Abstract
ISL1 is expressed in cardiac progenitor cells and plays critical roles in cardiac lineage differentiation and heart development. Cardiac progenitor cells hold great potential for clinical and translational applications. However, the mechanisms underlying ISL1 function in cardiac progenitor cells have not been fully elucidated. Here we uncover a hierarchical role of ISL1 in cardiac progenitor cells, showing that ISL1 directly regulates hundreds of potential downstream target genes that are implicated in cardiac differentiation, through an epigenetic mechanism. Specifically, ISL1 promotes the demethylation of tri-methylation of histone H3K27 (H3K27me3) at the enhancers of key downstream target genes, including Myocd and Mef2c, which are core cardiac transcription factors. ISL1 physically interacts with JMJD3, a H3K27me3 demethylase, and conditional depletion of JMJD3 leads to impaired cardiac progenitor cell differentiation, phenocopying that of ISL1 depletion. Interestingly, ISL1 is not only responsible for the recruitment of JMJD3 to specific target loci during cardiac progenitor differentiation, but also modulates its demethylase activity. In conclusion, ISL1 and JMJD3 partner to alter the cardiac epigenome, instructing gene expression changes that drive cardiac differentiation.
INTRODUCTION
Islet1 (insulin gene enhancer binding protein 1, ISL1), a LIM-homeodomain transcription factor, is involved in the development and differentiation of several organs including heart, pancreas, and motor neurons (1–3). In the course of cardiogenesis, ISL1-marked second heart field (SHF) progenitors give rise to the cells of the outflow tract, the majority of right ventricle and atria, as well as part of the left ventricle (2). ISL1-positive cells isolated from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), embryonic or postnatal heart tissue, harbor multipotent properties that can give rise to diverse cardiovascular cell types, such as cardiomyocytes, smooth muscle cells, endothelial cells and pacemaker cells (4–6). Notably, studies have shown that limited amounts of ISL1-positive cardiovascular progenitors are present in postnatal rat, mouse and human myocardium, raising the exciting possibility that these are endogenous cardiac progenitors that may potentially be used for cardiac regenerative applications (6–11).
ISL1 is primarily expressed in cardiac progenitors, and is downregulated upon terminal differentiation (2). ISL1 is known to cooperate with other transcriptional factors such as GATA4 (GATA binding protein 4), TBX20 (T-box 20) and forkhead transcription factor, FOXH1, to set in motion a cascade of transcriptional events involving downstream genes such as Mef2c, Myocd, Nkx2-5, Shh, Fgf10 and Bmp4, that collectively controls lineage specification of the cardiac progenitors (12,13). Prior studies have also shown that ISL1 binds to specific enhancers of Mef2c, Myocd and Nkx2-5, activating their expression in the cardiac progenitors and their derivatives (14–16). However, the precise mechanism underlying ISL1's functions in cardiac progenitors has not been fully elucidated.
During lineage specification, different signaling pathways, transcription factor networks, and chromatin regulators cooperate to bring about cell type-specific transcriptional programs (17). Genes are tightly packed into chromatin, which not only provides a packaging mechanism for DNA, but also plays a pivotal role in the control of gene expression. Chromatin is subjected to various post-translational modifications accomplished by DNA methylation, ATP-dependent chromatin remodeling and covalent histone modifications. Emerging evidence reveals that dynamic changes in epigenetic landscape accompanies cardiac specification, and that aberrant chromatin regulation contributes to congenital heart diseases (18–21). For example, mutations in the histone methyltransferase WHSC1 in humans cause congenital heart defects in Wolf-hirschhorn syndrome (22). Therefore, deciphering the mechanism underlying chromatin regulation during heart development may provide novel therapeutic strategies for the treatment of heart diseases.
It is well recognized that most epigenetic enzymes are ubiquitously expressed in different cell lineages, raising the question how specific gene expression patterns are imposed. One possible way is that these epigenetic enzymes may interact with lineage specific transcription factors in a context dependent manner. For example, UTX, a histone H3K27 demethylase, interacts with TBX5 to activate the cardiac development program, but partners with MYOD and SIX4 to regulate myogenesis (23,24). BAF60c, a subunit of the Swi/Snf-like Brg1/Brm-associated (BAF) chromatin remodeling complexes, also interacts with different transcription factors, such as GATA4, NKX2-5 and TBX5, to activate distinct group of genes during heart development (25). Thus, transcription factors may associate with specific sets of epigenetic enzymes to establish distinct transcription programs. Indeed, accumulating evidences indicate that ISL1 may play a role in epigenetic regulation (26,27).
JMJD3, a demethylase of histone H3K27me3, plays important roles in the organ development, immune response, neurodegenerative disease and cancer (28). Depletion of JMJD3 in ESCs impairs the formation of mesoderm and further profoundly suppresses endothelial and cardiac lineages differentiation (29). However, the precise mechanism underlying JMJD3 functions in cardiac differentiation are still unclear.
Here, we demonstrate that ISL1 exerts a key role in cardiac differentiation by directly regulating the expression of hundreds of downstream target genes. We further uncovered a novel epigenetic aspect of ISL1's function, showing that ISL1 impacts the epigenetic status of two critical cardiac-specific enhancers, Mef2c and Myocd, which are dynamically regulated during cardiac differentiation of ESCs. Notably, depletion of ISL1 prevents proper demethylation of H3K27me3 at the enhancers of its downstream target genes, leading to impaired expression of these genes. Mechanistically, ISL1 physically interacts with JMJD3, and are co-recruited to the enhancer regions of ISL1's downstream target genes. Conditional depletion of JMJD3 at the cardiac progenitor stage attenuates the expression of ISL1's downstream target genes, resulting in defective cardiac differentiation, similar to that of ISL1 depletion. Interestingly, we also found that ISL1 is not only responsible for the specific recognition of JMJD3 to these enhancers, but also moderately regulates the demethylase activity of JMJD3, further emphasizing a key role for ISL1-JMJD3 complex in cardiac differentiation.
MATERIALS AND METHODS
Cell culture and cardiac differentiation
HEK293T, HEK293FT and NIH 3T3 cells were grown in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) at 37°C, 5% CO2. Undifferentiated R1, E14 murine embryonic stem cells (ESCs) were maintained on 0.1% gelatin coated plates in DMEM supplemented with 15% fetal bovine serum (FBS, Invitrogen), 2 mM l-glutamine, 0.1 mM 2-mercaptoethanol (Sigma), 0.1 mM non-essential amino acids (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 4.5 mg/ml d-glucose, and 1000 U/ml of leukemia inhibitory factor (LIF ESGRO, Millipore). Cardiac differentiation experiments were performed as previously reported (30). Specifically, dissociated ES cells were cultured in bacterial culture plates at 1 × 105 cells/ml of ES cell medium, in the absence of LIF and supplemented with 10−4 M ascorbic acid. After 5 days in the suspension culture, the resulting EBs were transferred to 0.1% Gelatin coated tissue culture plates. The media were changed every the other day.
RT-PCR and real-time RT-PCR
Total RNA was extracted with TRIzol Reagent (Invitrogen) from EBs, according to the manufacturer's protocol. Complementary DNA (cDNA) was prepared from 1 μg RNA using the First Strand cDNA Synthesis Kit (TOYOBO). cDNA equivalent to 10 ng of the initial RNA input was used as templates for qPCR analysis. Relative mRNA expression was calculated from the comparative threshold cycle (ΔCt), and normalized to 18S rRNA. Primers are listed in Supplementary Table S5.
Plasmids and transfection
pCDNA3.1, pCDNA3.1-ISL1, pCMV-flag and pCMV-flag-ISL1 were stored by our lab. The construction of pCS2-Jmjd3-F was obtained from Addgene (plasmid # 17440). For construction of pGST-ISL1 full, pGST-LIM1, pGST-LIM2, pGST-HD, pGST-CT, pGST-TPR, pGST-JmjC, pGST-TCZ, cDNA was prepared from day 7 EBs of cardiac differentiation. PCR was used to amplify the fragments of interest before clone into the pGST-4T2 vector. Primers are listed in Supplementary Table S5. For the transfection, cells were seeded at a density of 2 × 106 cells/10 cm plates and transfected with 10–15 μg DNA using Lipofactamine 2000 (Invitrogen), according to the manufacturer instructions. After 48 h post-transfection, the cells were harvest for further analysis.
Lentiviral shRNAs
Production of lentiviral particles and transduction of ESCs was performed according to protocols described previously (31,32). Wild-type pLKO-tet-on and pLKO.1 were obtained from Addgene (#21915, #8453). Specifically, packaging plasmids were co-transfected into HEK293FT cells using Lipofectamine 2000 (Invitrogen) with short hairpin RNA constructs for targeting Isl1 or Jmjd3. Stable integrated into R1 or E14 ESCs was selected with 1 μg/ml puromycin. shRNA oligos are listed in Supplementary Table S5.
