Abstract
To improve the epigenomic analysis of tissues rich in 5-hydroxymethylcytosine (hmC), we developed a novel protocol called TAB-Methyl-SEQ, which allows for single base resolution profiling of both hmC and 5-methylcytosine by targeted next-generation sequencing. TAB-Methyl-SEQ data were extensively validated by a set of five methodologically different protocols. Importantly, these extensive cross-comparisons revealed that protocols based on Tet1-assisted bisulfite conversion provided more precise hmC values than TrueMethyl-based methods. A total of 109 454 CpG sites were analyzed by TAB-Methyl-SEQ for mC and hmC in 188 genes from 20 different adult human livers. We describe three types of variability of hepatic hmC profiles: (i) sample-specific variability at 40.8% of CpG sites analyzed, where the local hmC values correlate to the global hmC content of livers (measured by LC-MS), (ii) gene-specific variability, where hmC levels in the coding regions positively correlate to expression of the respective gene and (iii) site-specific variability, where prominent hmC peaks span only 1 to 3 neighboring CpG sites. Our data suggest that both the gene- and site-specific components of hmC variability might contribute to the epigenetic control of hepatic genes. The protocol described here should be useful for targeted DNA analysis in a variety of applications.
INTRODUCTION
Hydroxymethylcytosine (hmC) was first detected in mammalian DNA as early as in 1972, but did not get further attention until 2009, when it was rediscovered as a product of methylcytosine (mC) oxidation by the Ten-Eleven Translocation 1 (TET1) enzyme (1–3). This cytosine modification is well established as a transient intermediate in active enzymatic demethylation of DNA (4), however, it can also persist as a relatively stable epigenetic mark (5). Multiple studies have shown that, in contrast to mC, hmC is enriched in actively transcribed genes (6–11). In line with this observation, the composition of DNA binding proteins differs significantly between mC- and hmC-containing DNA, thereby mediating the opposite transcriptional influences of mC and hmC by the ‘epigenetic reader’ proteins (12). In particular, the hmC-sensitive binding of the liver-enriched transcription factor CEBPB to its cognate sequence may have important implications for the epigenetic regulation of hepatic genes (13). Interestingly, hmC is enriched in the flanks of demethylated regions such as CpG islands and enhancers, thereby further supporting its role in the regulation of transcription (6–11).
Currently, bisulfite sequencing (BS-Seq) is widely used for the analysis of DNA methylation with single base resolution. However, this methodology cannot discriminate between mC and hmC (14). In contrast, Tet1-assisted bisulfite sequencing (TAB-Seq) allows for the separate detection of all three cytosine states (unmodified C, mC and hmC), when combined with conventional BS-Seq data (15). An alternative modification of bisulfite sequencing, oxidative bisulfite sequencing (oxBS-Seq, or TrueMethyl) can also distinguish between mC and hmC (16). However, comprehensive cross-validation studies of TAB-Seq and TrueMethyl methods are still lacking.
The global hmC content of genomic DNA varies between different human tissues and cell types. Until recently, it was believed that hmC was abundant only in human and murine embryonic stem cells (ESCs) and neurons (17). However, we have shown that the adult human liver is also an hmC-rich tissue (10). Hydroxymethylcytosine is likely to be important for liver function, since the non-genotoxic carcinogen phenobarbital was demonstrated to cause genome-wide perturbations of the hepatic hmC profiles, accompanied by altered expression of multiple drug-metabolizing genes, in a mouse model (18).
Liver is a highly specialized organ responsible for the metabolism of numerous endogenous compounds (cholesterol, triglycerides, glycogen, amino acids, insulin etc). Moreover, hepatocytes express cytochromes P450 and other enzymes (n > 200) which are relevant for the metabolism of prescribed drugs and other xenobiotics. The ADME genes (which are responsible for Absorption, Distribution, Metabolism and Excretion of drugs) are distinguished by very high interindividual variability in expression, and such variation constitutes a major reason for interindividual differences in drug response and adverse drug reactions (ADRs) (19). It is estimated that ADRs cause up to 7% of all hospital admissions in the UK and are responsible for the withdrawal of 4% of new drugs from the market and up to 50% of drugs in development (20). Importantly, genetic factors were estimated to account for only 20–30% of the interindividual variation in drug response, thus a significant proportion of ADRs may be due to epigenetic regulation of genes involved in drug metabolism (21). Given the high abundance of hmC in hepatic DNA and its presumable role in liver function, we considered it of interest to study the mC and hmC distribution with single base resolution in 188 different ADME genes in relation to their expression.
For this purpose, we combined the TAB-Seq methodology for separate detection of cytosine modifications with the Agilent Methyl-SEQ platform for target enrichment, thus yielding the novel TAB-Methyl-SEQ protocol. We validated this method by two other TAB-Seq-based protocols (TAB-SeqCapEpi and TAB-450K) and two TrueMethyl-based ones (TrueMethyl-WGBS and TrueMethyl-450K), as well as by hydroxymethylation-sensitive restriction enzyme treatment followed by qPCR. Our data indicate that: (i) TAB-Methyl-SEQ and other TAB-Seq-based protocols are superior in terms of precision of hmC calls compared to TrueMethyl-based methods; (ii) the interindividual variability of hmC values is partially determined by the global hmC content of liver samples, (iii) the expression of hepatic genes positively correlates with the averaged level of hmC in their coding regions and (iv) at certain CpG sites, prominent hmC peaks with putative regulatory functions are observed, thus suggesting the targeted recruitment of DNA hydroxymethylation enzymes to specific genomic sequences. The results emphasize the necessity to use methods that can distinguish between mC and hmC with single base resolution for understanding the epigenetic gene regulation in hmC-rich tissues. In summary, our protocol offers a cost-effective way for the detection of mC and hmC with single base resolution at customizable gene panels of choice.
MATERIALS AND METHODS
Human liver samples
The twenty adult human liver samples used originate from organ donors who met accidental death. They were acquired from either Karolinska University Hospital (Huddinge, Sweden), or Sahlgrenska Hospital (Gothenburg, Sweden), or were purchased from the International Institute for the Advancement of Medicine (IIAM; NJ, USA) and from XenoTech (KS, USA). The use of tissue from adult livers in this study was approved by the Ethics Committees at Karolinska University Hospital.
Nucleic acid samples and expression profiling
Genomic DNA (gDNA) was isolated from 15–25 mg frozen liver tissue by DNA Mini kit (QIAgen, CA, USA). DNA was quantified using Quant-iT PicoGreen dsDNA Assay kit (Invitrogen, USA). RNA isolation and expression profiling by Illumina HumanHT-12 BeadChip were described previously (10,22). In this study, the specificity of Illumina HumanHT-12 probes that recognize ADME mRNAs was assessed by NCBI BLAST, and probes showing sequence homology to unrelated transcripts above 80%, were removed from further analysis (CYP3A5, CYP2B6, CYP2C9, CYP2C19, CYP2D6, GSTA1, GSTA5, GSTM1, GSTT2, SLC22A3, SULT1A3, SULT1A4, UGT2B7, UGT2B10, UGT2B11, UGT2B15, UGT2B28).
Liquid chromatography–mass spectrometry
For the quantification of global mC and hmC content by liquid chromatography-mass spectrometry (LC-MS), 200–400 ng of gDNA samples were converted to nucleoside monomers by the DNA Degradase Plus enzyme (ZymoResearch, CA, USA). The 5-Methylcytosine and 5-Hydroxymethylcytosine DNA Standard Set (ZymoResearch, CA, USA) was used to prepare quantification standards. To improve the accuracy, isotope-labeled compounds (2′-deoxyguanosine-13C15N2, 5-methyl-2′-deoxycytidine-d3 and 5-hydroxymethyl-2′-deoxycytidine-d3) were used as spike-in controls. The full LC-MS protocol is detailed in Supplementary Text S1 (see Supplementary File 1).
