary Figure S3). Switch-like behavior is also displayed by fumA transcripts as function of DTPA concentration (Figure 2B). To find out whether the switch-like change in μ is also observed upstream of sodB, we measured the promoter activity of ryhB in cell ensembles bearing plasmids with a promoter fusion, pryhB–YFP (Materials and Methods section). The promoter activity of ryhB, fitted with a Hill function is also shown in Figure 2A. Clearly, RyhB production also displays switch-like activation that mirrors the trend in the mean sodB transcript number, indicating that the origin for the switch lies upstream of ryhB transcription, i.e. in its regulation. Addition of iron to cells grown in M9 medium also resulted in a sharp reduction in the promoter activity of ryhB, and switch-like behavior of the mean sodB transcript number, with a Hill coefficient 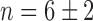 (Supplementary Figure S4). While this value of the Hill coefficient is smaller than when adding DTPA to LB
(Supplementary Figure S4). While this value of the Hill coefficient is smaller than when adding DTPA to LB 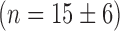 , the cooperative behavior is still significant. To test whether other genes whose transcription is regulated by Fur also display switch-like behavior independently of RyhB, we measured the mean transcript number of sufC, which like ryhB is directly repressed by Fur. Indeed, the mean sufC transcript number increases in a switch-like fashion over the same DTPA range as the mean sodB mRNA decreases (Supplementary Figure S5). In addition to stoichiometric degradation, RyhB can also down-regulate translation by blocking the ribosome binding site of a target transcript. We have tested the effects of RyhB on fepB, a target with which RyhB base-pairs in the region around the Shine-Dalgarno sequence (44). The observed increase in the mean transcript number is due to direct derepression by Fur, in agreement with previous observations (45) (Supplementary Figure S6). It is noteworthy that the mean transcript number of the shiA gene within the iron homeostasis network, whose transcripts’ translation is activated by binding to RyhB, increases with iron deprivation (Supplementary Figure S6). This observation is consistent with the increase in shiA transcript stability due to its interaction with RyhB (46). As expected, expression from chromosomal lac promoters, which are outside the iron homeostasis network, is independent of iron deprivation (25).
, the cooperative behavior is still significant. To test whether other genes whose transcription is regulated by Fur also display switch-like behavior independently of RyhB, we measured the mean transcript number of sufC, which like ryhB is directly repressed by Fur. Indeed, the mean sufC transcript number increases in a switch-like fashion over the same DTPA range as the mean sodB mRNA decreases (Supplementary Figure S5). In addition to stoichiometric degradation, RyhB can also down-regulate translation by blocking the ribosome binding site of a target transcript. We have tested the effects of RyhB on fepB, a target with which RyhB base-pairs in the region around the Shine-Dalgarno sequence (44). The observed increase in the mean transcript number is due to direct derepression by Fur, in agreement with previous observations (45) (Supplementary Figure S6). It is noteworthy that the mean transcript number of the shiA gene within the iron homeostasis network, whose transcripts’ translation is activated by binding to RyhB, increases with iron deprivation (Supplementary Figure S6). This observation is consistent with the increase in shiA transcript stability due to its interaction with RyhB (46). As expected, expression from chromosomal lac promoters, which are outside the iron homeostasis network, is independent of iron deprivation (25).
Noise of stoichiometrically degraded target RyhB transcripts increases with iron deprivation
The above measurements of transcript distributions allow us to test experimentally for the existence of theoretically predicted enhanced intrinsic fluctuations of target transcripts that undergo stoichiometric degradation with RyhB. To this end, we quantified the cell-to-cell variability of sodB and fumA transcripts as a function of iron deprivation by calculating the transcript noise, defined by the nondimensional ratio 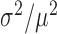 , where σ is the standard deviation and μ the mean of mRNA numbers per cell. Different values of μ were obtained by varying the DTPA concentration that cells were exposed to. Data were obtained from seven independent experimental runs in the case of sodB transcripts, color-coded to illustrate reproducibility, with four to seven different concentrations of DTPA per run, covering different regimes. Plots of the transcript noise of sodB and fumA as a function of their respective means μ are shown in Figure 3A and B. Transcript noise increases monotonically in both cases as the average number of transcripts decreases and as RhyB transcription increases. This contrasts sharply with measurements of SodB and FumA protein noise, which remain constant over a wide range of RyhB production rates (25). It is noteworthy that there is no signature of a theoretically predicted peak in transcript noise within the range of values of μ of our experiments (7,10). A plot of the ratios of the promoter activities of RyhB and those of sodB or fumA, as a function of the concentration of DTPA, shown in Supplementary Figure S7, verifies that our measurements access the regime in which enhanced stochastic fluctuations are expected, i.e. when the production rates of RyhB and either of the two genes are comparable (10).
, where σ is the standard deviation and μ the mean of mRNA numbers per cell. Different values of μ were obtained by varying the DTPA concentration that cells were exposed to. Data were obtained from seven independent experimental runs in the case of sodB transcripts, color-coded to illustrate reproducibility, with four to seven different concentrations of DTPA per run, covering different regimes. Plots of the transcript noise of sodB and fumA as a function of their respective means μ are shown in Figure 3A and B. Transcript noise increases monotonically in both cases as the average number of transcripts decreases and as RhyB transcription increases. This contrasts sharply with measurements of SodB and FumA protein noise, which remain constant over a wide range of RyhB production rates (25). It is noteworthy that there is no signature of a theoretically predicted peak in transcript noise within the range of values of μ of our experiments (7,10). A plot of the ratios of the promoter activities of RyhB and those of sodB or fumA, as a function of the concentration of DTPA, shown in Supplementary Figure S7, verifies that our measurements access the regime in which enhanced stochastic fluctuations are expected, i.e. when the production rates of RyhB and either of the two genes are comparable (10).
Figure 3.
