Abstract
Post-transcriptional regulation of transcription factors contributes to regulatory circuits. We created translational reporter fusions for multiple central regulators in Escherichia coli and examined the effect of Hfq-dependent non-coding RNAs on these fusions. This approach yields an ‘RNA landscape,’ identifying Hfq-dependent sRNAs that regulate a given fusion. No significant sRNA regulation of crp or fnr was detected. hns was regulated only by DsrA, as previously reported. Lrp and SoxS were both found to be regulated post-transcriptionally. Lrp, ‘leucine-responsive regulatory protein,’ regulates genes involved in amino acid biosynthesis and catabolism and other cellular functions. sRNAs DsrA, MicF and GcvB each independently downregulate the lrp translational fusion, confirming previous reports for MicF and GcvB. MicF and DsrA interact with an overlapping site early in the lrp ORF, while GcvB acts upstream at two independent sites in the long lrp leader. Surprisingly, GcvB was found to be responsible for significant downregulation of lrp after oxidative stress; MicF also contributed. SoxS, an activator of genes used to combat oxidative stress, is negatively regulated by sRNA MgrR. This study demonstrates that while not all global regulators are subject to sRNA regulation, post-transcriptional control by sRNAs allows multiple environmental signals to affect synthesis of the transcriptional regulator.
INTRODUCTION
Bacteria can adapt and survive under various conditions through adjustment of gene expression by changes in the amounts and activity of global transcriptional regulators. These regulators are characterized by their control over a diverse group of genes, all presumably helping the cell adjust to changing circumstances. Coordinating gene expression requires coordinating the expression and activity of the global regulators. Regulators are themselves regulated at every possible level, by regulation of transcription of the regulator itself, regulation of activity of the regulatory protein by small molecules or regulated modifications such as phosphorylation, regulation of regulator stability and regulation of the activity of inhibitory proteins (1–4).
In addition, some global regulators are subject to regulation by multiple Hfq-dependent sRNAs (5–11). Bacterial small RNAs regulate target messenger RNAs at the post-transcriptional level through base-pair interactions. RpoS, the stationary phase sigma factor, is positively regulated by three different sRNAs and negatively regulated, probably by competition for Hfq, by others (5–8,12). FlhDC, the regulator of flagellar synthesis, is both positively and negatively regulated by multiple sRNAs (9,10). CsgD, a key regulator of curli formation, has also been identified as the target of multiple sRNAs (11). Because each sRNA is itself subject to transcriptional control, the sRNA regulators in some cases reinforce the regulatory inputs that were already known to operate at the level of transcriptional control (see, for instance, (10)), and in others suggest new inputs to the regulatory circuit. However, transcriptional regulators are often expressed at low levels; therefore, their transcripts may not be abundant in the microarrays that are frequently used to define the changes in mRNA abundance after short-term overexpression of an sRNA (13,14), thus missing these important and interesting targets.
Our laboratory has developed a rapid method for screening genes of interest for translational regulation by Hfq-binding sRNAs; this was used to identify a new regulator of RpoS and the multiple sRNAs involved in FlhDC regulation (7,10). Here we applied this approach to a set of global transcriptional regulators. We chose five non-sigma and non-two component system transcriptional regulators with well-defined transcriptional start points, lrp, crp, fnr, soxS and hns. HNS (histone-like nucleoid structuring protein), a protein implicated in silencing of foreign DNA in Escherichia coli and Salmonella (15) and affecting global chromosome organization in bacteria (16), has previously been shown to be subject to DsrA-dependent repression (17). Lrp (leucine-responsive regulatory protein) regulates genes involved in amino acid biosynthesis and catabolism, nutrient transport, pili synthesis and other cellular functions (18–20); its activity is modulated by leucine as well as other amino acids (21,22). Translation of Lrp was reported to be negatively regulated by the sRNAs MicF and GcvB while our studies were being done (23,24). The other transcriptional regulators had not been identified as sRNA targets. Crp (cAMP receptor protein) regulates genes involved in the catabolism of secondary carbon sources (25). It is active for DNA binding only when the small molecule effector, cyclic adenosine mononucleotide phosphate (AMP), is made. Fnr is structurally related to Crp but acts to regulate genes in the transition from aerobic to anaerobic growth (26). SoxS plays a role in removal of superoxide and nitric oxide and protection from organic solvents and antibiotics (27–29). Our results demonstrate that some but not all of these regulatory proteins are subject to sRNA control. For SoxS and Lrp, our results demonstrate novel sRNA inputs into these global regulatory circuits. In particular, we find an unexpectedly complex mode of regulation of lrp by GcvB and identify an unexpected role for this highly conserved sRNA in regulation under oxidative stress.