CRISPR-Cas9 editing Jmjd3 in mESCs
Experimental procedures were essentially described previously (33). Wild-type Cas9 plasmid PX458 was obtained from Addgene (plasmid #48138). sgRNAs were synthesized and cloned into PX458. Doner for inserting a Flag-HA sequence at the N-terminal of Jmjd3 was a synthesized ultramer (Integrated DNA Technologies). Donor plasmid for knocking out Jmjd3 was prepared using Gibson Assembly (New England Bio Labs). For generating Jmjd3-NFH R1 mESCs, we transiently transfected PX458-Jmjd3 and donor ultramer by using Lipofectamine 2000 (Invitrogen). In 48 h post-transfection, GFP-positive cells were sorted and replated into 0.1% gelatin coated plates at the density of 10 000 cells per 10cm plate. After ∼7 days culture, clones were picked under microscope and screened by genomic PCRs and sequencing. For generating Jmjd3-knockout R1 mESCs, we transiently transfected PX458-Jmjd3 and donor plasmid by using Lipofectamine 2000. In 48 h post-transfection, cells were selected by using 400 μg/ml G418 for ∼7 days, and clones were screened by genomic PCRs and qRT-PCR. Jmjd3 knockout R1 mESCs were maintained without G418. Primers and the ultramer are summarized in Supplementary Table S5. Cassette sequences are available on request.
ChIP, ChIP-seq and RNA-seq
ChIP was performed as previously described (34). Briefly, cross-linked and isolated nuclei were sonicated using a Diagenode Bioruptor to an average size of ∼250 bp for ChIP-seq or ∼500 bp for ChIP-qPCR. After pre-clearing with BSA-blocked protein G Sepharose, chromatin was incubated with antibodies at 4°C overnight. The chromatin immunocomplexes were recovered with the same BSA-blocked protein G beads. For ChIP-seq library construction, ∼5 ng of DNA extracted from the chromatin immunocomplexes as described previously (35). Libraries were prepared according to manufacturer's instructions (Illumina) and as described (34). Briefly, immunoprecipitated DNA was first end-repaired using End-It Repair Kit (Epicentre), tailed with deoxyadenine using Klenow exo minus (NEB), and ligated to custom adapters with T4 Rapid DNA Ligase (Enzymatics). Fragments of 350 ± 50 bp were size-selected using Agencourt AMPure XP beads, and subjected to ligation-mediated PCR amplification (LM-PCR), using Q5 DNA polymerase (NEB). Libraries were quantified by qPCR using primers annealing to the adaptor sequence and sequenced at a concentration of 10 pM on an Illumina HiSeq 2000. For RNA-seq libraries, polyA+ RNA was isolated using Dynabeads Oligo (dT) 25 (Invitrogen) and constructed into strand-specific libraries using the dUTP method (36). Once dUTP-marked double-stranded cDNA was obtained, the remaining library construction steps followed the same protocol as described above for ChIP-seq libraries.
Data analysis
For ChIP-seq, sequenced reads were aligned to the mouse reference genome (assembly mm9) using Bowtie2 (37). Duplicated reads were removed with Samtools (38). ChIP-seq read density files were generated using Igvtools and were viewed in Integrative Genomics Viewer (IGV) (39). Reads were merged from two biological replicates, and then significantly (P < 1.0E-05 for ISL1 ChIP-seq, P < 1.0E−03 for JMJD3 ChIP-seq) enriched peaks for each ChIP-seq data set were identified with MACS (40). Genomic distribution of peaks and gene associated region annotations were obtained via PeakAnalyzer (41). ChIP-seq density heatmaps were generated by seqMINER (42). RNA-seq data were analyzed as previously descripted (43). Briefly, sequenced reads were aligned to the mouse reference genome (assembly mm9) using Tophat (44). Transcriptome was assembled using Cufflinks (43). Differential gene expression was calculated from two biological replicates with Cuffdiff, considering FPKM (fragments per kilobase of exon per million fragments mapped) ≥ 1 in either one of the 2 conditions and |fold-change Isl1 knockdown vs. ctrl| ≥1.5-fold as a cut-off. GO analysis was conducted with DAVID (Database for Annotation, Visualization, and Integrated Discovery (45)).
Nuclear extraction and immunoprecipitation
Nuclear extraction and immunoprecipitation experiments were performed as previously described (46,47). Specifically, 1 mg NE was incubated antibodies against 2 μg Isl1 (ab109517, Abcam) or 5 μg Flag (F3165, Sigma) in a volume of 400 μl Buffer C, supplemented with 200 μl Buffer BN (20 mM Tris–HCl, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mg/ml BSA, 0.1% NP-40, 0.5 mM PMSF, 1 μg/ml Pepstatin A, 1 μg/ml Leupeptin, 1 μg/ml Aprotinin). After overnight incubation at 4°C, 30 μl of protein A/G (1:1) beads were added for incubation at 4°C for 2 h. Beads were then washed with Buffer BN for 4 times and boiled in 2 × SDS sample buffer. The sample was analyzed by SDS-PAGE, followed by immunoblotting.
Immunofluorescence staining
Immunofluorescence staining was performed as previously described (19). Day 7 EBs of cardiac differentiation from JMJD3-NFH mESCs were cultured in 8 chamber slides, stained with DAPI, using antibodies against ISL1 and HA.
GST pull-down assays
GST-fused constructs were expressed in BL21 Escherichia coli. In vitro transcription and translation experiments were done with rabbit reticulocyte lysate (TNT systems, Promega) according to the manufacturer's recommendation. In GST pull-down assays, about 5 μg of the appropriate GST fusion proteins with 40 μl of glutathione-Sepharose beads was incubated with 20 μl of in vitro transcribed/translated products in binding buffer (20 mM Tris pH 7.4, 0.1 mM EDTA, 100 mM NaCl) at 4°C for 2 h in the presence of the protease inhibitor mixture. The beads were washed 5 times with binding buffer, resuspended in 30 μl of 2 × SDS-PAGE loading buffer, and detected by western blotting.
Histone extraction, Western blotting and CBB staining
Histone extraction was performed as described in Abcam protocols (www.abcam.com). Samples from histone extraction, nuclear extraction or total cell lysates were subjected to 8–15% SDS polyacrylamide gels as required. Coomassie brilliant blue (CBB) staining was followed the instruction of the Commassie blue fast staining kit (Beyotime).
Demethylase activity of JMJD3/UTX
Demethylase activities of JMJD3/UTX in day 7 EBs of cardiac differentiation and NIH3T3 cells were determined using JMJD3/UTX Demethylase Activity/Inhibition Assay Kit (Epigentek). Experiments were performed according to the manufacturer's protocol.
Statistical analysis
The data are expressed as mean ± standard deviation (SD). Comparisons between groups were analyzed using Student's t-test or ANOVA, and the Student–Newman–Kleuss method was used to estimate the level of significance. Differences were considered to be statistically significant at P < 0.05.