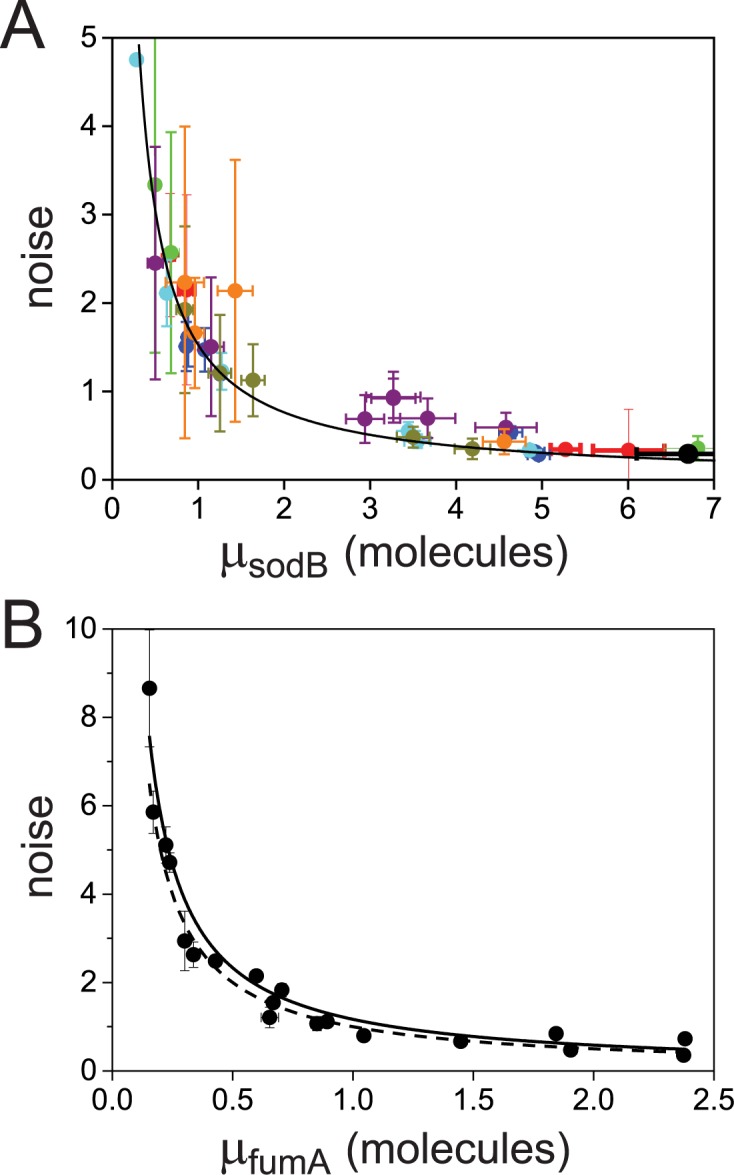
Noise of sodB and fumA transcripts as a function of their respective mean transcript number per cell. (A) Noise of sodB transcripts measured by the ratio 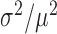 , where σ is the standard deviation and μ the mean of mRNA numbers per cell. The data were obtained from seven independent smFISH runs in which cells were exposed to different concentrations of DTPA for 3 h prior fixation, each color representing one experimental run, to illustrate the reproducibility of the data. Error bars were determined from 1000 bootstrap samples. The solid line is a fit to the experimental data using the expression for noise Equation (2-b) obtained from the negative binomial distribution Equation (1). The noise versus mean in a RyhB-deleted strain is shown with a black circle. Error bars are the standard error from three independent experiments. (B) Transcript noise as a function of
, where σ is the standard deviation and μ the mean of mRNA numbers per cell. The data were obtained from seven independent smFISH runs in which cells were exposed to different concentrations of DTPA for 3 h prior fixation, each color representing one experimental run, to illustrate the reproducibility of the data. Error bars were determined from 1000 bootstrap samples. The solid line is a fit to the experimental data using the expression for noise Equation (2-b) obtained from the negative binomial distribution Equation (1). The noise versus mean in a RyhB-deleted strain is shown with a black circle. Error bars are the standard error from three independent experiments. (B) Transcript noise as a function of 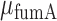 the mean number of fumA mRNA molecules per cell. Noise is defined by the ratio
the mean number of fumA mRNA molecules per cell. Noise is defined by the ratio 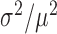 , where
, where 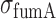 is the standard deviation of fumA mRNA molecules. The data were obtained from two independent smFISH runs shown separately, in which cells were exposed to different concentrations of DTPA for 3 h prior fixation. Error bars were determined from 1000 bootstrap samples. The solid line is a fit to the experimental data using the expression for noise Equation (2-b) obtained from the negative binomial distribution Equation (1), while the dashed line represents the noise of a Poisson distribution
is the standard deviation of fumA mRNA molecules. The data were obtained from two independent smFISH runs shown separately, in which cells were exposed to different concentrations of DTPA for 3 h prior fixation. Error bars were determined from 1000 bootstrap samples. The solid line is a fit to the experimental data using the expression for noise Equation (2-b) obtained from the negative binomial distribution Equation (1), while the dashed line represents the noise of a Poisson distribution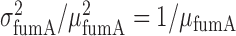 .
.
We reasoned that the narrowness of the window of DTPA concentrations spanning the switching region, over which RyhB production changes from values close to leaking production on one hand, to high production on the other (Figure 2A), may preclude the observation of enhanced fluctuations in transcript noise. In spite of repeated attempts with increasing resolution in changes of iron deprivation, the noise was observed to increase monotonously with iron stress for both sodB and fumA transcripts, as shown in Figure 3. We therefore constructed a mutant strain in which RyhB production is independent of iron deprivation and controlled from an inducible arabinose promoter. The dose response of this promoter fused to YFP, shown in Supplementary Figure S8, exhibits graded behavior. The effects of RyhB induction on the sodB mean transcript number in this strain are shown in Supplementary Figure S9. The measurements of sodB transcript noise in this strain as function of the sodB transcript number (Supplementary Figure S10), display fully monotonic behavior and largely overlap with those measured in the wild-type strain as function of DTPA concentration. There is no discernible evidence of a peak in noise due to enhanced stoichiometric fluctuations.
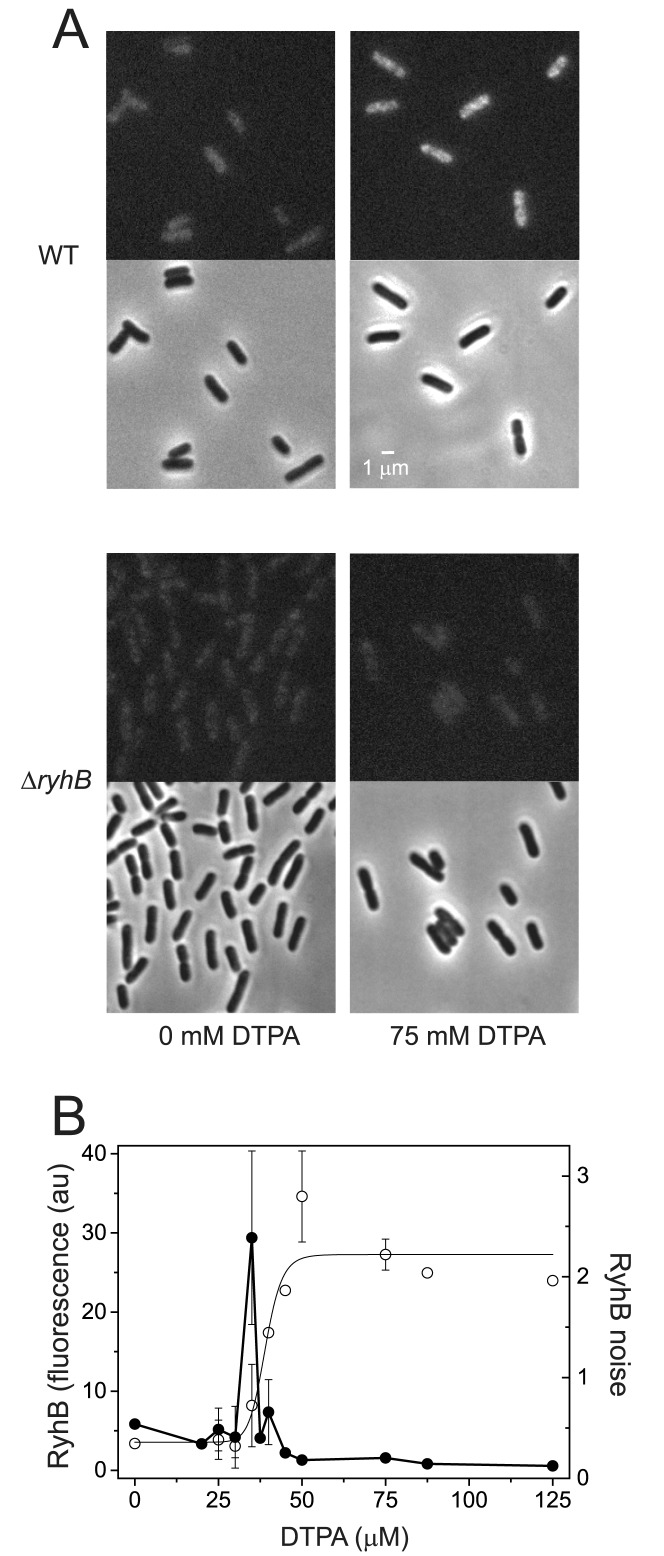
Direct measurement of RyhB and its noise as function of iron deprivation
Considerable insights can be obtained by monitoring RyhB and its cell–cell variability by detecting RyhB directly in individual cells as a function of iron deprivation, using FISH methods. FISH techniques are not used here in single-molecule modality, since RyhB molecules are tiled by maximum three oligomeric probes and therefore their intensity is close to that of the background. Furthermore, individual RyhB molecules cannot easily be resolved given their large number (Materials and Methods). Typical snapshots of cells displaying fluorescence from labeled RyhB under rich and poor iron conditions (0 and 75 μM DTPA, respectively) are shown in Figure 4A, for both the wild-type strain and a control ΔryhB mutant. Two features are noteworthy: first, RyhB production clearly increases with iron deprivation and second, a comparison with the control strain indicates that RyhB production can also be detected under iron-rich conditions. Consistent with this, sodB and fumA mean transcript numbers are higher in the ΔryhB mutant, as well as in other mutants in which components of the degradosome are impaired (Supplementary Figure S2). The mean fluorescence density of RyhB-defined as the total cell fluorescence divided by the cell's area, averaged over the cell population is shown in Figure 4B. The behavior of the mean fluorescence density of RyhB recapitulates the switch-like characteristics of the RyhB promoter activity. To estimate what fraction of the total RyhB production is detected in these experiments, we compared the mean fluorescence density of RyhB measured in an RNase E mutant strain in which stoichiometric degradation is abrogated, with the wild type, at 75 μM DTPA (Supplementary Figure S11). The ratio between the two is 1.8 ± 0.7 under conditions in which the production rate of RyhB is high, indicating that about half of the total RyhB produced is degraded with its targets and demonstrating that RyhB sRNA is not a limiting factor at low iron levels. We have also characterized the dependence of the noise in RyhB production as a function of DTPA concentration. A plot of RyhB noise is shown in Figure 4B, while distributions of RyhB in individual cells for selected DTPA concentrations are shown in Supplementary Figure S12. Significantly, a narrow, strong peak in RyhB noise is observed, exposing strong fluctuations in RyhB concentration within the switching regime. It is also noteworthy that RyhB noise is 5–10 times smaller than sodB transcript noise above the switch, a regime in which RyhB production is large. The noise of both RyhB and sodB are comparable below the switch. Control measurements of RyhB noise in an RNase E mutant strain in which stoichiometric degradation is abrogated, at 0 and 75 μM DTPA also display small noise levels.