TAB-Methyl-SEQ
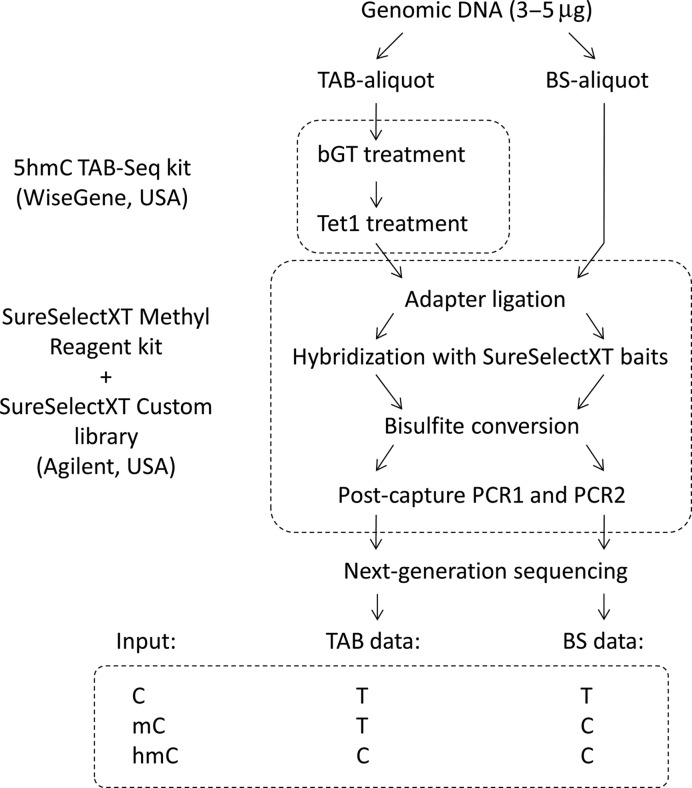
The TAB-Methyl-SEQ protocol requires the TAB-Seq kit (WiseGene, IL, USA) and the SureSelectXT Methyl Reagent kit together with a SureSelectXT Custom oligonucleotide library (Agilent, CA, USA). Sonicated genomic DNA samples (3–5 μg) were split into BS- and TAB-aliquots (1/3 and 2/3 of the input volume, respectively). The TAB-aliquots were treated with beta-glucosyltransferase (βGT) and then oxidized with the mTet1 enzyme from the TAB-Seq kit (mTet1 is a recombinant mouse homolog of the human TET1). Then both BS- and TAB-aliquots were subjected to the Methyl-SEQ protocol ver. B (Agilent G7530-90002), with the exception that the volume of the custom SureSelectXT oligonucleotide library in each hybrid capture reaction was half of the recommended volume (2.5 μl instead of 5 μl). The full TAB-Methyl-SEQ protocol is described in Figure 1 and detailed in Supplementary Text S3 (see Supplementary File 1).
Figure 1.
Description of the novel TAB-Methyl-SEQ workflow. Input genomic DNA is split into two aliquots (‘BS’ and ‘TAB’). The TAB-aliquot is treated by beta-glucosyltransferase (βGT) and then by recombinant mouse mTet1 enzyme from TAB-Seq kit. Both TAB- and BS-aliquots are subjected in parallel to the Agilent Methyl-SEQ protocol which enables the construction of target enriched NGS libraries. The sequencing of TAB-libraries allows detection of hmC separately from other cytosine states. By comparing TAB and BS data coming from the same input DNA sample, all three major cytosine states (C, mC and hmC) can be quantified.
TAB-SeqCapEpi
The TAB-SeqCapEpi protocol requires the TAB-Seq kit (WiseGene, IL, USA) and the SeqCap Epi Choice Enrichment library together with the compatible library prepation reagents (KAPA Lib Prep kit, SeqCap Adapter kit, SeqCap EZ Hybridization and Wash kit, SeqCap Epi Accessory kit, SeqCap HE-Oligo kit and SeqCap EZ Pure Capture Bead kit) (Roche Nimblegene, WI, USA). Sonicated genomic DNA samples (1–1.5 μg) were split into BS- and TAB-aliquots (1/3 and 2/3 of the input volume, respectively). The TAB-aliquots were treated with βGT and then oxidized with the mTet1 enzyme from the TAB-Seq kit. Then both BS- and TAB-aliquots were subjected to the SeqCap Epi Enrichment System protocol v1.0 with the exception that up to six aliquots (corresponding to three input gDNA samples) were pooled prior to hybrid capture. The full TAB-SeqCapEpi workflow is described in Supplementary Text S4 (Supplementary File 1).
TAB-450K
Sample preparation for the genome-wide analysis with the TAB-450K protocol requires the TAB-Array kit (WiseGene, IL, USA). Genomic DNA samples (400–800 ng) were split into BS- and TAB-aliquots (1/3 and 2/3 of the input volume, respectively). The TAB-aliquots were treated with TAB-Array kit following the manufacturer's instructions. Then both the BS- and TAB-aliquots were bisulfite modified using the EZ DNA Methylation kit (Zymo Research, CA, USA), according to the manufacturer's recommendations for the Illumina Infinium Assay. After purification, 4 μl of each bisulfite-converted DNA sample were used for hybridization on Infinium HumanMethylation450 BeadChips, following the Illumina Infinium HD Methylation protocol. The original IDAT files were extracted from the HiScanSQ scanner. Data pre-processing and quality control analysis were performed in R using the Bioconductor package minfi (23). ‘Raw’ pre-processing was used to convert the intensities from the red and the green channels into methylated and non-methylated signals. Beta values were computed using Illumina's formula [beta = M/(M + U + 100)]. The difference in the distribution of beta values for type I and type II probes was corrected using SWAN, a normalization method to deal with systematic changes between type I and type II probes (24). Detection P-values were obtained for every CpG probe in every sample. Only probes with detection P-values below 0.01 were used for downstream analysis.
TrueMethyl-450K
Genomic DNA isolated by DNA Mini kit (QIAgen, CA, USA) was additionally subjected to phenol–chloroform extraction followed by clean-up on AMPure XP beads to meet the TrueMethyl-specific requirements on input DNA purity. Around 1 μg of purified DNA was processed using the TrueMethyl Array kit (Cambridge Epigenetix, Cambridge, UK) according to the manufacturer's instructions. The efficiency of DNA oxidation was controlled by the color change during the oxidation reaction and also by gel electrophoresis of the spike-in Digestion Control, as outlined in the protocol for the TrueMethyl Array kit. Both controls revealed that the DNA oxidation efficiency was sufficient to continue with the microarray analysis. The ssDNA concentration in the BS and OxBS aliquots was determined by Qubit, and 7 μl corresponding to approximately 350 ng per aliquot were used for hybridization on Infinium HumanMethylation450 BeadChips following the Illumina Infinium HD Methylation protocol. Data processing was done in Illumina Genome Studio using Normalization controls and Subtract background setups. Only probes with detection P-values below 0.01 were used for downstream analysis.
TrueMethyl-WGBS
Whole genome bisulfite samples were generated using Cambridge Epigenetix (CEGX) TrueMethyl Whole Genome kit (Cambridge Epigenetix, Cambridge, UK), protocol version 2.1 (December 2015). DNA was sheared to approximately 700 bp using Covaris S2 prior to processing (Mode: Frequency Sweeping; Duty cycle: 5%; Intensity: 3; Cycles/burst: 200; Time: 82 s). Libraries were sequenced using an Illumina HiSeq X v2.0 paired end 150 bp run with a 1% spike-in of PhiX DNA. Both the BS and oxBS samples yielded 320 million raw reads. The efficiency of DNA oxidation by the TrueMethyl reagent was controlled by interrogation of the spike-in Digestion Control, as detailed in the TrueMethyl WholeGenome kit protocol. The BS aliquot yielded the following values: C = 0.59%, mC = 95.39%, hmC = 97.56%, whereas corresponding values in the oxBS aliquot were: C = 0.57%, mC = 95.91%, hmC = 6.16%, thus confirming the expected DNA oxidation efficiency.