RESULTS
Genome-wide downstream targets of ISL1 during cardiac progenitor differentiation
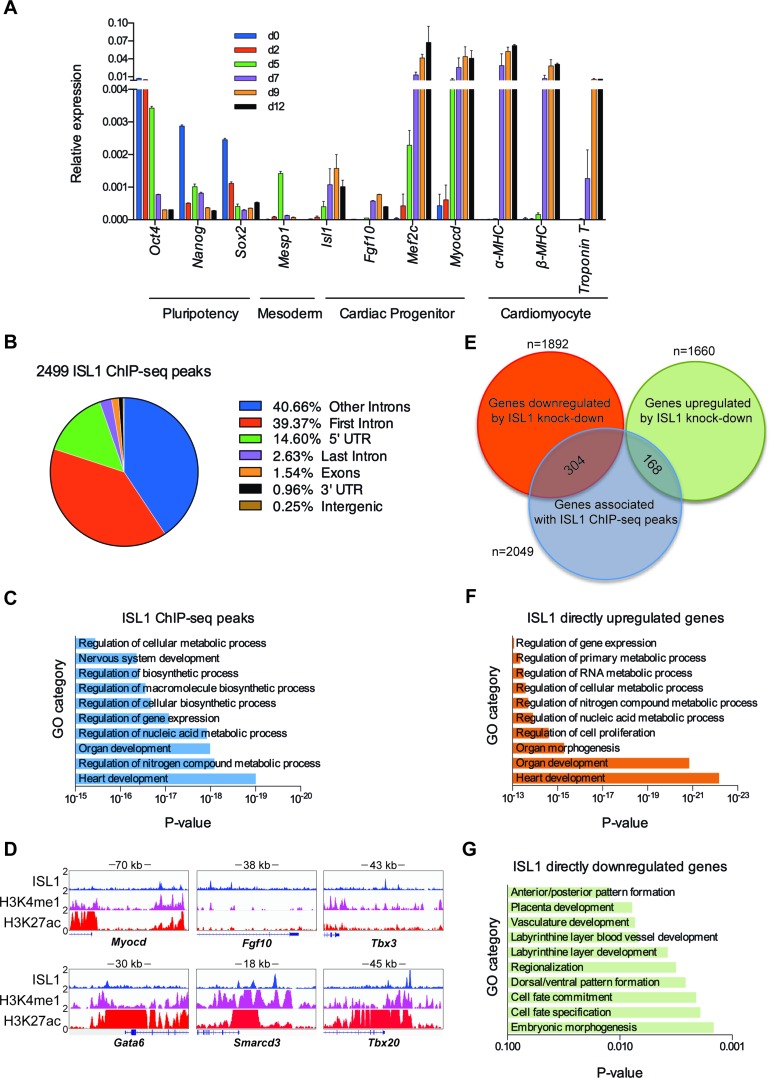
ISL1 is expressed in cardiac progenitors and is required for their proliferation, survival, and differentiation. To gain insights into the mechanisms underlying ISL1's function, we differentiated murine ESCs into cardiomyocytes by LIF withdrawal and in the presence of ascorbic acid to improve the efficiency of cardiac differentiation (30). To determine the kinetics of cardiac progenitor formation, we profiled the gene expression changes as a function of differentiation (day 0–day 12; Figure 1A). We found that pluripotency marker genes such as Oct4 and Nanog were dramatically downregulated as differentiation proceeds. On the other hand, Mesp1, a marker gene for mesoderm, was transiently expressed around day 5. Notably, cardiac progenitor marker gene, Isl1, was upregulated from day 5 onward, and downregulated after day 9. Following this dynamics, we observed the upregulation of downstream target genes of ISL1, such as Fgf10, Mef2c and Myocd, peaking around day 7. The cardiomyocyte marker genes α-MHC, β-MHC and Troponin T were dramatically upregulated after day 9. Taken together, our data indicates that cardiac progenitors are enriched in day 5–day 7 embryoid bodies (EBs).
Figure 1.
Genome-wide downstream targets of ISL1 during cardiac progenitor differentiation. (A) Relative mRNA expression of pluripotency (Oct4, Sox2 and Nanog), mesoderm (Mesp1), cardiac progenitor (Isl1, Fgf10, Mef2c and Myocd) and cardiomyocyte (α-MHC, β-MHC and Troponin T) markers in cardiac differentiation of ESCs was measured by qRT-PCR at indicated time points. 18S rRNA was used as an internal control. Data are mean ± SD, n = 4. (B) ChIP-seq ISL1-binding regions were mapped relative to their nearest downstream genes. Annotation includes whether a peak is in the first intron, other introns, 5′ UTR, last intron, exons (coding), 3′ UTR, intergenic. (C) GO functional clustering of genes associated with ISL1 ChIP-seq peaks (top 10 categories are shown). (D) The binding of ISL1 in EBs at day 7 of cardiac differentiation, H3K4me1 and H3K27ac in embryonic day 14.5 of heart tissue on representative ISL1 target genes, Myocd, Fgf10, Tbx3, Gata6, Smarcd3 and Tbx20. (E) Overlay of RNA-seq and ChIP-seq results revealed 472 genes as potential direct targets of ISL1 in day 7 EBs of differentiation. (F and G) GO functional clustering of genes allowed for identification of cellular functions directly regulated by ISL1 (top 10 categories are shown).
To unveil the potential direct downstream target genes of ISL1 during cardiac progenitor differentiation, we performed genome-wide Chromatin immunoprecipitation sequencing (ChIP-seq) for ISL1 on day 7 EBs. Analysis of the data identified 2499 ChIP-seq peaks for ISL1, with the majority of binding events (82.90%) occurring at intronic or intergenic sites (Figure 1B). The distribution for ISL1 genomic occupancy is consistent with other reported cellular specific binding patterns of transcription factors, localizing to distal regulatory regions (48,49). Our data indicate that ISL1 may preferentially localize to cardiac progenitor specific regulatory regions, impacting cardiac differentiation gene expression. Functional annotation of ISL1 binding sites reveals 2049 potential ISL1 targets including those involved in heart development, metabolic process, organ development and gene expression (Figure 1C; Supplementary Table S1). Notably, we observed the binding of ISL1 at the distal regulatory regions of Myocd, Fgf10 and Tbx3 (Figure 1D), key targets of ISL1 during cardiac differentiation, in good agreement with previous reports (5,16,50). We also identified new target genes, such as Gata6, Smarcd3 and Tbx20 (Figure 1D; Supplementary Figure S2), which are important for proper cardiac differentiation and heart development (14,25,51). To better annotate the feature of these regions bound by ISL1, we analyzed the ChIP-seq data sets of the histone modifications of embryonic day 14.5 (E14.5) heart tissue compiled as part of the ENCODE Project (GSM769025). Remarkably, most of these ISL1 bound regions are enriched with H3K4me1 and H3K27ac (Figure 1D), the hallmarks of enhancer, indicating ISL1 may preferentially localize to cardiac specific enhancers. To further investigate the transcriptional effects of ISL1 binding, RNA-seq analyses were performed on day 7 EBs in which Isl1 was constitutively knockdown (day 7 EBs transfected with non-specific shRNA were used as the control) (Supplementary Figure S1A, B). Our results revealed that 1892 genes were downregulated and 1660 genes were upregulated upon Isl1 knockdown (FPKM≥1, |fold change Isl1 knockdown vs. ctrl| ≥ 1.5). Intersection of ChIP-seq data with RNA-seq data identified 304 direct targets that are upregulated by ISL1, and 168 targets that are downregulated by ISL1 (Figure 1E). GO analyses revealed that the former category is overrepresented for genes involved in heart development, organ development, cell proliferation, metabolic process and gene expression (Figure 1F; Supplementary Table S2). The latter category included genes that are enriched for embryonic morphogenesis, cell differentiation and pattern formation (Figure 1G; Supplementary Table S3). Notably, a large number of genes upregulated or downregulated by depletion of ISL1 were not associated with ISL1 ChIP peaks, implying a potential hierarchical role of ISL1 in cardiac progenitor cells, which sets in motion a cascade of transcriptional events. Taken together, our genome-wide studies show that ISL1 controls a large cohort of genes implicated in cardiac progenitor differentiation, providing the basis for downstream mechanistic exploration of ISL1 function.
ISL1 perturbation impairs demethylation of H3K27me3 at the enhancers of its downstream cardiac-specific target genes
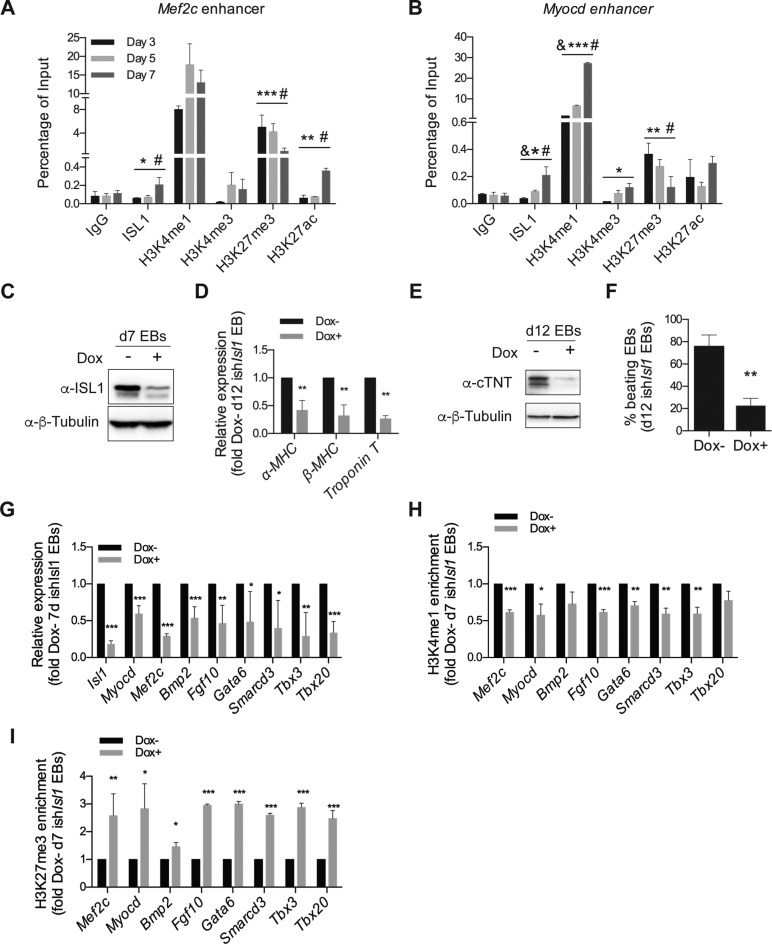
Our earlier findings showed that ISL1 might bind to regulatory regions of target genes to instruct gene expression changes. To gain further mechanistic insights, we performed ChIP-qPCR assays to study the dynamics of ISL1 target gene binding during differentiation, as well as to assess for any changes in epigenetic configuration. Specifically, we focused on changes that occur at the enhancers of Mef2c and Myocd, two well-known representative targets of ISL1, at three time points (day 3, day 5 and day 7, Figure 2A, B). Firstly, at day 3, we observed the presence of H3K4me1 and H3K27me3 at the enhancers of Mef2c and Myocd. This chromatin signature is indicative of enhancer inactivity, consistent with the lack of Mef2c and Myocd mRNA expression. Strikingly, at day 7 of differentiation, the level of H3K27me3 was significantly decreased at the enhancers of Mef2c and Myocd, concomitant with the occupation of ISL1. This was accompanied by a gain in H3K27ac, H3K4me1 and H3K4me3, hallmarks of gene activation. Taken together, our results indicate a possible epigenetic mechanism whereby the regulated binding of ISL1 at the enhancers of Mef2c and Myocd may orchestrate histone modification changes, priming the expression of ISL1's target genes.