Figure 4.
Direct measurement of RyhB in individual cells and its noise. (A) Top: typical snapshots of fluorescence from undegraded intracellular RyhB, tiled with labeled oligonucleotides under iron-rich (0 μM DTPA) and iron-poor (75 μM DTPA) conditions, in wild-type cells. The corresponding phase contrast images of the same fields of view are appended below each frame. Bottom: control images obtained in a ΔryhB background under the same conditions. (B) Empty circles: Switch-like behavior of the fluorescence density per cell from undegraded intracellular RyhB, tiled with labeled oligonucleotides, as function of DTPA concentration. The density is defined as the total cell fluorescence divided by the cell area. The line is a fit with a Hill function with 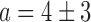 ,
, 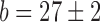 ,
, 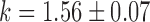 and
and 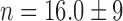 . Full circles: RyhB noise
. Full circles: RyhB noise 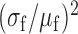 , where
, where 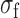 and
and 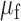 are the standard deviation and mean of the fluorescence density of undegraded intracellular labeled RyhB. Values of the fluorescence density and of the noise represent means over three independent experiments and the error bars represent standard errors. Crosses: control measurements of RyhB noise measured in an RNase E mutant strain in which stoichiometric degradation is abrogated, at 0 and 75 μM DTPA.
are the standard deviation and mean of the fluorescence density of undegraded intracellular labeled RyhB. Values of the fluorescence density and of the noise represent means over three independent experiments and the error bars represent standard errors. Crosses: control measurements of RyhB noise measured in an RNase E mutant strain in which stoichiometric degradation is abrogated, at 0 and 75 μM DTPA.
Kinetic parameters within the framework of a two-state model
We tested the applicability of the generalized two-state model (Materials and Methods) to describe the variability of targets whose degradation rate is not a constant, but is modulated by an sRNA. We assumed a mean field approximation in which the stochasticity of sRNA levels is smaller than the intrinsic fluctuations in mRNA production, and that Hfq is not a limiting factor. Therefore, degradation is set by the mean number of sRNA molecules in the cells.
The experimental noise data are well fitted by Equation (2-b), taking a fixed value of 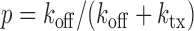 , where ktx is the transcription rate and koff is the rate of switching back to the off state, when the promoter is active. Fits for the noise of sodB and fumA transcripts are shown in Figure 3A and B for different experiments in which the concentration of DTPA is varied. In the case of sodB, the fit yields ± 0.01, from which we obtain
, where ktx is the transcription rate and koff is the rate of switching back to the off state, when the promoter is active. Fits for the noise of sodB and fumA transcripts are shown in Figure 3A and B for different experiments in which the concentration of DTPA is varied. In the case of sodB, the fit yields ± 0.01, from which we obtain 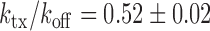 , indicating that transcription and the transition to an inactive promoter state occur on similar timescales, and that both processes are not appreciably affected by iron deprivation. This value of the ratio
, indicating that transcription and the transition to an inactive promoter state occur on similar timescales, and that both processes are not appreciably affected by iron deprivation. This value of the ratio 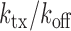 , falls within the range of variation expected from measurements carried out on seven different promoters (27). We checked that if the ratio
, falls within the range of variation expected from measurements carried out on seven different promoters (27). We checked that if the ratio 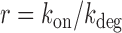 between the rates of gene activation
between the rates of gene activation 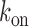 and degradation
and degradation 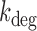 is taken as fixed, the fit to the data is quite poor, suggesting that
is taken as fixed, the fit to the data is quite poor, suggesting that 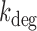 is altered when the DTPA levels are modified, as expected. Additionally, the transcript histograms are well fitted by the negative binomial distribution (Equation 1) over the whole range of DTPA concentrations, as illustrated in Figure 1B, using r as the only fitting parameter and the value of P obtained above. Similarly, in the case of fumA, a fit of Equation (2-b) to the data yields
is altered when the DTPA levels are modified, as expected. Additionally, the transcript histograms are well fitted by the negative binomial distribution (Equation 1) over the whole range of DTPA concentrations, as illustrated in Figure 1B, using r as the only fitting parameter and the value of P obtained above. Similarly, in the case of fumA, a fit of Equation (2-b) to the data yields 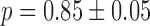 , a value that is closer to
, a value that is closer to  than for sodB transcripts, suggesting that the transcript noise of fumA has a more Poissonian character. While a Poisson distribution without fitting parameters describes the data well (Figure 3B), the negative binomial distribution is characterized by an extra degree of freedom that allows us to evaluate the RyhB-induced degradation rate. Fits of experimentally-measured fumA transcript histograms with negative binomial distributions are shown in Figure 1C.
than for sodB transcripts, suggesting that the transcript noise of fumA has a more Poissonian character. While a Poisson distribution without fitting parameters describes the data well (Figure 3B), the negative binomial distribution is characterized by an extra degree of freedom that allows us to evaluate the RyhB-induced degradation rate. Fits of experimentally-measured fumA transcript histograms with negative binomial distributions are shown in Figure 1C.