Validation of hmC values by qPCR
Validation of BS and hmC calls at selected CCGG sites was done by EpiMark 5-hmC and 5-mC Analysis Kit (New England Biolabs, MA, USA). Three aliquots per sample were processed from 1–1.5 μg of input DNA: (i) fully untreated (positive control); (ii) βGT/MspI-treated (hmC signal) and (iii) MspI-treated (negative control). Equal volumes of each aliquot (containing 10–25 ng enzyme treated DNA) were used for qPCR reactions. The coordinates of CCGG site and the sequences of corresponding qPCR primers are shown in Supplementary Table S1 (see Supplementary File 1). The cytosine modification values were calculated by the following algorithm. First, DNA modification ratios (R values) for ‘hmC’ and ‘negative’ aliquots were calculated relative to the ‘positive’ aliquot, using the delta Ct method. R values of ‘negative’ aliquots were around 0.05 in the majority of CCGG sites analyzed, thus demonstrating the high efficiency of MspI enzyme cutting nonglucosylated DNA. However, in roughly one out of ten CCGG sites analyzed the ‘negative’ aliquot showed unexpectedly high R values (above 0.1), thus suggesting either incomplete cutting of genomic DNA by MspI at given CCGG site, or low efficiency of qPCR amplification by given pair of primers, or both. To compensate for this undesired effect and thus to avoid possible overestimation of hmC calls, R values from ‘hmC’ aliquots were normalized, based on the assumption that R values from ‘positive’ and ‘negative’ aliquots correspond to 100 and 0% cytosine modification, respectively. The following formula was used: R_hmC_normalized = (R_hmC - R_negative)/(1 - R_negative). Technical reproducibility of this method was assessed using two aliquots of the same liver gDNA sample with primer pairs 1–24 (see Supplementary Table S1). One of these gDNA aliquots was subjected to phenol/chlorophorm purification step followed by AMPure XP bead clean-up prior to βGT/MspI treatment (to imitate the purity of DNA analyzed using the TrueMethyl protocols). The other aliquot was not subjected to any additional purification steps, thus imitating the purity of DNA analyzed by TAB-Seq-based protocols. The correlation between hmC values produced by qPCR analysis of these two aliquots was very high (r = 0.97, P < 0.0001), thus suggesting high accuracy of qPCR-based hmC quantification even without any additional purification of hepatic gDNA samples.
Bioinformatics
Next-generation sequencing data from TAB-Methyl-SEQ and TAB-SeqCapEpi experiments were processed as follows: raw paired-end bisulfite reads from Illumina HiSeq2500 were subjected to quality trimming and adapter removal by Trim Galore v0.3.7 (- -quality 20 - -adapter AGATCGGAAGAGC - -stringency 1 - -paired - -length 40). Pre-processed reads were then mapped to bisulfite converted human genome (Hg19) with Bismark v0.12.5/Bowtie2 (- -bowtie2 - -fastq -D 20 -R 3) (25). PCR duplicates were removed from sorted SAM files by Picard v1.80. Methylation calls from BS- and TAB-libraries were generated by bismark_methylation_extractor script (- -paired-end - -no_overlap). Methylation and hydroxymethylation states of individual CpG sites were determined by MLML software with default settings (26).
Next-generation sequencing data from the TrueMethyl-WGBS experiment were processed using the fastq_bismark Cluster Flow v0.4 pipeline (http://clusterflow.io). Reads were quality filtered using FastQC v0.11.2. Adapters were trimmed plus 6 bp from 5′ and 2 bp from 3′ of both reads using Trim Galore (- -paired - -gzip - -phred33 - -clip_r1 6 - -three_prime_clip_r1 2 - -clip_r2 6 - -three_prime_clip_r2 2). Reads were aligned (bismark - -multicore 3 - -bam - -phred33-quals), deduplicated (deduplicate_bismark -p - -bam) and methylation statuses called (bismark_methylation_extractor - -multi 4 - -ignore_r2 1 - -ignore_3prime_r2 2 - -bedGraph - -counts - -buffer_size 10G - -gzip -p - -no_overlap - -report) using Bismark v.014.4 (25).
Tertiary data analysis was done by in-house Python3 scripts, which are available on request. Data visualization was done by matplotlib. Correlation coefficients and P-values were calculated according to Spearman method. Multiple testing correction was done according to Benjamini-Hochberg method with FDR = 0.05. The design of custom SureSelectXT library for the 17.5 Mb region of interest (Supplementary File 2) was generated by Agilent eArray online software. Baits that do not cover any CpG sites were removed from the eArray output, and the final 6.6 Mb of target intervals (represented by merged coordinates of SureSelectXT baits) are listed in Supplementary File 3.
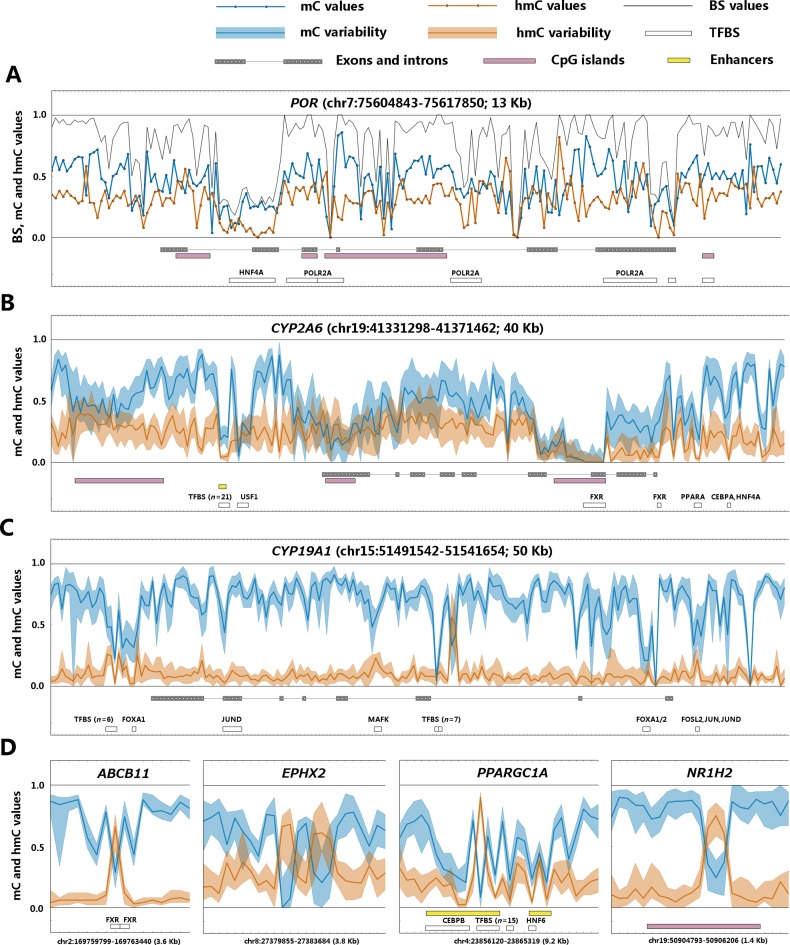
The coordinates of RefSeq genes, as well as the ChIP-Seq data on transcription factor binding sites in HepG2 cells were downloaded from UCSC Table Browser. Genomic CpG islands were obtained from (27), enhancers – from (28), ChIP-Seq data on binding sites for CEBPA, HNF4A, HNF6, HNF3G, PPARA and FXR in primary human hepatocytes – from (29–31). Coordinates of these genomic features in selected ADME genes are shown on Figure 6.
Figure 6.
Example cytosine modification profiles of ADME genes. (A) POR gene (P450 cytochrome oxidoreductase) in the most hmC-rich sample. The absolute bisulfite, mC and hmC values are shown by black, blue and orange lines, respectively. (B) CYP2A6 gene (hmC-rich, highly expressed). (C) CYP19A1 gene (hmC-poor, transcriptionally silent). (D) Selected short gene intervals with site-specific hmC peaks. On figures B–D, the blue and orange filled areas represent the range of inter-individual variation (from 5% quantile to 95% quantile) of mC and hmC signals in the 20 different livers analyzed by TAB-Methyl-SEQ. The blue and orange lines show the median mC and hmC values, respectively. This figure shows cytosine modification calls at individual CpG sites with single base resolution. The absolute BS, mC and hmC values were not averaged between adjacent CpG sites.