MATERIALS AND METHODS
Bacterial strains and plasmids
E. coli strains used in this study are derivatives of strain MG1655 and are listed in Supplementary Table S1. Plasmids were generally introduced into strains by TSS transformation, a one-step procedure for making E. coli competent in which cells are resuspended in Transformation and Storage Solution consisting of polyethylene glycol, dimethyl sulfoxide (DMSO) and Mg2+ (30). A transformed colony was cultured in LB (Luria Broth) with the appropriate antibiotic and a final concentration of 100 μM of isopropyl beta-D-1-thiogalactopyranoside (IPTG).
The wild-type and mutant lacZ transcriptional and translational fusions were constructed using the PM1205 or PM1805 system (31), which contains PBAD-catsacB upstream of lacZ at the chromosomal lacZ site. For translational fusions, the 5′ primer (PBAD-gene name-F) was homologous to the upstream region of PBAD to allow recombination. The downstream primer was lacZ-gene name-R. Polymerase chain reaction (PCR) products containing the wild-type 5′ UTR or mutant versions of the 5′ UTR and the initial 20 codons of each gene were constructed from template genomic DNA from strain MG1655, using the primer sets indicated in Supplementary Tables S1 and S2. Each product was recombined into the chromosome of strain PM1205 or PM1805 by lambda Red recombinase-mediated gene replacement to construct the lacZ translational fusion regulated by the araBAD promoter. For the transcriptional fusion to ilvIH, the PCR products were generated to amplify PCR products having the sequences from −327 to +33 of ilvIH (relative to transcription +1) and switching the PBAD-catsacB region for the region of the ilvIH promoter (see primers listed in Supplementary Table S2), resulting in HL1213. Each PCR product was recombined into the chromosome of either strain PM1205 or PM1805 by lambda Red recombinase-mediated gene replacement, selecting for sucrose resistance and screening for loss of chloramphenicol resistance. Sequences were confirmed by sequencing.
To construct the LRP-SPA-kan strain (where SPA is the sequential peptide affinity tag), the bacteriophage λ red recombination system was used as described previously (32). Briefly, PCR fragments were obtained by amplifying the SPA-kan cassette of strain BA315 stpA-SPA (where StpA is H-NS-like DNA-binding protein) (33) (primers Lrp-SPA-F and Lrp-SPA-R). The PCR fragments were then recombined into the chromosome of strain NM1100 (34). The transformed cells were selected on LB plates containing 25-μg/ml kanamycin at 37°C. Recombinant products were verified by PCR. The gene conferring resistance to kanamycin was then removed from the LRP-SPA-kan strain by using the pcp20 plasmid (35).
Site-directed mutants in pGcvB, pMicF, pDsrA and pSpot42 were constructed using the Quikchange II site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions with primers described in Supplementary Table S2.
Media and growth conditions
Strains were grown in LB media or in MOPS (1X MOPS mixture, 132-mM K2HPO4, 0.2% glycerol) (TEKNOVA). For the oxidative stress experiments, cells were grown in LB. When the cell OD600 reached 0.5, cultures were split and one portion was treated with 1.0-mM paraquat. Final antibiotic concentrations (in micrograms per milliliter) were as follows: ampicillin on plates, 50; ampicillin in liquid culture, 100; kanamycin, 25; chloramphenicol, 25; tetracycline, 25; zeocin, 25.
sRNA library screen
In our library screens, sRNAs are expressed in excess from a multicopy plasmid derived from pBR322, under control of an induced plac promoter. Translational reporters are expressed from a pBAD promoter, and levels of arabinose are adjusted to allow significant levels of expression of the reporter in the presence of the vector plasmid. The library screen was carried out in a 96-well microtiter plate. The library was introduced into a specific background strain by TSS transformation and spotted on an LB plate containing ampicillin as described previously (7). From those spots, microtiter wells were inoculated in LB, 100-μM IPTG, 100-μg/ml ampicillin and arabinose (levels indicated in legends) and grown at 37°C to stationary phase (OD 2) and assayed (see below).