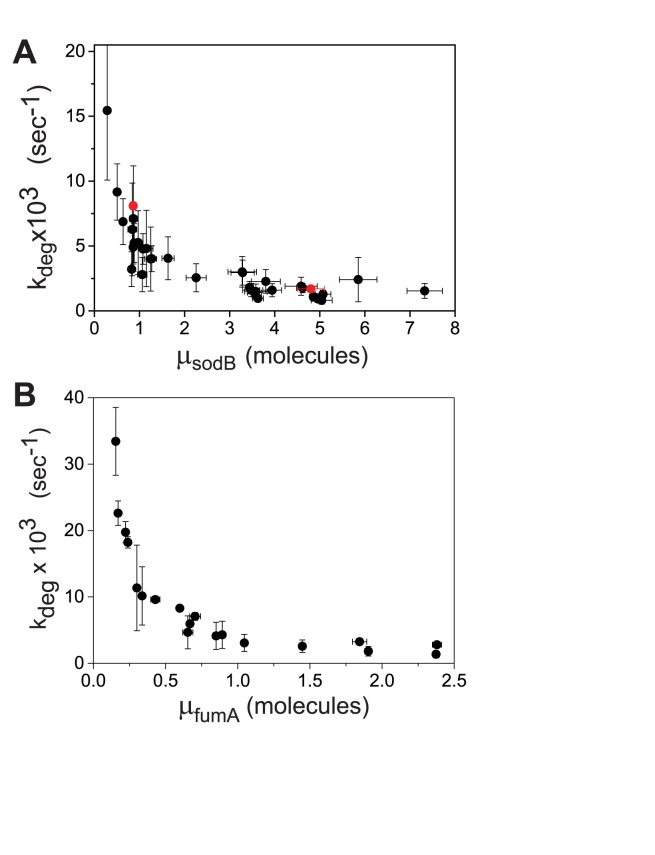
Using the values of p for sodB and fumA transcripts, one can now extract the respective in vivo degradation rates and their dependence on the degree of iron deprivation. 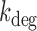 can be obtained from the relation
can be obtained from the relation 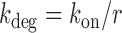 using Equation (2-a). To determine kon in the case of sodB, we carried out measurements of the half-life of sodB transcripts for 0 and 75 μM DTPA using qRT-PCR and a transcription inhibitor (Materials and Methods). These concentrations of DTPA are representative of low and high expression levels of RyhB. The values we obtained,
using Equation (2-a). To determine kon in the case of sodB, we carried out measurements of the half-life of sodB transcripts for 0 and 75 μM DTPA using qRT-PCR and a transcription inhibitor (Materials and Methods). These concentrations of DTPA are representative of low and high expression levels of RyhB. The values we obtained, 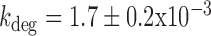 s−1 and 8.1 ± 0.3 × 10−3 s−1, respectively (plotted in Figure 5A), are in good agreement with the values obtained from our analysis based on the two-state model when choosing
s−1 and 8.1 ± 0.3 × 10−3 s−1, respectively (plotted in Figure 5A), are in good agreement with the values obtained from our analysis based on the two-state model when choosing 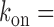 0.01 s−1, a value that falls well within the range of values measured for different promoters in E. coli, which can display approximately a ten-fold variation (21). The dependence of
0.01 s−1, a value that falls well within the range of values measured for different promoters in E. coli, which can display approximately a ten-fold variation (21). The dependence of 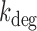 on the sodB mean transcript number μ is shown in Figure 5A. The degradation rate increases about 8-fold relative to the value under iron-rich conditions, as the mean transcript number decreases over the full range of values of DTPA we tested. From the
on the sodB mean transcript number μ is shown in Figure 5A. The degradation rate increases about 8-fold relative to the value under iron-rich conditions, as the mean transcript number decreases over the full range of values of DTPA we tested. From the 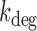 values obtained from the qRT-PCR measurements we calculated the corresponding half-lives under these conditions: t1/2 = 6.74 ± 0.7 min and t1/2 = 1.43 ± 0.1 min, respectively (Supplementary Figure S13). These values are in agreement with half-life measurements of sodB transcripts reported previously (19). Recently the association/dissociation of the SgrS sRNA interacting with two of its targets has been studied using super-resolution microscopy and mean-field kinetic equations, which neglect any fluctuations including bursty transcription and possible limitation of Hfq (47). The degradation rates obtained in that study are comparable to those we observed under conditions in which RyhB production is high (e.g. at 75 μM DTPA).
values obtained from the qRT-PCR measurements we calculated the corresponding half-lives under these conditions: t1/2 = 6.74 ± 0.7 min and t1/2 = 1.43 ± 0.1 min, respectively (Supplementary Figure S13). These values are in agreement with half-life measurements of sodB transcripts reported previously (19). Recently the association/dissociation of the SgrS sRNA interacting with two of its targets has been studied using super-resolution microscopy and mean-field kinetic equations, which neglect any fluctuations including bursty transcription and possible limitation of Hfq (47). The degradation rates obtained in that study are comparable to those we observed under conditions in which RyhB production is high (e.g. at 75 μM DTPA).
Figure 5.
Degradation rates 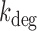 of sodB and fumA transcripts calculated from an analysis of noise data with a generalized two-state model of bursty transcription. (A) Degradation rate
of sodB and fumA transcripts calculated from an analysis of noise data with a generalized two-state model of bursty transcription. (A) Degradation rate 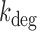 of sodB mRNA as function of the mean sodB transcript number. The values of
of sodB mRNA as function of the mean sodB transcript number. The values of 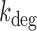 for sodB transcripts were obtained from Equation (2-a), with the value of
for sodB transcripts were obtained from Equation (2-a), with the value of 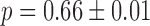 obtained from a fit to the noise (Figure 3A) and
obtained from a fit to the noise (Figure 3A) and 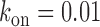 s−1. Error bars were determined from propagation of the noise data (Figure 3A), via Equation (2-a). Red points represent independent measurements of
s−1. Error bars were determined from propagation of the noise data (Figure 3A), via Equation (2-a). Red points represent independent measurements of 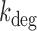 using qRT-PCR at 0 and 75 μM DTPA (Supplementary Figure S13). (B) Degradation rate
using qRT-PCR at 0 and 75 μM DTPA (Supplementary Figure S13). (B) Degradation rate 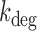 of fumA mRNA as function of the mean fumA transcript number. The values of
of fumA mRNA as function of the mean fumA transcript number. The values of 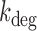 were obtained from Equation (2-a) using the value of
were obtained from Equation (2-a) using the value of 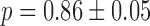 obtained from a fit to the noise data (Figure 3B) and
obtained from a fit to the noise data (Figure 3B) and 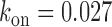 s−1. Error bars were determined from 1000 bootstrap samples of the noise data and propagated via Equation (2-a).
s−1. Error bars were determined from 1000 bootstrap samples of the noise data and propagated via Equation (2-a).