RESULTS
Development and benchmarking of the TAB-Methyl-SEQ protocol
For detailed analysis of mC and hmC distribution with single base resolution, we developed a novel TAB-Methyl-SEQ workflow, which combines the TAB-Seq methodology for separate detection of mC and hmC (15) with the Agilent Methyl-SEQ protocol for target enrichment of genomic DNA (Figure 1). The TAB-Methyl-SEQ protocol was used together with a custom SureSelectXT library which contains 55 000 baits and covers 6.6 Mb of genomic sequences in coding and regulatory regions of 188 ADME genes. The quality metrics obtained in the TAB-Methyl-SEQ experiment with 20 adult human liver samples are shown in Table 1. The region of interest encompasses 203 248 CpG sites, and 124 269 (61.1%) of these CpG sites can be analyzed by Agilent SureSelect target enrichment libraries. The 109 454 CpG sites on target, covered by at least 10 reads in both BS- and TAB-aliquots in at least 10 out of 20 samples, were considered for the subsequent analysis.
Table 1. Quality metrics in TAB-Methyl-SEQ and TAB-SeqCapEpi experiments.
| TAB-Methyl-SEQ (20 samples) | TAB-SeqCapEpi (8 samples) | |||
|---|---|---|---|---|
| BS aliquots | TAB aliquots | BS aliquots | TAB aliquots | |
| Raw reads | 5.1–13.1 M | 4.3–12.7 M | 14.7–37.0 M | 15.4–41.1 M |
| % mapped reads | 83.0–91.9% | 82.8–91.8% | 86.2–92.0% | 87.8–90.8% |
| % unique reads | 85.1–95.6% | 76.6–94.3% | 52.8–98.4 M | 55.3–98.7% |
| % reads on target | 64.4–79.9% | 71.9–80.4% | 30.7–38.6% | 29.6–37.6% |
| Representative reads on target | 3.0–7.4 M | 2.3–6.9 M | 4.1–7.8 M | 4.2–8.6 M |
| Target size | 6.6 Mb | 40.6 Mb | ||
| Median read depth on target | 40–104× | 32–97× | 9–16× | 11–18× |
The TAB-Methyl-SEQ data for one liver sample were validated using a set of different TAB-Seq-based and TrueMethyl-based protocols (Figure 2):
TAB-SeqCapEpi combines the TAB-Seq approach with the Roche Nimblegen SeqCap Epi platform for target enrichment of genomic DNA followed by NGS (32). We tested this platform with a custom SeqCap Epi library which covers 702 000 CpG sites in 40.6 Mb genomic sequences including 451 protein-coding genes, 118 miRNA genes and 6 lncRNA genes (Supplementary File 4). Besides of 20 Kb gene flanking intervals, the region of interest included also CpG islands and hepatic enhancers located within 50 Kb from these genes. Quality metrics obtained in our pilot TAB-SeqCapEpi experiments are shown in Table 1. For analysis of TAB-SeqCapEpi performance in comparison to TAB-Methyl-SEQ, see Supplementary Text S2 (Supplementary File 1). DNA (hydroxy)methylation data from TAB-SeqCapEpi were merged between forward and reverse DNA strands to achieve higher read depth at the cost of losing information on the possible strand-specific hmC deposition. Only CpG sites with merged read depth above 25× were used for cross-validation purposes;
TAB-450K combines TAB-Seq with Illumina Infinium HumanMethylation450 BeadChip assay (33,34). The latter is a microarray-based technique which allows to interrogate a defined set of CpG sites (n = 485 463);
TrueMethyl-450K employs the same Illumina HumanMethylation450 microarray platform together with the oxidative bisulfite (OxBS-Seq) methodology (35,36). This alternative approach for hmC detection involves the oxidation of input DNA by potassium perruthenate followed by bisulfite conversion (16);
Regarding TrueMethyl-WGBS (whole-genome oxidative bisulfute sequencing), we sequenced one liver gDNA sample with 4× median read depth. DNA (hydroxy)methylation data were merged between forward and reverse DNA strands, and only CpG sites covered by at least 25 reads were used for cross-validation analysis.
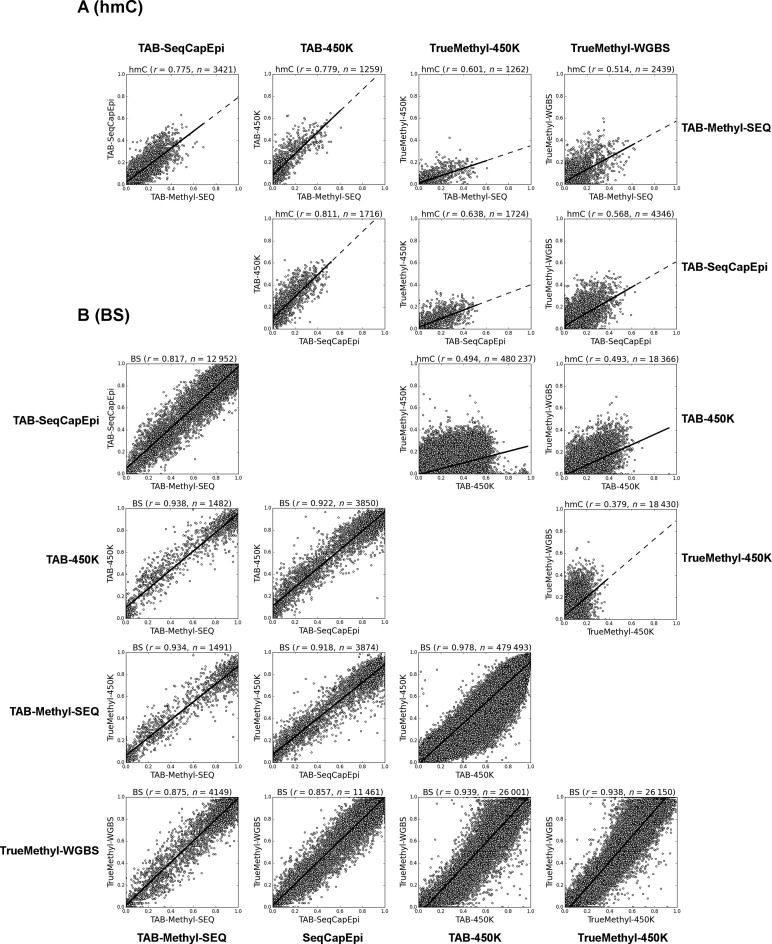
Figure 2.
Validation of TAB-Methyl-SEQ data. The hmC (A) and bisulfite (B) calls from one liver sample were compared between TAB-Methyl-SEQ, TAB-450K, TrueMethyl-450K and TrueMethyl-WGBS methods. Only CpG sites analyzed with read depth not less than 25× in NGS experiment were included into the analysis. Spearman correlation coefficient, as well as the number of CpG sites analyzed are indicated on top of each scatter plot. The axes scales are from 0.0 (no modified cytosines at given CpG site) to 1.0 (all cytosine residues are modified).
The comparison of these methods for single base resolution profiling of cytosine modifications clearly demonstrates that the best correlations of hmC calls are observed between the three TAB-Seq-based protocols (0.77 < r < 0.81; see Figure 2A). In contrast, correlation between TrueMethyl-450K and TrueMethyl-WGBS datasets was substantially lower (r = 0.38; see Figure 2A), thus suggesting that the TrueMethyl platform is less precise compared to TAB-based methodologies. When TrueMethyl-based protocols are compared with TAB-Seq data, the correlation coefficients are intermediate (0.49 < r < 0.64; see Figure 2A). In addition, the slope of the regression line in TrueMethyl versus TAB-Seq comparisons suggests a systematic bias between these two platforms for hmC detection. Bisulfite calls, on the other hand, correlated well between all protocols compared (see Figure 2B).