Figure 2.
ISL1 perturbation impairs demethylation of H3K27me3 at the enhancers of its downstream cardiac-specific target genes. (A and B) ChIP of nuclear extracts from day 3, 5 and 7 EBs differentiated from ESCs using anti-ISL1, anti-H3K4me1, anti-H3K4me3, anti-H3K27me3 and anti-H3K27ac or IgG as the control. qRT-PCRs were performed using primers flanking conserved ISL1 binding sites in the Mef2c enhancer (A) and the Myocd enhancer (B). Data are mean ± SD, n = 3. &P < 0.01 d5 VS d3, *P < 0.05 d7 versus d3, **P < 0.01 d7 versus d3, ***P < 0.001 d7 versus d3, #P < 0.01 d7 versus d5. (C) Western blot analysis of total protein extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. β-Tubulin served as a loading control. (D) Relative mRNA expression of cardiomyocyte genes (α-MHC, β-MHC and Troponin T) in day 12 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (E) Western blot analysis of total protein extracts of day 12 EBs differentiated from ishIsl1 in the presence or absence of Dox. β-Tubulin served as a loading control. (F) Percentage of beating EBs in day 12 EBs differentiated from ishIsl1 in the presence or absence of Dox. (G) Relative mRNA expression of Isl1 and its downstream target genes (Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20 of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (H and I) ChIP of nuclear extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox using anti-H3K4me1 (H) and anti-H3K27me3 (I). qRT-PCRs were performed using primers targeting ISL1 binding sites in the enhancers of Mef2c, Myocd, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20. ChIP enrichments are normalized to Input and are represented as fold change relative to Dox- EBs. Data in D and F-I are mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
To further clarify an epigenetic role of ISL1, we generated stable doxycycline inducible knockdown Isl1 (ishIsl1) mESCs. To deplete ISL1 specifically at the cardiac progenitor stage, doxycycline (Dox) was added at day 3 of differentiation and the media were changed every two days. As shown in Figure 2C, ISL1 was significantly reduced by Dox induction. Importantly, the efficiency of cardiac differentiation was also significantly impaired upon ISL1 knockdown, further attesting to the functional importance of ISL1 in cardiomyocyte formation. We observed a decrease in the expression of cardiomyocyte markers (α-MHC, β-MHC, Troponin T), accompanied by a reduction in the percentage of beating embryonic bodies (Figure 2D–F). As expected, the diminution in ISL1 level also affected the expression of Myocd and Mef2c as well as other ISL1's downstream target genes, unveiled by our ChIP-seq and RNA-seq data, including Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20 in day 7 EBs (Figure 2G).
Next, we performed ChIP assay to address whether ISL1 is responsible for the changes in histone modification pattern at day 7 of differentiation (Figure 2A, B). In contrast to our earlier findings, we observed a significant increase in H3K27me3 levels at the enhancers of all tested ISL1's downstream targets, including Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20 (Figure 2I) specific to the ISL1 depleted cells. On the other hand, the level of H3K4me1 remained low (Figure 2H). Taken together, these results highlight a novel epigenetic role of ISL1, implicated in H3K27me3 demethylation.
JMJD3 physically interacts with ISL1 at cardiac progenitor stage
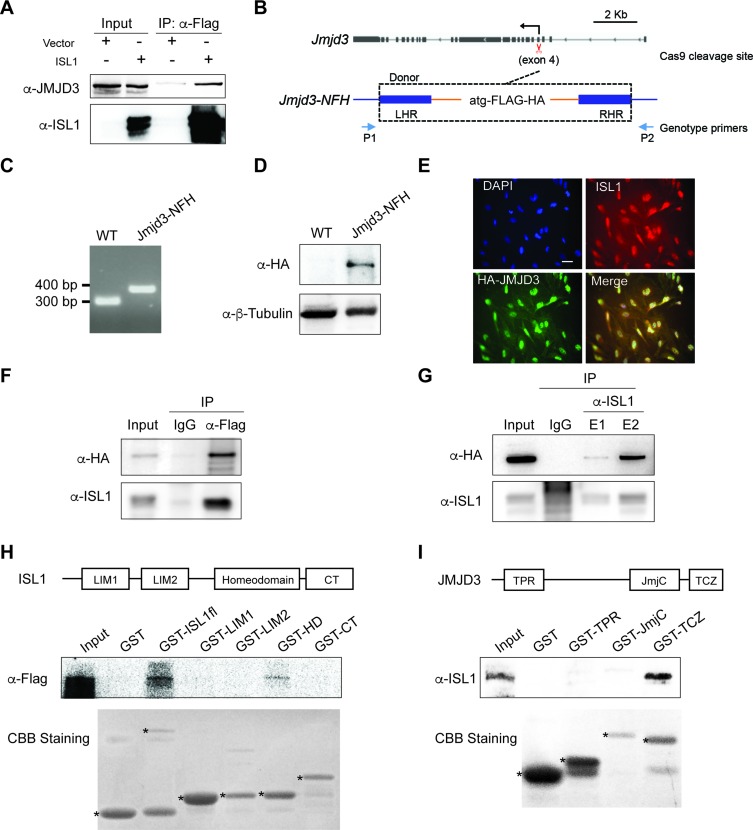
To further investigate how ISL1 may regulate H3K27me3 demethylation, we have performed a proteomic screening to identify putative partners of ISL1 in day 7 EBs differentiated from wild type ESCs (unpublished data). Amongst the list of interactors recovered, we noted the presence of JMJD3, a H3K27me3 demethylase. To validate the interaction between ISL1 and JMJD3, we transfected HEK293T cells with either a Flag construct or a Flag tagged ISL1 expression plasmid, followed by immunoprecipitation (IP) with an anti-Flag antibody and immunoblotting (IB) with antibodies against ISL1 or JMJD3. Importantly, JMJD3 was efficiently co-immunoprecipitated by ISL1 (Figure 3A). Next, we wanted to confirm if ISL1 could also interact with JMJD3 in the cardiac progenitors. Given that there is no available commercial JMJD3 antibody suitable for immunoprecipitation, we used CRISPR/Cas9 methodology (33) to engineer a Flag-HA epitope tag at the N-terminus of JMJD3 in mESCs (thereafter referred to as JMJD3-NFH ESCs; Figure 3B). Correctly targeted JMJD3-NFH ESCs line was validated by genotyping, and IB with an antibody against HA (Figure 3C, D). Whilst ISL1 is a nuclear protein, previous studies indicate that JMJD3 can shuttle between the cytoplasm and the nucleus (52). Hence, we first performed immunofluorescence experiments in day 7 EBs during cardiac differentiation, to ascertain that ISL1 and JMJD3 were co-localized in the nucleus (Figure 3E). Thereafter, to validate a physiological interaction between ISL1 and JMJD3, we performed endogenous co-IP experiments using nuclear extracts prepared from day 7 JMJD3-NFH EBs. IP with anti-Flag antibody followed by IB with antibodies against ISL1 or HA demonstrated that ISL1 was efficiently co-immunoprecipitated with JMJD3 (Figure 3F). Reciprocally, IP with anti-ISL1 antibody followed by IB with antibodies against HA or ISL1 also showed that JMJD3 was efficiently co-immunoprecipitated by ISL1 (Figure 3G). These results confirm that ISL1 physically interacts with JMJD3 in the cardiac progenitors.