To check what the degradation rate of sodB transcripts is in the absence of RyhB production, we carried out smFISH measurements of sodB transcripts in a ryhB-deleted strain under iron-rich conditions. We obtained a mean sodB transcript number of 6.7 ± 0.6, within the range of values obtained for the RyhB wild type strain under iron-rich conditions (4.9 ± 0.6 transcripts per cell). The noise 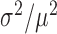 as a function of mean for this strain is shown in Figure 3A. The data point falls within experimental error on the same curve as the data for the wild-type strain, allowing us to use the same parameters as for the wild-type in the determination of
as a function of mean for this strain is shown in Figure 3A. The data point falls within experimental error on the same curve as the data for the wild-type strain, allowing us to use the same parameters as for the wild-type in the determination of 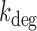 using the two-state model. We obtained
using the two-state model. We obtained 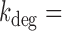 1.0 ± 0.1 × 10−3 s−1 (t1/2 = 11.5 ± 1.2 min), in fair agreement with values for the wild type under iron-rich conditions, consistent with the literature (27). The slightly higher degradation rate values for the wild type could reflect RyhB promoter leakage.
1.0 ± 0.1 × 10−3 s−1 (t1/2 = 11.5 ± 1.2 min), in fair agreement with values for the wild type under iron-rich conditions, consistent with the literature (27). The slightly higher degradation rate values for the wild type could reflect RyhB promoter leakage.
The degradation rate of fumA transcripts for different levels of iron deprivation can be evaluated following a similar procedure. Using 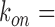 0.01 s−1 as for sodB transcripts yields t1/2∼6 min for the half-life of fumA transcripts under iron-rich conditions, a value which is higher than measured values by ∼50% (39). To match the measured half-life of fumA transcripts (∼4 min), a higher value,
0.01 s−1 as for sodB transcripts yields t1/2∼6 min for the half-life of fumA transcripts under iron-rich conditions, a value which is higher than measured values by ∼50% (39). To match the measured half-life of fumA transcripts (∼4 min), a higher value, 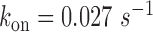 is needed. Using the latter yields the results plotted in Figure 5B. The RyhB-dependent degradation rate of fumA transcripts is higher than for sodB transcripts.
is needed. Using the latter yields the results plotted in Figure 5B. The RyhB-dependent degradation rate of fumA transcripts is higher than for sodB transcripts.
Estimate of the in vivo effective interaction strength between RyhB and its target sodB and fumA transcripts
A simpler mean-field kinetic model for sRNA-mediated silencing that neglects bursty mRNA production and fluctuations leads to the characteristic threshold-linear switching behavior of stoichiometric regulation (8). According to this model, cellular numbers of an sRNA (s) and a target mRNA (m) obey the following rate equations:
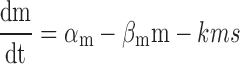 |
(3) |
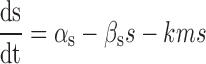 |
where 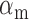 and
and 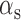 are the effective – constant – production rates of the target and sRNA, respectively,
are the effective – constant – production rates of the target and sRNA, respectively, 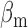 and
and 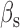 the respective degradation rates, and the second order kinetic constant k is a measure of the effective strength of the sRNA–mRNA interaction. Under steady-state conditions, the solution to Equation (3) is:
the respective degradation rates, and the second order kinetic constant k is a measure of the effective strength of the sRNA–mRNA interaction. Under steady-state conditions, the solution to Equation (3) is:
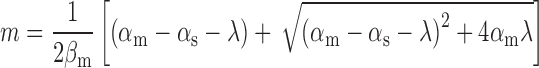 |
(4) |
where 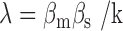 . We have plotted the mean transcript per cell
. We have plotted the mean transcript per cell 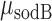 and
and 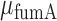 as a function of the promoter activity
as a function of the promoter activity 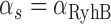 in Figure 6. Fits of Equation (4) to both sets of data with λ as a fitting parameter and fixed values of the respective promoter activities
in Figure 6. Fits of Equation (4) to both sets of data with λ as a fitting parameter and fixed values of the respective promoter activities 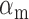 (Materials and Methods) are shown in Figure 6. From the values of λ derived from the best fits we obtain
(Materials and Methods) are shown in Figure 6. From the values of λ derived from the best fits we obtain 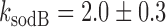 (x10−3 min−1) and
(x10−3 min−1) and 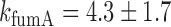 (x10−3 min−1).
(x10−3 min−1).
Figure 6.
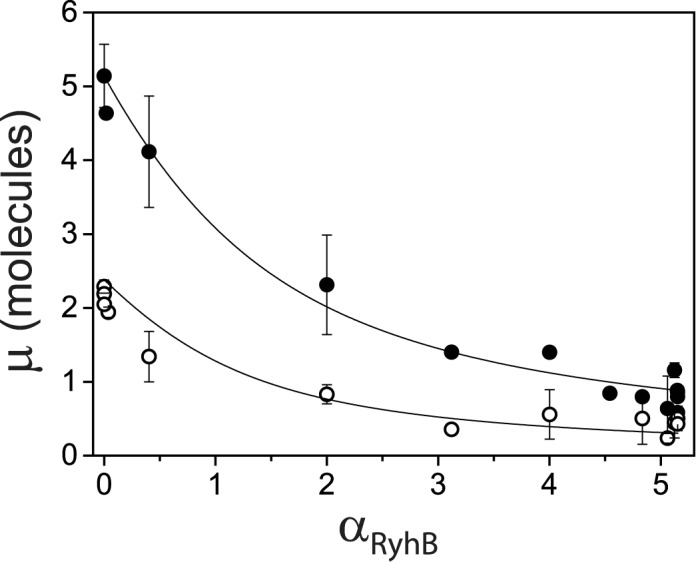
Threshold-linear switching and in vivo effective interaction strength between RyhB and its target sodB and fumA transcripts. Mean transcript per cell 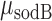 (full circles) and
(full circles) and 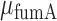 (empty circles) as function of
(empty circles) as function of 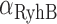 , the promoter activity of RyhB (Figure 2A). The lines represent best fits of Equation (4) to both sets of data, with either
, the promoter activity of RyhB (Figure 2A). The lines represent best fits of Equation (4) to both sets of data, with either 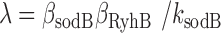 or
or 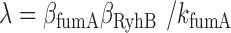 as a fitting parameters, where
as a fitting parameters, where 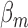 are the degradation rates of either of the two gene transcripts,
are the degradation rates of either of the two gene transcripts, 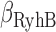 that of RyhB, and k is the effective strength of interaction between RyhB and either of the transcripts sodB or fumA. Error bars represent standard errors over at least three experimental runs.
that of RyhB, and k is the effective strength of interaction between RyhB and either of the transcripts sodB or fumA. Error bars represent standard errors over at least three experimental runs.