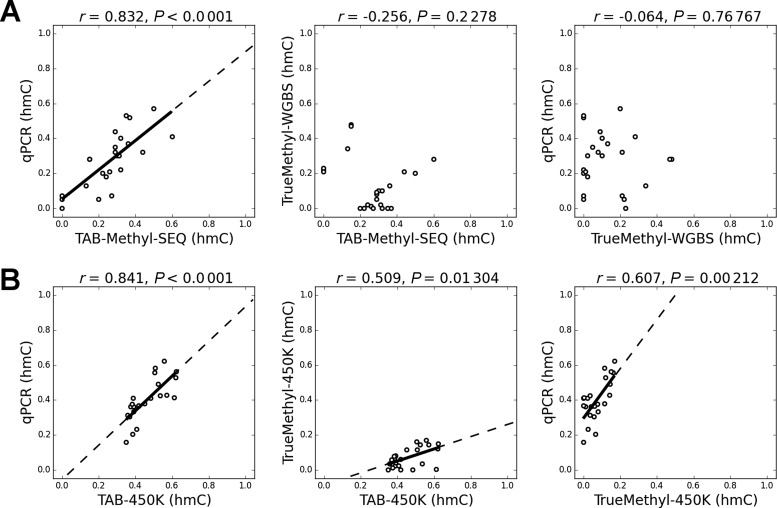
To determine which platform for hmC analysis provides the most accurate hmC calls, we validated a subset of TAB-Seq and TrueMethyl data by an additional unrelated method which allows for quantification of cytosine modifications in CCGG sites. This third methodology implemented in NEB EpiMark 5-hmC and 5-mC Analysis Kit is based on the selective protection of hmC residues by βGT, which is followed by cutting unprotected CCGG sites by MspI restriction enzyme (37). DNA that remains uncut is quantified by qPCR with primers flanking the CCGG site of interest. At the first step of qPCR validation we considered CCGG sites which were strongly discordant between TAB-Methyl-SEQ and TrueMethyl-WGBS datasets. Among 2439 CpG sites which were sequenced with at least 25× coverage by both methods, hmC values were different by more than 0.2 in 125 CpG sites (5.1%). In 90 of these discordant CpG sites (72%), the hmC values were higher in TAB-Methyl-SEQ than in TrueMethyl-WGBS. Discordant CpG sites located in CCGG context (n = 24) were analyzed by qPCR (see primer pairs 1–24 in Supplementary Table S1). As evident from Figure 3A, the hmC values obtained from qPCR are in agreement with the TAB-Methyl-SEQ data but not with TrueMethyl-WGBS.
Figure 3.
Validation of discordant CCGG sites by EpiMark 5-hmC and 5-mC Analysis kit. (A) CCGG sites with hmC values discordant for more than 0.2 between TAB-Methyl-SEQ and TrueMethyl-WGBS data were validated by qPCR (n = 24). All sites were covered by at least 25 reads in both NGS experiments. (B) CCGG sites with hmC values discordant for more than 0.2 between TAB-450K and TrueMethyl-450K data were validated by qPCR (n = 23). For the coordinates of these 47 CpG sites and the corresponding primers sequences see Supplementary Table S1. The axes scales are from 0.0 (no hmC at given CCGG site) to 1.0 (all cytosine residues are represented by hmC).
At the next step, we validated CpG sites that were discordant between TAB-450K and TrueMethyl-450K datasets. Among 480 237 CpG sites having detection P-values below 0.01 in both datasets, hmC values were different by more than 0.2 in 75,976 CpG sites (15.8%), and by more than 0.3—in 20,531 CpG sites (4.3%). In both cases the vast majority of discordant hmC calls (>99%) was higher in TAB-450K than in TrueMethyl-450K, thus confirming the systematic hmC bias observed between TAB-Seq and TrueMethyl platforms. We chose 23 discordant CpG sites in CCGG context for qPCR validation (see primer pairs 25–47 in Supplementary Table S1). The results indicate that qPCR data are in better agreement with TAB-450K than with TrueMethyl-450K (Figure 3B).
Analysis of mC and hmC in 20 different human livers
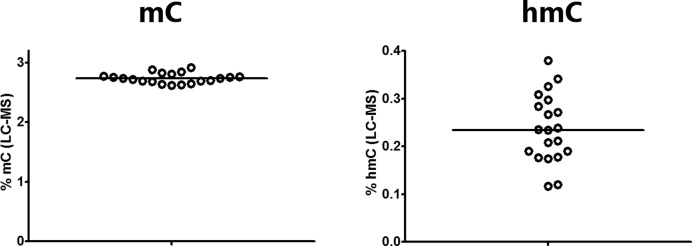
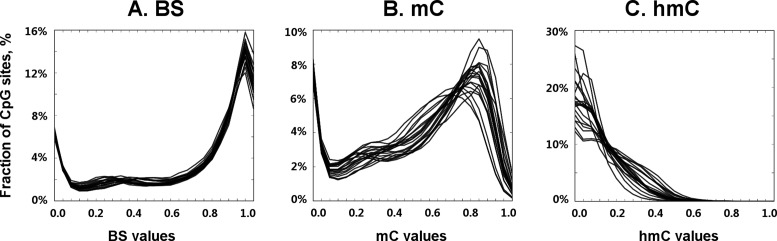
To study the role of hmC in the epigenetic regulation of ADME genes, we analyzed cytosine modification profiles in genomic DNA samples isolated from adult human livers (n = 20) by TAB-Methyl-SEQ. In parallel, we quantified the global mC and hmC content of these DNA samples by LC-MS and found that the hmC content varied about 3-fold (0.12 to 0.38% of total cytosine), whereas the global mC content varied only about 1.1-fold between individual livers (2.62 to 2.91% of total cytosine; Figure 4). The frequency distributions of mC and hmC values in TAB-Methyl-SEQ data were highly variable, whereas the distributions of cytosine modification values as revealed by the bisulfite technique were highly consistent between these samples (Figure 5). Importantly, a significant fraction of CpG sites in hmC-rich samples have a strong hmC signal. For example, the percentage of CpG sites with hmC values above 0.25 varies from 3% to 34% among individual livers (median 16%). Moreover, the percentage of CpG sites where hmC values exceed the corresponding mC values (with the latter filtered with a cutoff of > 0.1) varies from 1% to 11% in liver samples (median 4%), depending on their global hmC content. A representative example of cytosine modification profile of an ADME gene in the most hmC-rich liver sample is shown in Figure 6A. As seen, the abundance of hmC is comparable to that of mC along the whole gene encoding cytochrome P450 oxidoreductase, whereas at certain CpG sites the hmC signal does even exceed the corresponding mC signal.
Figure 4.
The global hmC and mC content of genomic DNA. Genomic DNA samples from 20 different adult livers were analyzed by LC-MS. The global mC and hmC content values are expressed as % of modified cytosine molecules, considering the total cytosine ( = C + mC + hmC) as 100%.
Figure 5.
Frequency distributions of cytosine modification values in 20 liver samples. BS (A), mC (B) and hmC (C) values from 109 454 CpG sites analyzed in TAB-Methyl-SEQ experiment are shown as frequency distribution curves (separately for each liver sample). X-axis: cytosine modification values ranging from 0.0 (unmodified cytosine only) to 1.0 (full cytosine modification). Y-axis: the fraction of CpG sites with given cytosine modification value.
The TAB-Methyl-SEQ method revealed that hmC is detectable at virtually every CpG site across the coding and regulatory regions of ADME genes (with the exception of strongly demethylated intervals such as CpG islands, where only unmodified cytosines were observed; see Figure 6A–D). The abundance of hmC in the coding regions varies up to 15-fold between different ADME genes in the same liver sample. Two representative examples from sequencing of the CYP2A6 and CYP19A1 genes are given in Figure 6B and C. As seen, the actively transcribed CYP2A6 is relatively hmC-rich (the median hmC value in its coding region varies from 0.11 to 0.30 in individual samples). By contrast, the median hmC value in the transcriptionally silent CYP19A1 gene varies from 0.04 to 0.10 in the different liver samples, and only two adjacent CpG sites in its intron 2 are covered by an hmC peak that exceeds 0.5 (see Figure 6C). A list of ADME genes ranked by their relative hmC abundance is given in Supplementary Table S2. As evident from this table, the median hmC values in the most hmC-poor genes, such as CYP26A1, which is not expressed in the human liver, do not exceed 0.02 even in the most hmC-rich sample.
The hmC values were also found to be highly variable between adjacent CpG sites, thus suggesting that the detection of hmC with single base resolution is important for the identification of hmC peaks. Several examples of hmC peaks spanning only 1 to 3 CpG sites are shown in Figure 6D. Importantly, such hmC peaks cannot be detected by the conventional bisulfite technique due to the reciprocity of mC and hmC values.