Figure 3.
JMJD3 physically interacts with ISL1 at cardiac progenitor stage. (A) Co-immunoprecipitation of nuclear extracts from HEK293T cells transfected with a Flag construct or Flag tagged of ISL1 expression plasmid using anti-Flag were analysis by anti-JMJD3 and anti-ISL1 immunoblotting. Five percent of total lysates were loaded as input. (B) Schematic diagram of the endogenous tagging N-ternimal of JMJD3 with Flag-HA epitope (JMJD3-NFH) using Crispr/Cas9. CRISPR-Cas9 cleavage site (exon4, 5.5 kb downstream of the TSS) is shown by scissor. Positions of the genotype primers used in C are shown by arrows. Donor ultramer utilized for targeting Jmjd3 locus is shown: left homolog arm (LHR), Flag-HA sequence, and right homolog arm (RHR). (C) PCR genotyping of wild type (WT) and Jmjd3-NFH ESCs. The size of PCR product of wild type ESCs is 299 bp, and the Jmjd3-NFH ESCs is 368 bp. (D) Western blot analysis of total protein extracts of WT and JMJD3-NFH ESCs. β-Tubulin served as a loading control. (E) ISL1 (red) and HA (green) immunostaining on day 7 EBs differentiated from JMJD3-NFH ESCs. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bar, 20 μm. (F) Co-immunoprecipitation of nuclear extracts of day 7 EBs differentiated from JMJD3-NFH ESCs using anti-FLAG (F) or anti-ISL1 (G) or IgG as a control were analysis by anti-HA and anti-ISL1 immunoblotting. Five percent of total lysates were loaded as input. E1 and E2 represented two times elution with glycine. (H and I) Schematic representation of ISL1 (H) or JMJD3 (I) and its domains and GST pull-down assays. GST pull-down assays with purified GST-fused proteins and in vitro transcribed/translated JMJD3-Flag (H) or ISL1 (I) were analyzed with anti-FLAG (H) or ISL1 (I) immunoblotting. Five percent of total in vitro transcribed/translated JMJD3-Flag (H) or ISL1 (I) was loaded as input. Coomassie brilliant blue staining (CBB staining) was applied to detect the purified GST-fused protein expression, * indicate the predicted proteins.
To further characterize the molecular interactions between ISL1 and JMJD3, we performed interaction studies using different protein fragments of ISL1 and JMJD3. In vitro transcribed/translated JMJD3 protein was incubated with different fragments of GST-fused ISL1, namely full-length (1-348 AA, GST-ISL1fl), LIM1 domain (15-77 AA, GST-LIM1), LIM2 domain (78-139 AA, GST-LIM2), Homeodomain (179-239 AA, GST-HD), or the C-terminal fragment (240-349 AA, GST-CT) of ISL1. Our results showed that the homeodomain of ISL1 is responsible for its interaction with JMJD3 (Figure 3H). Reciprocal GST pull-down experiments using different fragments of GST-JMJD3, namely TPR domain (98–150 AA, GST-TPR), JmjC domain (1337-1500 AA, GST-JmjC), or the TCZ domain (1501-1641 AA, GST-TCZ)) indicated that the TCZ domain of JMJD3 is responsible for its interaction with ISL1 (Figure 3I).
JMJD3 regulates cardiac differentiation
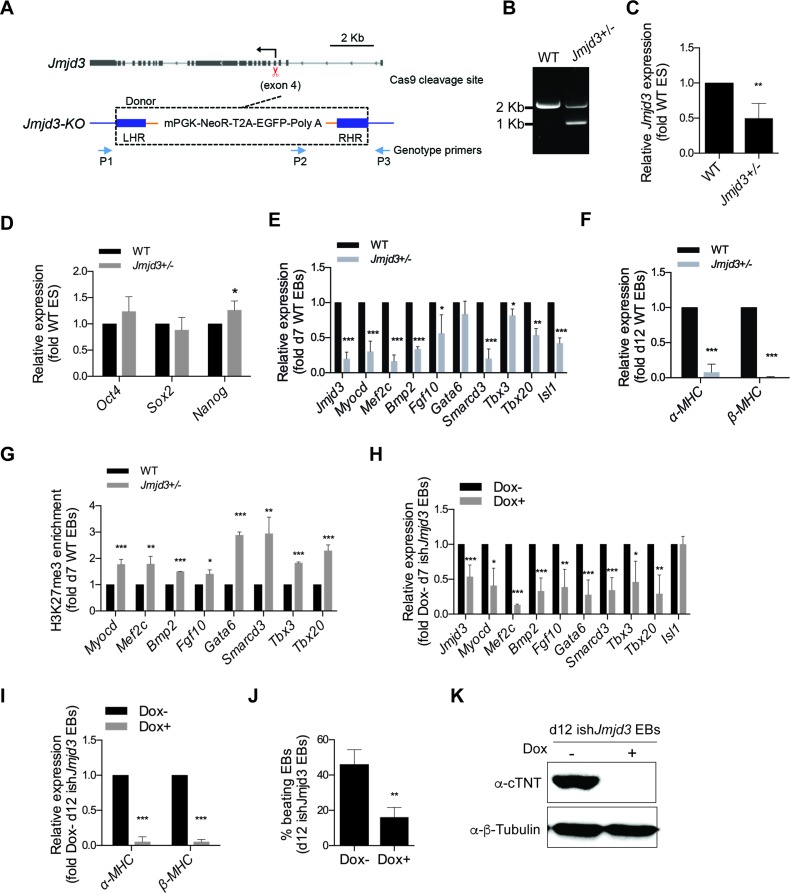
Since JMJD3 directly interacts with ISL1 at cardiac progenitor stage, we wanted to investigate whether JMJD3 is required for the expression of downstream target genes of ISL1 during cardiac differentiation. To this end, we generated Jmjd3 knockout ESCs by utilizing the CRISPR/Cas9 methodology. In this case, we inserted a cassette containing mPGK-NeoR-EGFP-Poly A sequence, into exon 4 of Jmjd3 to constitutively knockout Jmjd3 expression. Interestingly, following drug selection, we only recovered Jmjd3 heterozygotes (Jmjd3+/−) ESCs, likely due to the non-viability and/or selective growth disadvantage of Jmjd3 complete knockout ESCs (Figure 4A, B). Nevertheless, relative to wild type cells, the expression level of Jmjd3 was reduced by over 50% in Jmjd3+/− ESCs and day 7 Jmjd3+/− EBs (Figure 4C, E; Supplementary Figure S3). Importantly, whereas the expression of pluripotency markers (Oct4 and Sox2) in Jmjd3+/− ESCs remained largely comparable to wild type ESCs (Figure 4D), a striking impairment in cardiomyocyte marker expression (α-MHC and β-MHC) was observed in day 12 Jmjd3+/− EBs relative to wild type EBs (Figure 4F). In particular, the expression of numerous downstream targets of ISL1 was also significantly downregulated in day 7 Jmjd3+/− EBs (Figure 4E). These data strongly indicate that JMJD3 is required for the cardiac differentiation.
Figure 4.