DISCUSSION
In the present work, we have studied how the post-transcriptional downregulation of gene expression by the sRNA RyhB affects various aspects of the cellular response to graded changes in iron availability. Our experiments were carried out at the RNA level, where RyhB naturally exerts its effects. At the RNA level, fluctuations are dominated by intrinsic noise sources (27), in contrast to the dominance of extrinsic fluctuations when protein levels are assessed (25,26). We discuss below the impact of RyhB on various aspects of the cellular response to iron scarcity.
Our analysis of the dependence of transcript distributions on iron availability shows that the two-state model, which has been used successfully to describe bursts of transcription created by infrequent initiations by RNA polymerase in individual cells (27), can be generalized to include RyhB-mediated transcript degradation. Remarkably, this analysis demonstrates that the resulting negative binomial distribution describes well the experimentally measured transcript distributions over the whole range of DTPA concentrations by changing only one parameter, namely the in vivo rate of degradation 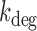 . RyhB increases the degradation rates of sodB and fumA mRNA, by a factor of five or more when comparing iron-rich to iron-poor conditions.
. RyhB increases the degradation rates of sodB and fumA mRNA, by a factor of five or more when comparing iron-rich to iron-poor conditions.
Analysis of mean transcript numbers of both sodB and fumA as the levels of iron deprivation vary, in terms of a mean-field threshold-linear model behavior has allowed us to estimate k, the effective in vivo strength of interaction between RyhB and each of its two targets sodB and fumA transcripts. Remarkably, the value of 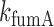 is about twice larger than that of
is about twice larger than that of 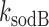 . While it has been considered that sodB mRNA is the target whose expression RyhB affects most strongly (20), the relative reductions we observe in the abundances of sodB and fumA transcripts as function of iron availability are comparable. Moreover they are similar to those observed in other experiments (48,49), and at the level of the respective proteins (17). It is noteworthy that the predicted hybridization energies between RyhB and either sodB or fumA transcripts differ by only ∼10%, including corrections due to molecular unfolding (50,51). We stress however that in addition to these factors, k includes contributions from Hfq-mediated binding and the degradosome (52), as well as effects due to possible different spatial colocalizations of sodB or fumA transcripts with RyhB (47).
. While it has been considered that sodB mRNA is the target whose expression RyhB affects most strongly (20), the relative reductions we observe in the abundances of sodB and fumA transcripts as function of iron availability are comparable. Moreover they are similar to those observed in other experiments (48,49), and at the level of the respective proteins (17). It is noteworthy that the predicted hybridization energies between RyhB and either sodB or fumA transcripts differ by only ∼10%, including corrections due to molecular unfolding (50,51). We stress however that in addition to these factors, k includes contributions from Hfq-mediated binding and the degradosome (52), as well as effects due to possible different spatial colocalizations of sodB or fumA transcripts with RyhB (47).
Transcript distributions for both genes are unimodal for the range of DTPA concentrations in our experiments. While iron deprivation induces the expression of RyhB through derepression by Fur, RyhB was reported to downregulate Fur, albeit indirectly (21), thereby establishing a double-negative feedback loop. A double-negative feedback motif may give rise to bistable behavior in the case of two transcription factors negatively regulating each other (22), suggesting similar behavior could be observed for the mixed sRNA-transcription factor motif (24,53). The notion of bimodal behavior has also been considered in the case of the sRNA MicF, which forms a double negative feedback loop with the global transcription factor Lrp (54). The unimodal character of the distributions we observe does not support this notion. Consistent with our observations, Gillespie simulations of a mixed loop involving a transcription factor and an sRNA show a limited region in parameter space where such a loop exhibits bistability (24). While the absence of bistability does not contradict the existence of a mixed double-negative feedback loop involving Fur and RyhB, we note that other biochemical studies have failed to reproduce this motif (48).
The predominance of extrinsic noise sources in controlling cell–cell variations at the protein level was suggested in our earlier work as a possible factor masking the theoretically predicted enhanced fluctuations in the crossover regime, which separates the strongly repressed, from the expressing regime characteristic of intrinsic noise in stoichiometrically dominated systems (25). Extrinsic noise stems from cell–cell variations in factors such as RNA polymerases, ribosomes and metabolites that affect the expression of all genes equally. In contrast, intrinsic noise originates from the stochastic nature of biochemical processes such as transcription and translation, and manifests itself as differences in expression between two copies of the same promoter, each labeled with a different reporter, within the same cell (14,55). The present experiments, conducted at the transcript level in which intrinsic noise is dominant, reveal no evidence of a peak in noise due to enhanced stoichiometric fluctuations, within a range of RyhB production rates in which the mean transcript number decreases for almost its full span. We surmised that the very narrow range of iron concentrations over which switch-like activation of RyhB production takes place in the wild-type strain prevented us from observing enhanced near-critical fluctuations, and therefore constructed a strain in which RyhB production is graded, by expressing it from an inducible promoter. No enhanced fluctuations were observed in this strain either. A number of factors, which we now discuss, may account for their absence: the possible limiting nature of Hfq (56,57); the presence of auxiliary weak RyhB targets (6), the presence of highly expressed 3′ external transcribed spacer (3′ ETS) fragments of pre-tRNAs (58) and finally, inefficient binding and degradation of RyhB with its targets (8,10).
Previous calculations of enhanced fluctuations did not take any of these factors into account, except for inefficient binding (7,9,10). Hfq limitation may be significant under certain conditions (56,57), leading to a reduction in target transcript-RyhB coupling and thus target degradation, thereby limiting the range of possible fluctuations for given production rates of RyhB and its targets transcripts. The fact that sodB transcript numbers do not decay to zero at high concentrations of DTPA for which undegraded RyhB is directly detected, is consistent with Hfq limitation. Calculations indicate that the presence of auxiliary weak targets diminishes the correlation between sRNA numbers and those of main transcripts, reducing significantly fluctuations (6). The significant numbers of predicted RyhB targets that remain to be validated (18,44,50,51), precludes experiments in which mutations in both RyhB and sodB would make the latter a sole target and thus provide a more stringent test of this notion (38). Lastly, numerical calculations indicate that the strength of enhanced fluctuations depends strongly on k (see Equation 3), the effective strength of the sRNA–mRNA interaction (10). As k grows, enhanced fluctuations are larger and at the same time, threshold-linear switching becomes sharper (59). The rounded behavior of the threshold-linear response of both sodB and fumA transcripts we observed (Figure 5) yields instead moderate values k for both transcripts. This suggests that enhanced fluctuations may not be large enough to be observable. It has recently been reported that the abundant 3′ ETSleuZ fragment of the glyW-cysT-leuZ polycistronic tRNA precursor base pairs with RyhB, sequestering the latter and thus acting as a sponge that may buffer noise (58). Disruption of this pairing increases RyhB activity significantly, even under iron-rich conditions. Interestingly, Hfq mediates the interaction between the polycistronic tRNA transcript and RyhB (58,60). Finally, the presence of auxiliary targets, together with the switch-like production of RyhB, contrive on one hand to attenuate the presence of enhanced fluctuations in the principal RyhB target, and on the other hand, minimize conditions under which these fluctuations could appear. This may confer an evolutionary advantage in populations of wild-type cells, ensuring that all cells activate the iron homeostasis network in response to iron stress. Effects of auxiliary targets on noise have also been proposed in the case of microRNAs in eukaryotes (61).