As evident from Figure 6B–D, the hmC signal is also variable between individual livers at the majority of the CpG sites. We found that the interindividual variability of hmC values to some extent correlates with the global hmC content of the liver samples measured by LC-MS. Indeed, the hmC values at 40.8% CpG sites positively correlate with the global hmC content of corresponding liver samples, whereas at 10.5% CpG sites the mC values positively correlate with the global mC content, after Benjamini–Hochberg correction (FDR = 0.05). In contrast, bisulfite values correlate with the LC-MS data only at a negligible fraction of individual CpG sites (Table 2).
Table 2. Frequencies of CpG sites where cytosine modification values correlate with the LC-MS data.
| Liquid chromatography – mass spectrometry (LC-MS) | ||||
|---|---|---|---|---|
| Global hmC content values | Global mC content values | |||
| TAB-Methyl-SEQ | r > 0 | r < 0 | r > 0 | r < 0 |
| hmC values | 40.8% | 0.4% | 0.4% | 8.6% |
| mC values | 1.0% | 25.7% | 10.5% | 0.4% |
| BS values | 2.8% | 0.7% | 0.9% | 0.6% |
Cytosine modifications in relation to gene expression
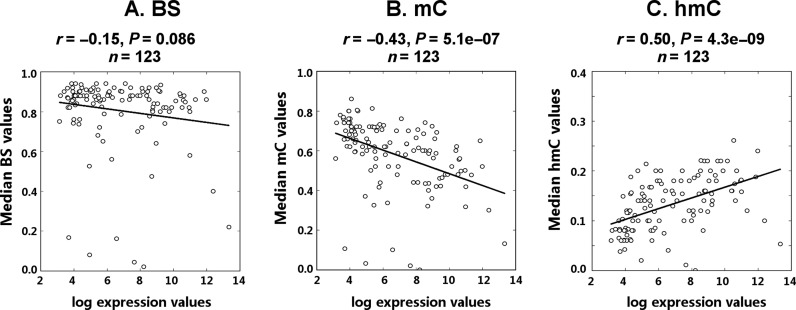
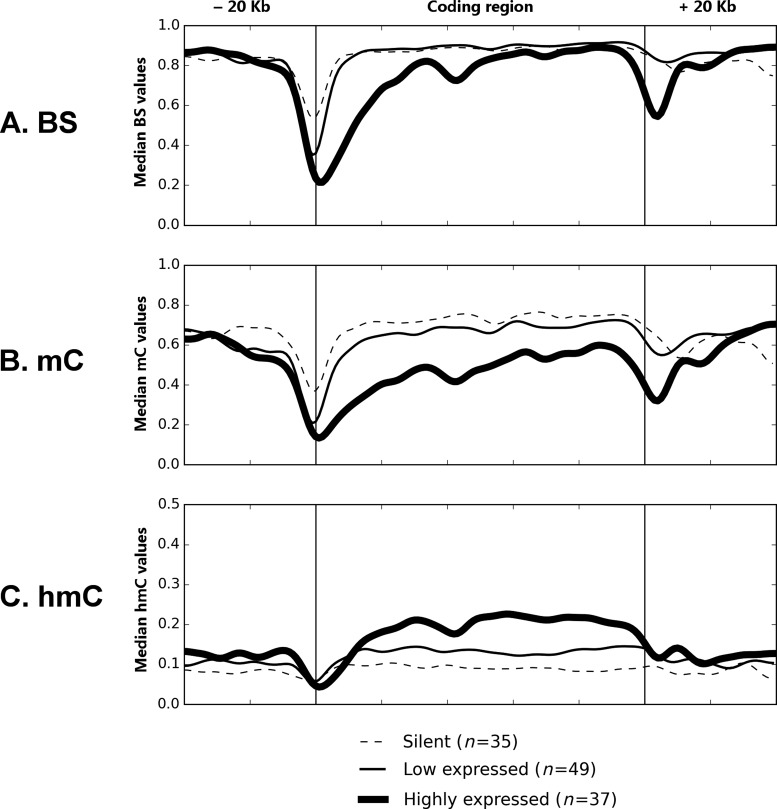
It is widely accepted that hmC is enriched in actively transcribed genes in neurons and ESCs, however the link between hmC and gene expression has not been sufficiently studied in hepatocytes. Thus, we evaluated the relative abundance of cytosine modifications of different ADME genes in relation to their expression. The expression levels of genes did not correlate with the median bisulfite values in their coding regions (P = 0.086; Figure 7A). However, gene expression correlated negatively with the median mC values (r = −0.43, P = 5E-07; Figure 7B) and positively with median hmC values under the same testing conditions (r = 0.50, P = 4E-09; Figure 7C). These findings were further confirmed by subdividing genes into silent, low and highly expressed groups. The cytosine modification profiles of genes were averaged within each expression group and plotted against their coding and regulatory regions (Figure 8). It is clear that the averaged mC and hmC profiles are lower and higher, respectively, in the group of actively transcribed genes. Within genes, the differences in mC/hmC abundance are seen along the whole coding region, but not in the 5′- and 3′-flanking regions (see Figure 8).
Figure 7.
Median cytosine modification values of ADME genes correlate with their expression. Genes (n = 123) were selected according to the following criteria: (i) they were quantified by reliable probes in Illumina HumanHT-12 assay; (ii) at least 100 CpG sites were analyzed by TAB-Methyl-SEQ in the coding region of each gene. The median BS, mC and hmC values were taken from CpG sites in the whole coding region of each gene. The resultant median BS (A), mC (B) and hmC (C) values were plotted against the log2-transformed expression levels of corresponding genes. Both median cytosine modification values and gene expression levels were averaged between all 20 samples.
Figure 8.
The averaged cytosine modification profiles of ADME genes, grouped by their expression levels. Genes (n = 121) were selected according to the following criteria: (i) they were quantified by reliable probes in Illumina HumanHT-12 assay; (ii) at least 300 CpG sites were analyzed by TAB-Methyl-SEQ in the coding and 5′- and 3′-flanking regions of each gene. These genes were split into three categories: highly expressed genes (with median linear expression values above 400), genes with low expression (between 30 and 400) and silent genes (expression below 30). At that, gene expression values were averaged between 20 samples. BS (A), mC (B) and hmC (C) values at individual CpG sites were averaged between 20 samples and between genes in given expression category, smoothed by the Gaussian formula and plotted against the genomic intervals corresponding to the coding and regulatory regions of ADME genes.
DISCUSSION
The genomic distribution of hmC has so far been studied mainly in human and murine ESCs and post-mitotic neurons, and it has been suggested that the functional role of hmC might be fundamentally different from that of mC (6–9,11). However, the traditional methods for analysis of DNA modifications, including the widely used bisulfite sequencing, do not allow for the discrimination between mC and hmC, thus potentially confounding the results of epigenomic profiling in hmC-rich tissues (14). In a previous study, we demonstrated that adult human livers contain variable but considerably high genomic levels of hmC, and affinity capture of hmC-containing DNA fragments followed by NGS revealed that the distribution of hmC-enriched intervals in the whole hepatic epigenome correlates with actively transcribed genes (10). However, this method does not provide absolute cytosine modification values and also lacks the single base resolution (38). Therefore, we developed a novel TAB-Methyl-SEQ protocol, which combines the TAB-Seq methodology (15) with the Agilent Methyl-SEQ target enrichment system, and used this method for epigenetic analysis of 188 ADME genes in 20 adult human livers. The protocol allows for the profiling of absolute mC and hmC values with single base resolution in genomic intervals of interest that can encompass up to hundreds of thousands of CpG sites. The NGS quality metrics of the target enriched libraries prepared according to the TAB-Methyl-SEQ workflow do not differ from their regular Methyl-SEQ counterparts, thus suggesting that the additional enzymatic treatment of the input DNA does not affect library performance (see Table 1).