JMJD3 regulates cardiac differentiation. (A) Schematic diagram of the knockout Jmjd3 using Crispr/Cas9. CRISPR-Cas9 cleavage site (exon4, 5.5 kb downstream of the TSS) is shown by scissor. Positions of the genotype primers used in B are shown by arrows. Donor plasmid utilized for targeting Jmjd3 locus is shown: left homolog arm (LHR), mPGK, neomycin resistance (NeoR), T2A, EGFP, polyA sequence, and right homolog arm (RHR). (B) PCR genotyping of WT and Jmjd3+/− ESCs. The size of PCR product of wild type ESCs is ∼2 kb, and is ∼1 kb of the Jmjd3-KO ESCs. (C) Relative mRNA expression of Jmjd3 in wild type and Jmjd3+/− ESCs using primers specific target to exon 9–10. (D) Relative mRNA expression of pluripotency genes (Oct4, Sox2 and Nanog) in wild type and Jmjd3+/− ESCs. (E) Relative mRNA expression of Jmjd3, Isl1 and ISL1's downstream genes (Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20) in day 7 EBs differentiated from wild type and Jmjd3+/− ESCs. (F) Relative mRNA expression of cardiomyocyte genes (α-MHC and β-MHC) in day 12 EBs differentiated from wild type and Jmjd3+/− ESCs. (G) ChIP of nuclear extracts of day 7 EBs differentiated from wild type and Jmjd3+/− ESCs using anti- H3K27me3. qRT-PCRs were performed using primers targeting ISL1 binding sites in the enhancers of Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20. ChIP enrichments are normalized to Input and are represented as fold change relative to wild type EBs. (H) Relative mRNA expression of Jmjd3, Isl1 and ISL1's downstream genes (Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20) in day 7 EBs differentiated from ishJmjd3 in the presence or absence of Dox. Doxycycline was supplemented to the culture medium since day 5. (I) Relative mRNA expression of cardiomyocyte genes (α-MHC and β-MHC) in day 12 EBs differentiated from ishJmjd3 in the presence or absence of Dox. Doxycycline was supplemented to the culture medium since day 5. (J) Percentage of beating EBs in day 12 EBs differentiated from ishJmjd3 in the presence or absence of Dox. (K) Western blot analysis of total protein extracts of day 12 EBs differentiated from ishJmjd3 in the presence or absence of Dox. β-Tubulin served as a loading control. Data in C–J are mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
To explore whether JMJD3 is responsible for the removal of tri-methylation of H3K27 at the enhancers of ISL1's downstream target genes, we performed ChIP assays for H3K27me3 in day 7 EBs. Notably, we found that H3K27me3 level was significantly increased on all of the tested enhancers specifically in day 7 Jmjd3+/− EBs relative to wild type controls (Figure 4G). Interestingly, we also noticed that the expression of Isl1 was decreased in day 7 Jmjd3+/− EBs (Figure 4E). This raised the possibility that the reduction in the expression of ISL1's downstream genes might be a secondary effect attributed to a role of JMJD3 in mesoderm regulation as previous reported (29). To critically assess this possibility, we generated doxycycline inducible knockdown Jmjd3 (ishJmjd3) mESCs that will enable us to precisely regulate Jmjd3 level at specific time points in differentiation.
To deplete Jmjd3 specifically at the cardiac progenitor stage, differentiation medium was supplemented with doxycycline at day 5 EBs and changed every two days. Jmjd3 expression was reduced more than half-fold in day 7 EBs (Figure 4H). Importantly, in this system, the expression levels of all the tested ISL1's downstream targets were significantly decreased with no overt changes in Isl1 expression. Consistently, Jmjd3 depleted cells exhibited impaired terminal cardiomyocyte differentiation efficiency, as evident from the reduced expression of cardiomyocyte specific markers (α-MHC, β-MHC and cTNT) and EBs beating efficiency in day 12 EBs (Figure 4I–K). Taken together, these results establish that JMJD3 promotes cardiac progenitor differentiation by regulating the expression of the ISL1 target genes via demethylation of H3K27me3 at their enhancers.
ISL1 and JMJD3 form a transcriptional regulatory complex
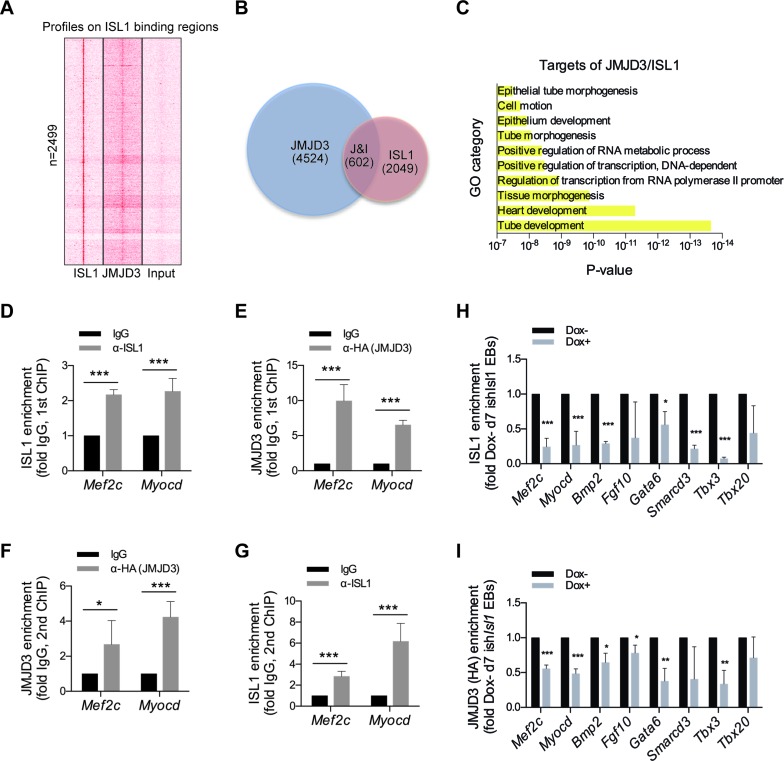
To gain insight into the functional significance of the physical association between JMJD3 and ISL1, we explored the genome-wide direct targets of the JMJD3 in day 7 JMJD3-NFH EBs by ChIP-seq using anti-HA antibody. Analysis of the characteristic enrichment of JMJD3 revealed that JMJD3 was remarkably enriched in the regions surrounding the ISL1 genomic binding sites (Figure 5A). Functional annotation of JMJD3 binding sites, reveals 4524 potential JMJD3 targets, among which 602 direct target genes were overlapped by JMJD3 and ISL1 including those involved in tube development, heart development, tissue morphogenesis (Figure 5B, C; Supplementary Table S4). To definitively show that ISL1 co-opts JMJD3 to regulate its downstream target genes expression in cardiac progenitors, we performed ChIP/re-ChIP experiments for ISL1 and JMJD3. Chromatin prepared from day 7 JMJD3-NFH EBs was sequentially immunoprecipitated with either anti-ISL1 or anti-HA (JMJD3) antibodies (and vice versa) (Figure 5D–G). ChIP experiments confirmed that JMJD3 and ISL1 occupied the enhancers of Mef2c and Myocd in day 7 EBs of cardiac differentiation (Figure 5D, E). Importantly, re-ChIP experiments confirmed that both ISL1 and JMJD3 exist as one protein complex at the enhancers of Mef2c and Myocd (Figure 5F and G).
Figure 5.
ISL1 and JMJD3 form a transcriptional regulatory complex. (A) ChIP-seq density heatmaps of ISL1 and JMJD3 on ISL1 binding sites, Input served as a negative control. (B) Overlay of JMJD3 and ISL1 ChIP-seq results revealed 602 genes as potential direct targets of ISL1/JMJD3 in EBs at day 7 of differentiation. (C) GO functional clustering of genes allowed for identification of cellular functions directly regulated by ISL1/JMJD3 (top 10 categories are shown). (D and E) ChIP of nuclear extracts of day 7 EBs differentiated from JMJD3-NFH ESCs using anti-ISL1 (D) and anti-HA (E) or IgG as control. qRT-PCRs were performed using primers targeting ISL1 binding sites on Mef2c and Myocd enhancers. ChIP enrichments are normalized to Input and are represented as fold change relative to IgG. (F and G) re-ChIP of material from anti-ISL1 ChIP elutions using anti-HA (F), or from anti-HA ChIP elutions using anti-ISL1 (G). IgG served as control. qRT-PCRs were performed using primers targeting ISL1 binding sites on Mef2c and Myocd enhancers. ChIP enrichments are normalized to Input and are represented as fold change relative to IgG. (H and I) ChIP of nuclear extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox using anti-ISL1 (H) or anti-HA (I). qRT-PCRs were performed using primers targeting ISL1 binding sites in the enhancers of Mef2c, Myocd, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20. ChIP enrichments are normalized to Input and are represented as fold change relative to wild Dox- EBs. Data in D–I are mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, using Jmjd3-NFN ESCs engineered to express a doxycycline-inducible shRNA that targets Isl1, we explored a possible hierarchical targeting of ISL1 and JMJD3 to the enhancers of ISL1's downstream target genes. Indeed, our ChIP experiments showed that depletion of ISL1 resulted in a striking reduction of JMJD3 recruitment to the enhancers of ISL1's downstream target genes in day 7 EBs (Figure 5H and I). Taken together we present two lines of evidence to show conclusively that ISL1 promotes the recruitment of JMJD3 to key target loci during cardiac progenitor differentiation.