Iron, as an essential trace element, is known to affect cell growth in E. coli through several pathways (62). Consistently, the cell doubling time increases with iron deprivation in our experiments (Supplementary Figure S14). Within the iron homeostasis network, RyhB inhibits cell growth in a RelA-dependent fashion under limiting iron conditions (63). It was proposed that RelA, a central regulator of the bacterial stringent response, carries out this function by enhancing the amount of the protein chaperone Hfq that facilitates the association of sRNAs with their target transcripts. These experiments, as others in the past (20), typically used extreme conditions of iron abundance: either media rich in iron, or media in which large concentrations of iron chelators were added.
Our experiments challenging cells with high-resolution changes in external iron availability either by the addition of an iron-selective chelator to a rich medium (LB) or iron to a minimal medium (M9) have revealed that far from showing a graded response, the iron homeostasis network exhibits switch-like activation. This is observed most clearly both in the change of the mean transcript number of RyhB targets and in RyhB's promoter activity as function of the extent of iron limitation. The apparent difference in cooperativity quantified by the Hill coefficients in both growth conditions may derive from global physiological differences caused by the growth media, since in addition to iron, these media differ in many other components (62). It is likely that the presence of multiple binding sites of Fur on the RyhB promoter that allow Fur to interact cooperatively (64), together with the polymerization of Fur on DNA substrates (64,65), may be behind the switch-like activation we observe. Consistently with this notion, when RyhB is produced from a Fur-independent promoter e.g. as in the strain with arabinose-induced production, no switch-like behavior is observed. Furthermore, the promoter of the suf operon contains a single predicted Fur binding site (66). Cooperative binding of proteins on DNA substrates occurs in other contexts in which precision is desired, e.g. in the case of Bicoid, providing a mechanism for threshold-dependent gene activation in the Drosophila embryo, allowing this transcription factor to generate sharp boundaries of gene expression (67); another example is furnished by the octamerization of the lambda repressor in the lysogenic induction switch (23). Switch-like activation of the iron homeostasis network may on one hand buffer small fluctuations in iron availability, reducing the cost of the considerable metabolic remodeling involved network activation and on the other, ensure full activation when iron deprivation becomes significant. Switch-like activation may be important in the case of pathogens, in which Fur plays a key role sensing the limited iron environment in the host, regulating many virulence factors and altering gene expression to promote pathogen survival and transmission (68).
The experiments in which we tile RyhB molecules with labeled oligomeric probes to study directly the statistics of RyhB production as a function of iron deprivation reveal a number of important features. First, the observed increase in fluorescence as a function of DTPA concentration suggests that RyhB is produced in sufficiently large amounts so as not to be a limiting factor. The promoter of RyhB is indeed known to be strong (69). Consistent with this and when the RyhB promoter is induced by iron deprivation, experiments in which RNase E was truncated and lost its ability to interact with Hfq (34), yield nearly twice the amount of RyhB compared to the strain with wild-type RNase E. This leads us to suggest that the low noise levels or RyhB we measure are unrelated to any subsequent degradation of RyhB and are due primarily to transcriptional sources. Second, the peak of RyhB noise captures sensitively the cell–cell fluctuations in RyhB availability within the switching region. Third, the fact that noise of RyhB is 5–10 times smaller than the transcript noise of sodB in the regime in which RyhB production is significant, justifies our approach of neglecting bursts in RyhB production and regarding RyhB in an effective way by enhancing the degradation rate of RyhB targets in the two-state model. It is important to bear in mind that the fluorescence measurements report only on the RyhB subpopulation of molecules that have not undergone mutual degradation with their targets, and that are not blocked to binding by FISH probes, e.g. by RyhB sponges (58).
Our methods to obtain in vivo kinetic parameters of interest of sRNA function are based on measurable changes of target transcript numbers in response to changes in sRNA production, and can therefore be applicable to other sRNAs that undergo stoichiometric degradation with their targets, e.g. SgrS and MicC (70). However they will be of limited applicability in cases in which the number of transcripts does not change such as when an sRNA activates gene expression, e.g. RyhB acting on shiA transcripts (46), or when an sRNA acts catalytically (71–73).
To sum up, our study has shown that the transcript noise of target genes that undergo stoichiometric degradation with an sRNA increases in a fashion consistent with intrinsic small-number fluctuations and bursty transcription, allowing the generalization of a two-state model to include sRNA-induced transcript degradation as a nonfluctuating parameter. The generalized two-state model describes our data quantitatively over a large range of iron levels, allowing us to determine the in vivo degradation rate of RyhB's target transcripts. Notably, the degradation rate is the only parameter that needs to be varied in order to describe quantitatively transcript distributions over a large range of levels of iron availability. The absence of enhanced cell–cell fluctuations, a hallmark of stoichiometric degradation, is consistent with the coordinate action of RyhB on a multiplicity of weak, auxiliary targets, with possible Hfq limitation, imperfect binding of RyhB to its targets and with the presence of RNA sponges sequestering RyhB. These factors provide efficient mechanisms for noise suppression. Lastly, our study has revealed that activation of the iron homeostasis network in E. coli by chelation of external iron is switch-like, enabling the bacterial cell to filter out small iron fluctuations and to ensure metabolic remodeling only when iron stress becomes significant.
Supplementary Material
Acknowledgments
We thank O. Biham and E. Levine for useful conversations, S. Gottesman, E. Masse, J. Cronan, T. Ha and H. Aiba for strains and P. Choppakatla for carrying out some of the experiments.