β-galactosidase activity measurements
In the library screens in Figure 1, the β-galactosidase activity of strains carrying lacZ fusions was assayed on a SpectraMax 250 (Molecular Devices) microtiter plate reader as described previously (5). Specific activities are represented as the Vmax divided by the OD600 and are about 25-fold lower than Miller units. sRNA plasmids showing significant regulation from this screen (close to a two-fold change) were retested individually using cultures grown in tubes to OD 2 and assayed using the standard assay described by Miller (36). If results were not confirmed in these tests, no further investigation was done. This was the case, for instance, for MicC and MicF regulation of crp. All other assays of β-galactosidase activity of transcriptional and translational fusions were determined using the standard assay described by Miller (36).
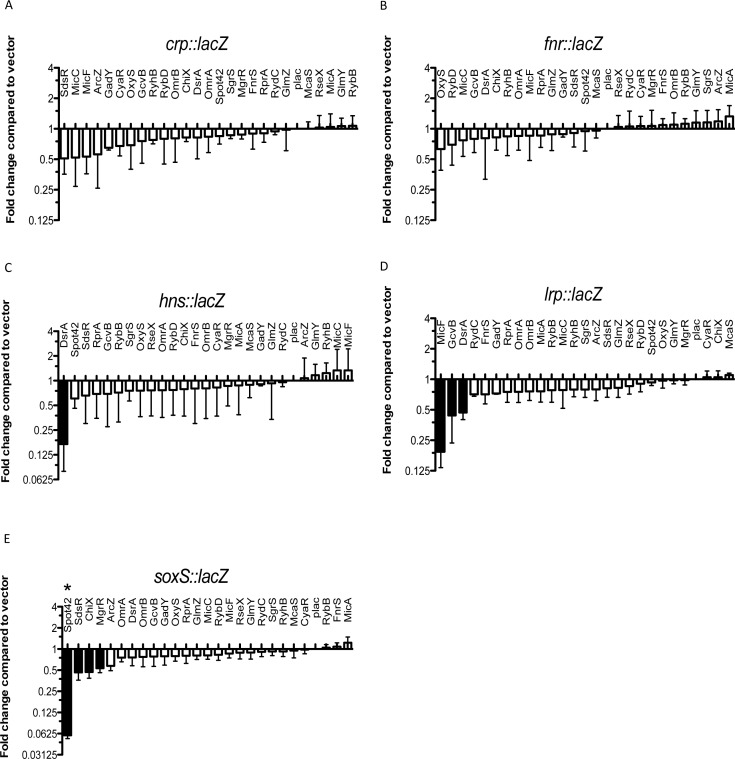
Figure 1.
Use of a library of small RNAs to study translational regulation of transcription factors. Screening of the sRNA library on the PBAD-crp-lacZ fusion (HL1070) (A), the PBAD-fnr-lacZ fusion (HL1069) (B), the PBAD-hns-lacZ fusion (HL1061) (C), the PBAD-lrp-lacZ fusion (HL1044) (D) and the PBAD-soxS-lacZ fusion (HL1064) (E). Cells were grown on LB medium containing 100-μg/ml ampicillin, 100-μM IPTG and 0.001% arabinose (for soxS-lacZ fusion; 0.0001% arabinose). The effect of the overexpression of each sRNA on each fusion was plotted as a function of the fold change compared with the basal activity of the same fusion containing a pBRplac control vector. Basal specific activities for each fusion (in machine units, about 25x less than Miller units) are 145 units for crp; 90 units for fnr, 1899 units for hns, 314 units for lrp and 352 units for soxS. Fold changes greater than two were considered significant (black bars). Assays were done in quadruplicate; error bars indicate standard deviation. * Spot 42 has been found to block arabinose induction of the pBAD promoter at the very low arabinose concentrations used for the soxS fusion (J. Chen, in preparation).
RNA isolation and northern blot analysis
Overnight cultures of the strains to be analyzed were grown in LB, diluted 100-fold in fresh medium and incubated at 37°C with agitation. At the indicated OD600, 800 μl samples were taken from each culture and RNA was extracted from the samples using the hot phenol method (37). Northern blots were performed with 10-μg total RNA as described previously (38). Detection was performed with the biotinylated probes in Supplementary Table S2.