ISL1 modulates the demethylase activity of JMJD3
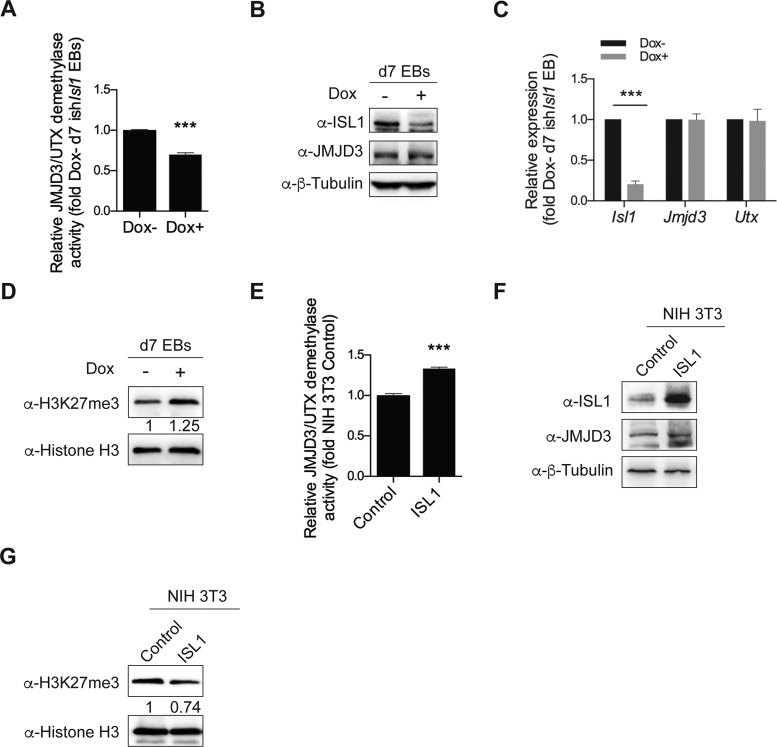
Since ISL1 and JMJD3 physically interact in cardiac progenitors, we wondered if the activity of JMJD3 might be modulated by ISL1 interaction. Interestingly, we found that the demethylase activity of JMJD3/UTX in nuclear extracts from day 7 EBs was inhibited following ISL1 depletion, that occurred in the absence of any changes in the expression levels of JMJD3 and UTX (Figure 6A–C). Consistently, we further observed a striking increase in the overall level of H3K27me3 in ISL1 depleted EBs (Figure 6D), similar to that of JMJD3 depletion (Supplementary Figure S4A-B). In excellent support, overexpression of ISL1 in NIH 3T3 cells led to a significant reduction in level of H3K27me3, with no overt changes in JMJD3 expression (Figure 6E–G). Taken together, these results indicate that ISL1 potentially regulates the demethylase activity of JMJD3, impacting transcriptional regulation of ISL1 targets during cardiac differentiation.
Figure 6.
ISL1 modulates the demethylase activity of JMJD3. (A) Relative JMJD3/UTX demethylase activity of nuclear extracts from day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (B) Western blot analysis of total cell lysates of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. β-Tubulin served as a loading control. (C) Relative mRNA expression of Isl1, Jmjd3 and Utx of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (D) Western blot analysis of histone extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. Histone H3 served as a loading control. (E) Relative JMJD3/UTX demethylase activity of nuclear extracts from NIH-3T3 cells transient transfected with control or ISL1 expression construct. (F) Western blot analysis of total cell lysates of NIH-3T3 cells transient transfected with control or ISL1 expression construct. β-Tubulin served as a loading control. (G) Western blot analysis of histone extracts of NIH-3T3 cells transient transfected with control or ISL1 expression construct. Histone H3 served as a loading control. Optical densities of protein bands were quantified by Image J software and relative expression levels of H3K27me3 to Histone H3 were shown in D and G. Data in A, C and E are mean ± SD, n = 3. ***P < 0.001.
DISCUSSION
ISL1 is a LIM-Homeodomain transcription factor that marks cardiac progenitors in second heart field (2), and plays a critical role in regulating the expression of multiple downstream targets to control the proliferation, survival and differentiation of cardiac progenitors. However, the precise mechanism underlying ISL1 function in cardiac progenitors has not been fully elucidated. In our study, taking advantage of next generation sequencing methodologies, we have identified genome-wide targets of ISL1 in cardiac progenitors and further identified JMJD3 as a novel partner of ISL1.
Emerging evidence indicates that ISL1 may play a role in epigenetic regulation (26,27). It is well recognized that H3K27me3-H3K4me3 bivalent marks are co-enriched at the poised promoter regions, whereas H3K27me3-H3K4me1 marks are enriched at the poised enhancer regions (53–55). In our study, we analyzed the histone modifications at the enhancers of Mef2c and Myocd, two representative downstream targets of ISL1 during cardiac differentiation. Our date revealed that the occupation of ISL1 at these enhancers triggers dynamic modeling of histone modification profiles, leading to the expression of Mef2c and Myocd. We showed that H3K27me3 was gradually replaced by H3K27ac specifically at cardiac progenitor stage, coincidental with ISL1 binding. This event marks the transition of the inactive/poised enhancer to an active state. Notably, previous report suggests that ISL1 can interact with P300 during heart development, potentially accounting for the accumulation of H3K27ac at these enhancers (26).
In our study, we found that the demethylation of H3K27me3 was significantly impaired at the enhancers of ISL1's downstream targets upon depletion of ISL1. However, we also observed that H3K4me1 was differentially affected on these target loci, indicating that ISL1 may possess different regulation mechanisms for these targets. Recently, our group found that ISL1 interacts with SET7/9, a H3K4me1 methyltransferase, and together with PDX1 forms a complex to regulate pancreatic islet cells proliferation (56). Whether SET7/9 could physically interact with ISL1 to regulate H3K4 methylation in the context of cardiac differentiation remains to be tested.
JMJD3 plays important roles in development and disease (28). In our study, we uncovered a novel function for JMJD3 in cardiac progenitor differentiation. Specifically, JMJD3 is responsible for the proper removal of tri-methylation of H3K27 at the enhancers of ISL1's downstream targets at cardiac progenitor stage, and that JMJD3 depleted ESCs are defective for cardiac differentiation. It has been reported that JMJD3 can interact with β-catenin to regulate mesoderm genes (29). We also previously showed that ISL1 is regulated by WNT/β-catenin/LEF1 signaling during the early cardiac differentiation stage (57). Considering these, the constitutive depletion of JMJD3 may impair mesoderm differentiation, thereby affecting the expression of ISL1. With these in mind, and to better elucidate the role of JMJD3 at cardiac progenitors stage, we conditionally depleted JMJD3 without affecting mesoderm differentiation and the expression of ISL1. Notably, we showed that the expression of all tested downstream genes of ISL1 was still significantly impaired, accounting for the defective cardiac differentiation. Hence, JMJD3 not only plays an important role for mesoderm differentiation, but also for the cardiac progenitors formation. Physically, ISL1 and JMJD3 form a transcription regulatory complex that co-occupies at the enhancers of ISL1's downstream targets at cardiac progenitors stage, and that ISL1 is required for the recruitment of JMJD3 to these enhancers. These results indicate that JMJD3 exerts a context-dependent effect during development by interacting with different partners.
However, the biological relevant of JMJD3 in heart development is still unclear. It has been reported that Jmjd3 KO mice die perinatally due to the respiratory failure and premature development of lung tissues (58,59). And the heart development is still normal in these mice. In contrast to these findings, Ohtani et al group reported that Jmjd3 KO mice die at embryonic day 6.5 (29), which is consistent with the requirement of Jmjd3 for blastocyst development (60). Unfortunately, the early embryonic lethality of these mice, prior to heart development, precluded the further analysis of the effect of Jmjd3 on heart development. The discrepancy between the phenotypes of Jmjd3 KO mice is still unclear but probably due to the different strategies that were used to generate the Jmjd3 KO mice. Nevertheless, Jmjd3 KO mESCs are defective for the mesoderm formation and cardiac differentiation (29), which is consistent with our findings, inferring that JMJD3 might play important role in heart development. However, to definitively unveil the function of JMJD3 in heart development, cardiac specific (or conditional) Jmjd3 KO mice might be required.
Last but not least, we found that ISL1 could modulate the demethylase activity of JMJD3, regulating the global level of histone H3K27me3. JMJD3 is a jumonji C (JmjC) domain containing protein, that belongs to the α-ketoglutarate (αKG)-dependent oxygenase super family. The demethylase activity of JMJD3 is regulated by its substrates, such as oxygen, iron (Fe) and αKG (61). Increasing intracellular αKG promotes H3K27me3 demethylation (62). It remains unclear whether ISL1 could regulate the activity of JMJD3 indirectly through modulating its substrates. Interestingly, our genome-wide analyses identified several downstream targets of ISL1 that are involved in metabolic processes. These include Plod2, Ogfod2, Aldh4a1 and Gfpt2, potentially involved in the regulation of intracellular αKG availability. Alternatively, our data show that ISL1 can directly interact with the TCZ domain of JMJD3, which is adjacent to the JmjC domain. It is thus plausible that the interaction between ISL1 and JMJD3 may lead to allosteric changes in the catalytic domain of JMJD3, in this case, directly modulating the activity of JMJD3. This is an attractive possibility that requires further experimental validation.