A representative subset of cytosine modification calls from the TAB-Methyl-SEQ dataset were validated by two different TAB-Seq-based protocols (TAB-SeqCapEpi and TAB-450K), as well as by two TrueMethyl-based protocols (TrueMethyl-450K and TrueMethyl-WGBS; see Figure 2). To our knowledge, this is the first study where the performance of TAB-Seq and TrueMethyl platforms for single base hmC profiling was directly compared on genomic DNA. Our cross-validation data suggest that all three TAB-Seq-based methods are in a good agreement, whereas the TrueMethyl-based protocols systematically yield lower hmC values. The accuracy of the TAB- and TrueMethyl-based methodologies was validated by a third method (EpiMark 5-hmC and 5-mC Analysis Kit) which is based on MspI restriction enzyme treatment of glucosylated DNA followed by qPCR. Focusing on strongly discordant CpG sites, this method revealed better agreement with the hmC calls from TAB-Seq-based datasets than with those from TrueMethyl-based ones (see Figure 3). We suppose that the differences seen are not due to suboptimal activity of the T4 beta-glucosyltransferase (βGT) enzyme, since then one would expect to get systematically lower hmC values in βGT-dependent protocols (TAB-Seq and qPCR) compared to the βGT-independent TrueMethyl, whereas we do see the opposite. Instead, the observed underestimation of hmC calls in the TrueMethyl workflow might be explained by suboptimal efficiency of hmC oxidation by the TrueMethyl Oxidant Solution at certain CpG sites. On the other hand, interrogation of the synthetic spike-in controls in TrueMethyl-450K and TrueMethyl-WGBS workflows revealed that in both cases the hmC oxidation efficiency was sufficiently high to pass the quality check procedure suggested by the manufacturer (see Materials and methods). One can assume that the frequency of hmC conversion in the spike-in controls may not serve as a reliable indicator of the oxidation efficiency of genomic DNA sample, probably due to the variable nucleotide context of genomic CpG sites. Taken together, our data therefore suggest that the TAB-based methods are more precise and accurate than both TrueMethyl-based protocols. Further studies are however required for a comprehensive benchmarking of the TrueMethyl methodology in comparison to TAB-Seq.
According to the technical validation data, the TAB-Methyl-SEQ protocol appears to be robust and reliable. Hence, mC/hmC profiles of ADME genes generated by this protocol can be considered for further analysis, with the ultimate goal to characterize the role of cytosine modifications in the epigenetic regulation of hepatic genes. In earlier studies, the human and mouse hepatic hydroxymethylomes were analyzed by affinity capture of hmC-containing DNA fragments followed by hmC peak calling, and hmC peaks were shown to be associated with actively transcribed hepatic genes (10,18). Importantly, the affinity capture methodology used in these studies allows only for qualitative detection of hmC peaks with kilobase resolution. In contrast, the development of TAB-Methyl-SEQ protocol allowed us to quantitatively analyze the mC/hmC profiles of human livers with single base resolution, although at a limited number of genes. Another unique feature of the present study is that multiple samples from the same tissue were analyzed in parallel, thus allowing for the first time to characterize the DNA (hydroxy)methylome in relation to the variable global hmC content of individual samples.
Using the TAB-Methyl-SEQ protocol, we found that the local abundance of hmC can be similar to or even higher than that of mC (see Figure 6). Thus, hmC has to be considered as the second major cytosine modification in human liver, besides mC. These findings have important conceptual implications, as the conventional bisulfite technique implemented in the regular Methyl-SEQ protocol would considerably overestimate the true DNA methylation values at a significant fraction of CpG sites in hmC-rich samples, unless corrected for the hmC signal by TAB-Methyl-SEQ.
The abundance of hmC also differs up to 15-fold between genes within the same sample (see Supplementary Table S2). As evident from Figure 7, the local hmC content of ADME genes positively correlates with their expression (r = 0.50), whereas the median mC values correlate negatively with expression (r = −0.43). At the same time, we detected no correlation between the median bisulfite values and gene expression (P = 0.086; see Figure 7A), although a decrease of the bisulfite signal is observed in the beginning of the coding region of highly expressed genes (see Figure 8). The observed positive correlation between active gene transcription and the abundance of hmC in coding intervals is in agreement with the previously reported data in mammalian ESCs and in nervous tissue using either hmC immunoprecipitation (6–9) or whole-genome TAB-Seq (11,39). Here, we demonstrate that mC and hmC values averaged throughout the whole gene, but not the corresponding BS values, can discriminate between low- and highly expressed genes, thus underlining that the separation of mC and hmC signals is required to truly predict the gene transcription.
The correlation between mC/hmC abundance and gene expression (see Figure 7), as well as the precise mapping of the observed (hydroxy)methylation differences between highly expressed and silent genes to their coding regions (see Figure 8), suggest a functional link between DNA hydroxymethylation and the transcriptional machinery. However, these data do not allow us to judge if the increased hmC level determines the active gene transcription, or vice versa. In the latter case, the open chromatin conformation of highly expressed genes might promote the recruitment of TET enzymes, thus increasing the probability of DNA hydroxymethylation in the transcribed chromatin. These two alternatives are not mutually exclusive, because the increased hmC level, even being a consequence of gene activation by transcription factors, might reinforce the active state of expressed genes. Further studies in cell cultures are required to shed light onto the functional significance of the gene-specific hmC abundance in the transcriptional regulation of hepatic genes.
The results presented also imply a profound variability of the global hmC content (from 0.12 to 0.38% of total cytosine) between DNA samples isolated from different human livers, whereas the global mC content was considerably less variable, as measured by LC-MS (2.62 to 2.91% of total cytosine; see Figure 4). The reason for the observed interindividual variability of the global hmC content in livers remains to be identified. However, it might be suggested that alterations in the activity of enzymes involved into the biochemical transitions of hmC contribute to this phenomenon. In particular, the production of hmC due to the activity of TET family enzymes is influenced by the intracellular levels of their co-factor Fe2+ and co-substrate alpha-ketoglutarate (αKG), with the latter being produced from isocitrate by IDH (isocitrate dehydrogenase) enzymes (40). In addition to IDH, other components of the Krebs cycle such as SDH (succinate dehydrogenase) and FH (fumarate hydratase) have also been shown to competitively inhibit the activity of TET enzymes by changing the levels of succinate and fumarate, respectively (40). Furthermore, oxidative stress has been proposed to affect TET enzyme activity through the Sirt3-dependent acetylation of IDH2 (41). The exposure to chemicals such as phenobarbital, diethylstilbestrol or hydroquinone, has also been implicated in the alteration of global hmC content in different kinds of exposed cells, most probably by modulating the activity of TET enzymes (41). In particular, the vitamin C was recently shown to act as an additional co-factor of TET enzymes, most probably through the reduction of Fe3+ to Fe2+. In line with these findings, the treatment of melanoma cells with physiological concentrations of ascorbate increases the global hmC content and partially restores the normal epigenetic landscape of cells, thus attenuating their malignant phenotype (42). Based on these observations, one can speculate that TET enzymes might serve as epigenetic sensors of different metabolic liver states. Thus, the interindividual variability of the global hepatic hmC content might reflect the differences in lifestyle and/or xenobiotic exposures of the individuals.
In this study we demonstrate that the inter-individual variability of hmC values correlates with the global hmC content of liver samples at 40.8% of the analyzed CpG sites in ADME genes (Table 2). Hence, the hepatic hydroxymethylome at a subset of CpG sites may dynamically respond to the physiological conditions and environmental exposures through the altered rate of hmC production by TET enzymes. Such non-specific alterations of hmC profiles might influence the gene expression in certain cases, e.g. when a dynamically hydroxymethylated CpG site by chance overlaps with the binding site for an hmC-sensitive protein such as CEBPB (13). However, a statistically significant association with the global hmC content is not seen at the remaining 59.2% of the CpG sites analyzed. Thus, other factors must be considered as important local determinants of the hepatic hydroxymethylome. In particular, the level of transcription correlates with the local hmC abundance in the coding intervals of hepatic genes (see Figures 7C and 8C), and this effect is observed in all individual livers, irrespective of whether their global hmC content is high or low (Supplementary Figure S1). In addition, we observed prominent hmC peaks that are likely to be caused by local determinants such as histone modifications at separate nucleosomes or sequence-specific DNA binding proteins (see Figure 6D). However, the functional role of local epigenetic modifiers in shaping the hepatic hydroxymethylome remains to be determined in future studies.