Author contributions: R.A.G., A.D. and B.P. performed the experiments. R.A.G. analyzed the data. A.T. contributed analysis algorithm tools. D.L.C. and N.C. contributed the recombineered strains. J.M.G. analyzed data numerically. R.A.G., A.T. and D.L.C. contributed to the experimental design and interpretation. J.S. conceived the project and wrote the manuscript. All authors provided feedback on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation [514415 to J.S.]; Feinberg Foundation Visiting Faculty Program ( to J.M.-G.); MICINN (Spain) [FIS2012-32349 to J.M.-G.]; Intramural Research Program of the National Institutes of Health (to D.L.C.); National Cancer Institute (to D.L.C.); Center for Cancer Research (to D.L.C.); Siegfried and Irma Ullman Professorial Chair ( to J. S.). Funding for open access charge: Israel Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Storz G., Vogel J., Wassarman K.M. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman S., Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner E.G.H., Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Beisel C.L., Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards G.R., Vanderpool C.K. Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta. 2011;1809:525–531. doi: 10.1016/j.bbagrm.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jost D., Nowojewski A., Levine E. Regulating the many to benefit the few: role of weak small RNA targets. Biophys. J. 2013;104:1773–1782. doi: 10.1016/j.bpj.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elf J., Paulsson J., Berg O.G., Ehrenberg M. Mesoscopic kinetics and its applications in protein synthesis. In: Alberghina L, Westerhoff HV, editors. Topics in Current Genetics: Systems Biology: Definitions and Perspectives. Berlin: Springer-Verlag; 2005. pp. 95–116. [Google Scholar]
- 8.Levine E., Zhang Z., Kuhlman T., Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia Y., Liu W., Li A., Yang L., Zhan X. Intrinsic noise in post-transcriptional gene regulation by small non-coding RNA. Biophys. Chem. 2009;143:60–69. doi: 10.1016/j.bpc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Mehta P., Goyal S., Wingreen N.S. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol. Syst. Biol. 2008;4:221. doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eldar A., Elowitz M.B. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balazsi G., van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raser J.M., O'Shea E.K. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 15.Swain P.S., Elowitz M.B., Siggia E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arbel-Goren R., Tal A., Stavans J. Phenotypic noise: effects of post-transcriptional regulatory processes affecting mRNA. Wiley Interdiscip. Rev. RNA. 2014;5:197–207. doi: 10.1002/wrna.1209. [DOI] [PubMed] [Google Scholar]
- 17.Cai L., Friedman N., Xie X.S. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 18.Salvail H., Masse E. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA. 2012;3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 19.Masse E., Escorcia F.E., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse E., Vanderpool C.K., Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vecerek B., Moll I., Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 23.Oppenheim A.B., Kobiler O., Stavans J., Court D.L., Adhya S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- 24.Nitzan N., Shimoni Y., Rosolio O., Margalit H., Biham O. Stochastic analysis of bistability in coherent mixed feedback loops combining transcriptional and posttranscriptional regulations. Phys. Rev. E. 2015;91:52706. doi: 10.1103/PhysRevE.91.052706. [DOI] [PubMed] [Google Scholar]
- 25.Arbel-Goren R., Tal A., Friedlander T., Meshner S., Costantino N., Court D.L., Stavans J. Effects of post-transcriptional regulation on phenotypic noise in Escherichia coli. Nucleic Acids Res. 2013;41:4825–4834. doi: 10.1093/nar/gkt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniguchi Y., Choi P.J., Li G.W., Chen H., Babu M., Hearn J., Emili A., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.So L.H., Ghosh A., Zong C., Sepulveda L.A., Segev R., Golding I. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peccoud J., Ycart B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 2008;48:222–234. [Google Scholar]
- 31.Shahrezaei V., Swain P.S. Analytical distributions for stochastic gene expression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17256–17261. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:8. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masse E., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita T., Maki K., Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amir A., Meshner S., Beatus T., Stavans J. Damped oscillations in the adaptive response of the iron homeostasis network of E. coli. Mol. Microbiol. 2010;76:428–436. doi: 10.1111/j.1365-2958.2010.07111.x. [DOI] [PubMed] [Google Scholar]
- 36.Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones D.L., Brewster R.C., Phillips R. Promoter architecture dictates cell-to-cell variability in gene expression. Science. 2014;346:1533–1536. doi: 10.1126/science.1255301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bremer H., Dennis P.P. In: Escherichia coli and Salmonella typhimurium?: cellular and molecular biology. 2nd edn. Neidhardt FC, Curtiss IR, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Washington D.C.: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 39.Lin H.H., Lin C.H., Hwang S.M., Tseng C.P. High growth rate downregulates fumA mRNA transcription but is dramatically compensated by its mRNA stability in Escherichia coli. Curr. Microbiol. 2012;64:412–417. doi: 10.1007/s00284-012-0087-6. [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Shiroguchi K., Ge H., Xie X.S. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 2015;11:781. doi: 10.15252/msb.20145794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skinner S.O., Sepúlveda L.A., Xu H., Golding I. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 2013;8:1100–1113. doi: 10.1038/nprot.2013.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacques J.F., Jang S., Prevost K., Desnoyers G., Desmarais M., Imlay J., Masse E. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang J., Rennie W., Liu C., Carmack C.S., Prévost K., Caron M.-P., Massé E., Ding Y., Wade J.T. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 2015;43:10308–10320. doi: 10.1093/nar/gkv1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hook-Barnard I.G., Brickman T.J., McIntosh M.A. Identification of an AU-rich translational enhancer within the Escherichia coli fepB leader RNA. J. Bacteriol. 2007;189:4028–4037. doi: 10.1128/JB.01924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prevost K., Salvail H., Desnoyers G., Jacques J.F., Phaneuf E., Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 47.Fei J., Singh D., Zhang Q., Park S., Balasubramanian D., Golding I., Vanderpool C.K., Ha T. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science. 2015;347:1371–1374. doi: 10.1126/science.1258849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvail H., Lanthier-Bourbonnais P., Sobota J.M., Caza M., Benjamin J.-A.M., Mendieta M.E.S., Lépine F., Dozois C.M., Imlay J., Massé E. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15223–15228. doi: 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao Y., Zhang Z.J., Erickson D.W., Huang M., Huang Y., Li J., Hwa T., Shi H. Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12473–12478. doi: 10.1073/pnas.1100432108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–W123. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright P.R., Richter A.S., Papenfort K., Mann M., Vogel J., Hess W.R., Backofen R., Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prevost K., Desnoyers G., Jacques J.F., Lavoie F., Masse E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D., Chang X., Liu Z., Chen L., Wang R. Bistability and oscillations in gene regulation mediated by small noncoding RNAs. PLoS One. 2011;6:e17029. doi: 10.1371/journal.pone.0017029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmqvist E., Unoson C., Reimegard J., Wagner E.G. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- 55.Maheshri N., O'Shea E.K. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu. Rev. Biophys. Biomol. Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 56.Moon K., Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hussein R., Lim H.N. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lalaouna D., Carrier M.-C., Semsey S., Brouard J.-S., Wang J., Wade J.T., Massé E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell. 2015;58:393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 59.Jost D., Nowojewski A., Levine E. Small RNA biology is systems biology. BMB Rep. 2011;44:11–21. doi: 10.5483/BMBRep.2011.44.1.11. [DOI] [PubMed] [Google Scholar]
- 60.Lalaouna D., Simoneau-Roy M., Lafontaine D., Masse E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta. 2013;1829:742–747. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdul-Tehrani H., Hudson A.J., Chang Y.S., Timms A.R., Hawkins C., Williams J.M., Harrison P.M., Guest J.R., Andrews S.C. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Argaman L., Elgrably-Weiss M., Hershko T., Vogel J., Altuvia S. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4621–4626. doi: 10.1073/pnas.1113113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escolar L., Perez-Martin J., de Lorenzo V. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 65.Le Cam E., Frechon D., Barray M., Fourcade A., Delain E. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11816–11820. doi: 10.1073/pnas.91.25.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J.-H., Yeo W.-S., Roe J.-H. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 2004;51:1745–1755. doi: 10.1111/j.1365-2958.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 67.Burz D.S., Rivera-Pomar R., Jackle H., Hanes S.D. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker K.W., Skaar E.P. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014;38:1235–1249. doi: 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitarai N., Benjamin J.A., Krishna S., Semsey S., Csiszovszki Z., Masse E., Sneppen K. Dynamic features of gene expression control by small regulatory RNAs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10655–10659. doi: 10.1073/pnas.0901466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caron M.P., Lafontaine D.A., Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 71.Figueroa-Bossi N., Valentini M., Malleret L., Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Overgaard M., Johansen J., Møller-Jensen J., Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 73.Feng L., Rutherford S.T., Papenfort K., Bagert J.D., van Kessel J.C., Tirrell D.A., Wingreen N.S., Bassler B.L. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.