In conclusion, our study highlights a novel role for ISL1 in enhancer patterning of key cardiac progenitor genes and provides mechanistic insights into how ISL1/JMJD3 complex orchestrates histone modification changes, coordinating gene expression driving cardiogenesis.
ACCESSION NUMBERS
All sequencing data have been deposited to the Gene Expression Omnibus under accession number GSE 79701.
Supplementary Material
Acknowledgments
We thank Pedro Lee (New York University Langone School of Medicine), and Weewei Tee (Agency for Science, Technology and Research) for thoughtful discussions and revision of the manuscript. We also thank the New York University Genome Technology Center for help with sequencing and Ricardo Saldana-Meyer (New York University Langone School of Medicine) for the generous help with data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81071675, 81370236, 81371889, 81472022, 81170713]; Natural Science Foundation of Beijing [5122021]; Leading Academic Discipline Project of Beijing Education Bureau, the 111 Project of China [B07001]; China Scholarship Council (to Y. W.). Funding for open access charge: the National Natural Science Foundation of China [81370236, 81371889].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ericson J., Thor S., Edlund T., Jessell T.M., Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;256:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- 2.Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlgren U., Pfaff S.L., Jessell T.M., Edlund T., Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 4.Moretti A., Caron L., Nakano A., Lam J., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martinpuig S. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N.J., Jiang C., Cao X., Zhao X., Zhang X., et al. Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. invest. 2015;125:3256–3268. doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretti A., Bellin M., Jung C.B., Thies T.M., Takashima Y., Bernshausen A., Schiemann M., Fischer S., Moosmang S., Smith A.G., et al. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J. 2010;24:700–711. doi: 10.1096/fj.09-139477. [DOI] [PubMed] [Google Scholar]
- 7.Genead R., Danielsson C., Andersson A.B., Corbascio M., Franco-Cereceda A., Sylven C., Grinnemo K.H. Islet-1 cells are cardiac progenitors present during the entire lifespan: from the embryonic stage to adulthood. Stem Cells Dev. 2010;19:1601–1615. doi: 10.1089/scd.2009.0483. [DOI] [PubMed] [Google Scholar]
- 8.Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S., et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 10.Weinberger F., Mehrkens D., Friedrich F.W., Stubbendorff M., Hua X., Muller J.C., Schrepfer S., Evans S.M., Carrier L., Eschenhagen T. Localization of Islet-1-positive cells in the healthy and infarcted adult murine heart. Circ Res. 2012;110:1303–1310. doi: 10.1161/CIRCRESAHA.111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuentes T.I., Appleby N., Tsay E., Martinez J.J., Bailey L., Hasaniya N., Kearns-Jonker M. Human neonatal cardiovascular progenitors: unlocking the secret to regenerative ability. PLoS One. 2013;8:e77464. doi: 10.1371/journal.pone.0077464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laugwitz K.-L., Moretti A., Lam J., Gruber P., Chen Y., Woodard S., Lin L.-Z., Cai C.-L., Lu M.M., Reth M., et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black B.L. Transcriptional pathways in second heart field development. Semin. Cell De Dov. Biol. 2007;18:67–76. doi: 10.1016/j.semcdb.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi J.K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A.B., Mori A.D., Arruda E.P., Gertsenstein M., Georges R., Davidson L., Mo R., et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 15.Dodou E. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 16.Kwon C., Qian L., Cheng P., Nigam V., Arnold J., Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat. Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Son C.G., Bilke S., Davis S., Greer B.T., Wei J.S., Whiteford C.C., Chen Q.R., Cenacchi N., Khan J. Database of mRNA gene expression profiles of multiple human organs. Genome Res. 2005;15:443–450. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wamstad J.A., Alexander J.M., Truty R.M., Shrikumar A., Li F., Eilertson K.E., Ding H., Wylie J.N., Pico A.R., Capra J.A., et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.P., Bruneau B.G. Epigenetics and cardiovascular development. Ann. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 21.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K., et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergemann A.D., Cole F., Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet.: TIG. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Lee S., Lee J.W., Lee S.K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakroun I., Yang D., Girgis J., Gunasekharan A., Phenix H., Kaern M., Blais A. Genome-wide association between Six4, MyoD and the histone demethylase Utx during myogenesis. FASEB J. 2015;29:4738–4755. doi: 10.1096/fj.15-277053. [DOI] [PubMed] [Google Scholar]
- 25.Lickert H., Takeuchi J.K., Von Both I., Walls J.R., McAuliffe F., Adamson S.L., Henkelman R.M., Wrana J.L., Rossant J., Bruneau B.G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z., Kong J., Pan B., Sun H., Lv T., Zhu J., Huang G., Tian J. Islet-1 may function as an assistant factor for histone acetylation and regulation of cardiac development-related transcription factor Mef2c expression. PLoS One. 2013;8:e77690. doi: 10.1371/journal.pone.0077690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caputo L., Witzel H.R., Kolovos P., Cheedipudi S., Looso M., Mylona A., van I.W.F., Laugwitz K.L., Evans S.M., Braun T., et al. The Isl1/Ldb1 complex orchestrates genome-wide chromatin organization to instruct differentiation of multipotent cardiac progenitors. Cell Stem Cell. 2015;17:287–299. doi: 10.1016/j.stem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burchfield J.S., Li Q., Wang H.Y., Wang R.F. JMJD3 as an epigenetic regulator in development and disease. Int. J. Biochem. Cell Biol. 2015;67:148–157. doi: 10.1016/j.biocel.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtani K., Zhao C., Dobreva G., Manavski Y., Kluge B., Braun T., Rieger M.A., Zeiher A.M., Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circulation Res. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi T., Lord B., Schulze P.C., Fryer R.M., Sarang S.S., Gullans S.R., Lee R.T. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 31.Wee S., Wiederschain D., Maira S.M., Loo A., Miller C., deBeaumont R., Stegmeier F., Yao Y.M., Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc. Nat. Academy U.S.A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C., et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 33.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis N.J., Kingston R.E., Woodcock C.L. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 36.Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nature Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon-Divon M., Dvinge H., Tammoja K., Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye T., Krebs A.R., Choukrallah M.A., Keime C., Plewniak F., Davidson I., Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis G. Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated Discovery. Genome bBol. 2003;4:P3. [PubMed] [Google Scholar]
- 46.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golzio C., Havis E., Daubas P., Nuel G., Babarit C., Munnich A., Vekemans M., Zaffran S., Lyonnet S., Etchevers H.C. ISL1 directly regulates FGF10 transcription during human cardiac outflow formation. PloS one. 2012;7:e30677. doi: 10.1371/journal.pone.0030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly M.L., Astsaturov A., Rhodes J., Chernoff J. A Pak1/Erk signaling module acts through Gata6 to regulate cardiovascular development in zebrafish. Devel. Cell. 2014;29:350–359. doi: 10.1016/j.devcel.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamikawa Y.F., Donohoe M.E. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics. 2014;9:834–841. doi: 10.4161/epi.28524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 55.Bulger M., Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z., Zhang Q., Lu Q., Jia Z., Chen P., Ma K., Wang W., Zhou C. ISL-1 promotes pancreatic islet cell proliferation by forming an ISL-1/Set7/9/PDX-1 complex. Cell Cycle. 2015;14:3820–3829. doi: 10.1080/15384101.2015.1069926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu H., Li Y., Wang Y., Liu Y., Wang W., Jia Z., Chen P., Ma K., Zhou C. Wnt-promoted Isl1 expression through a novel TCF/LEF1 binding site and H3K9 acetylation in early stages of cardiomyocyte differentiation of P19CL6 cells. Mol. Cell. Biochem. 2014;391:183–192. doi: 10.1007/s11010-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 58.Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 59.Burgold T., Voituron N., Caganova M., Tripathi P.P., Menuet C., Tusi B.K., Spreafico F., Bevengut M., Gestreau C., Buontempo S., et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2012;2:1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Canovas S., Cibelli J.B., Ross P.J. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2400–2405. doi: 10.1073/pnas.1119112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cloos P.A., Christensen J., Agger K., Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carey B.W., Finley L.W., Cross J.R., Allis C.D., Thompson C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.