In conclusion, our results show that the global hmC levels are highly variable among individual livers and that the genomic distribution of hmC is strikingly non-uniform along the chromosomes. Moreover, hmC is unevenly distributed among ADME genes, with hmC-rich genes being actively transcribed, whereas hmC-poor genes are either transcribed at low levels or silent. In addition, specific genomic intervals were found to be highly enriched in hmC with putative regulatory functions. The TAB-Methyl-SEQ protocol described allows for separate detection of mC and hmC with single base resolution and thus should be useful for targeted analysis of cytosine modification profiles in a variety of applications.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr Carolina Johansson for help in the preparation of the manuscript; Drs. Emily LeProust, Frida Oliv, Christofer Flood and Johanna Hasmats (Agilent Technologies, CA, USA) for their assistance in designing the custom SureSelect libraries; Drs. Hans von Stedingk, Davide Tamburro, Rui Branca and Mattias Vesterlund (Dept. of Oncology-Pathology, Karolinska Institutet, Stockholm, Sweden) for their great help with the LC-MS quantification of cytosine modifications; Dr Michael Wilson (SickKids Research Institute, Toronto, Canada), Dr Patrick McMullen (The Hamner Institutes for Health Sciences, Institute for Chemical Safety Sciences, Research Triangle Park, NC, USA) and Dr Le Zhan (Rutgers University, Piscataway, NJ, USA) for providing the original ChIP-Seq data from their papers. The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council; European Community's Seventh Framework Programme (FP7/2007-2013) [267038, in part]; Cosmetics Europe and the Innovative Medicine Initiative project MIP-DILI [115336]; Estonian Science Foundation [ETF9293]; European Union through the European Social Fund [MJD71]; Estonian Research Council [IUT20-60]; Marie-Curie [CIG 322283]; Carl Tryggers [CTS15:41]. V.M.L. was supported by a Marie Curie IEF fellowship for career development in the context of the European FP7 framework programme. Funding for open access charge: Governmental grant from Karolinska Institutet (C331601172).
Conflict of interest statement. None declared.
REFERENCES
- 1.Penn N.W., Suwalski R., O'Riley C., Bojanowski K., Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S., et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroud H., Feng S., Morey Kinney S., Pradhan S., Jacobsen S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K., et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov M., Kals M., Kacevska M., Barragan I., Kasuga K., Rane A., Metspalu A., Milani L., Ingelman-Sundberg M. Ontogeny, distribution and potential roles of 5-hydroxymethylcytosine in human liver function. Genome Biol. 2013;14:R83. doi: 10.1186/gb-2013-14-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen L., Li X., Yan L., Tan Y., Li R., Zhao Y., Wang Y., Xie J., Zhang Y., Song C., et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F., et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Khund Sayeed S., Zhao J., Sathyanarayana B.K., Golla J.P., Vinson C. C/EBPbeta (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim. Biophys. Acta. 2015;1849:583–589. doi: 10.1016/j.bbagrm.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 17.Globisch D., Munzel M., Muller M., Michalakis S., Wagner M., Koch S., Bruckl T., Biel M., Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson J.P., Hunter J.M., Lempiainen H., Muller A., Terranova R., Moggs J.G., Meehan R.R. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res. 2013;41:5639–5654. doi: 10.1093/nar/gkt232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingelman-Sundberg M. Personalized medicine into the next generation. J. Intern. Med. 2015;277:152–154. doi: 10.1111/joim.12325. [DOI] [PubMed] [Google Scholar]
- 20.Sim S.C., Ingelman-Sundberg M. Pharmacogenomic biomarkers: new tools in current and future drug therapy. Trends Pharmacol. Sci. 2011;32:72–81. doi: 10.1016/j.tips.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov M., Kacevska M., Ingelman-Sundberg M. Epigenomics and interindividual differences in drug response. Clin. Pharmacol. Ther. 2012;92:727–736. doi: 10.1038/clpt.2012.152. [DOI] [PubMed] [Google Scholar]
- 22.Bonder M.J., Kasela S., Kals M., Tamm R., Lokk K., Barragan I., Buurman W.A., Deelen P., Greve J.W., Ivanov M., et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics. 2014;15:860. doi: 10.1186/1471-2164-15-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maksimovic J., Gordon L., Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu J., Zhou M., Song Q., Hong E.E., Smith A.D. MLML: consistent simultaneous estimates of DNA methylation and hydroxymethylation. Bioinformatics. 2013;29:2645–2646. doi: 10.1093/bioinformatics/btt459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H., Caffo B., Jaffee H.A., Irizarry R.A., Feinberg A.P. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballester B., Medina-Rivera A., Schmidt D., Gonzalez-Porta M., Carlucci M., Chen X., Chessman K., Faure A.J., Funnell A.P., Goncalves A., et al. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014;3:e02626. doi: 10.7554/eLife.02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMullen P.D., Bhattacharya S., Woods C.G., Sun B., Yarborough K., Ross S.M., Miller M.E., McBride M.T., LeCluyse E.L., Clewell R.A., et al. A map of the PPARalpha transcription regulatory network for primary human hepatocytes. Chem. Biol. Interact. 2014;209:14–24. doi: 10.1016/j.cbi.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Zhan L., Liu H.X., Fang Y., Kong B., He Y., Zhong X.B., Fang J., Wan Y.J., Guo G.L. Genome-wide binding and transcriptome analysis of human farnesoid X receptor in primary human hepatocytes. PLoS One. 2014;9:e105930. doi: 10.1371/journal.pone.0105930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q., Suzuki M., Wendt J., Patterson N., Eichten S.R., Hermanson P.J., Green D., Jeddeloh J., Richmond T., Rosenbaum H., et al. Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Res. 2015;43:e81. doi: 10.1093/nar/gkv244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazor K.L., Boland M.J., Bibikova M., Klotzle B., Yu M., Glenn-Pratola V.L., Schell J.P., Coleman R.L., Cabral-da-Silva M.C., Schmidt U., et al. Application of a low cost array-based technique - TAB-Array - for quantifying and mapping both 5mC and 5hmC at single base resolution in human pluripotent stem cells. Genomics. 2014;104:358–367. doi: 10.1016/j.ygeno.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chopra P., Papale L.A., White A.T., Hatch A., Brown R.M., Garthwaite M.A., Roseboom P.H., Golos T.G., Warren S.T., Alisch R.S. Array-based assay detects genome-wide 5-mC and 5-hmC in the brains of humans, non-human primates, and mice. BMC Genomics. 2014;15:131. doi: 10.1186/1471-2164-15-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart S.K., Morris T.J., Guilhamon P., Bulstrode H., Bachman M., Balasubramanian S., Beck S. oxBS-450K: a method for analysing hydroxymethylation using 450K BeadChips. Methods. 2015;72:9–15. doi: 10.1016/j.ymeth.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Field S.F., Beraldi D., Bachman M., Stewart S.K., Beck S., Balasubramanian S. Accurate measurement of 5-methylcytosine and 5-hydroxymethylcytosine in human cerebellum DNA by oxidative bisulfite on an array (OxBS-array) PLoS One. 2015;10:e0118202. doi: 10.1371/journal.pone.0118202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinney S.M., Chin H.G., Vaisvila R., Bitinaite J., Zheng Y., Esteve P.O., Feng S., Stroud H., Jacobsen S.E., Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J. Biol. Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D., et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroeze L.I., van der Reijden B.A., Jansen J.H. 5-Hydroxymethylcytosine: an epigenetic mark frequently deregulated in cancer. Biochim. Biophys. Acta. 2015;1855:144–154. doi: 10.1016/j.bbcan.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Dao T., Cheng R.Y., Revelo M.P., Mitzner W., Tang W. Hydroxymethylation as a novel environmental biosensor. Curr. Environ. Health Rep. 2014;1:1–10. doi: 10.1007/s40572-013-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young J.I., Zuchner S., Wang G. Regulation of the epigenome by vitamin C. Annu. Rev. Nutr. 2015;35:545–564. doi: 10.1146/annurev-nutr-071714-034228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.