Protein electrophoresis and western blot analysis
Samples were analyzed using Nu-PAGE 12% bis-Tris gels (Invitrogen, CA, USA), transferred to a nitrocellulose membrane and probed with a 1:1000 dilution of anti-Flag antiserum (Sigma-Aldrich). Blots were developed with the Lumi-Phos western blotting chemiluminescent substrate (Thermo Scientific) using a luminescence image analyzer (LAS-4000 Mini; Fujifilm). Quantification was performed by using ImageQuant software (GE Healthcare Life Sciences). Values presented are the mean of at least three independent assays.
Gel mobility shift assay
In vitro transcripts were generated with the T7 Megashortscript Kit (Ambion) using DNA templates. DNA templates carrying a T7 promoter sequence for in vitro transcription were generated by PCR (for lrp1, lrp2 and GcvB) or by annealing oligonucleotides (for MicF and DsrA). Primers and sequences of the T7 transcripts are included in Supplementary Table S2. A total of 20 pmol of RNA was 5′-end-labeled and purified as described previously (39). For all the binding reactions, 0.1 pmol of the labeled transcripts, 100 ng of yeast tRNA, five-fold excess unlabeled sRNA, and with or without 100 nM of purified Hfq were mixed in 10 μl of 1x binding buffer (10-mM Tris-Cl, pH 7.5, 50-mM NH4Cl, 0.2-mM EDTA, 10% glycerol) and 1x structure buffer (10-mM Tris, pH 7.0, 100-mM KCl, 10-mM MgCl2; Ambion). The reactions were incubated at 37°C for 60 min, and 1 μl of 10x loading buffer (1x TBE, 50% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol) was added. The samples were analyzed on a 6% native polyacrylamide gel run in 0.5x TBE on ice. Hfq was a gift of the Sarah Woodson Lab.
In vitro structure probing assay
Structure probing was performed using 0.1 pmol of mRNA (lrp1 and lrp2) in 10 μl reactions as previously described by Sharma et al. (40). 5′-end-labeled RNA was denatured for 1 min at 95°C followed by incubation on ice for 5 min and hybridized with five-fold excess of cold sRNA for 60 min at 37°C in the presence of 1 μg of yeast tRNA, 100-nM Hfq protein and 1x structure buffer. The RNA mixture was digested with a final concentration of 5-mM lead(II) (Fluka) or 0.002 units of RNase T1 (Ambion) for 1.5 or 3 min at 37°C. An RNase T1 ladder was generated by incubating 0.2 pmol of denatured mRNA with 0.1 units of RNase T1 in 1x sequencing buffer (Ambion) for 5 min at 37°C. The OH ladder was obtained by incubating 0.2 pmol of mRNA in alkaline hydrolysis buffer (Ambion) for 5 min at 95°C. Reactions were stopped by adding 12 μl loading buffer II (95% v/v formamide, 18-mM EDTA, 0.025% SDS, xylene cyanole, bromophenol blue; Ambion). Samples were denatured for 3 min at 95°C and run on 6% polyacrylamide/7M urea sequencing gels in 1x TBE buffer at 65 W for 120 min for lrp1 and 60 min for lrp2. Gels were dried and analyzed using Storm 860 Molecular Imager and ImageQuant software (GE Healthcare).
RESULTS
Screening of the sRNA library for translational regulation of transcriptional regulators
To study the translational regulation of transcriptional regulators, we constructed lacZ reporter fusions under the control of the arabinose-inducible PBAD promoter, by recombineering in E. coli PM1205, a strain designed to simplify construction of translational fusions (31). Because the vast majority of sRNA-dependent regulation takes place at the 5′ UTR and initial translated region of the ORF (open reading frame), fusions were made that contained the 5′ UTR (untranslated region) of the regulator, from the mapped transcriptional start site as defined in EcoCyc (41), and the coding region for the first 20 amino acids, fused in frame to lacZ. Fusions are at the chromosomal lac site.
A library of plasmids, derivatives of pBR322 and each encoding one of the known Hfq-dependent sRNAs known when this study began, expressed from an induced plac promoter, has previously been constructed (7,10). A plasmid expressing the newly described sRNA McaS (9) was added to the library. Each of the sRNA-expressing plasmids or a vector control was used to transform strains carrying the PBAD-transcriptional regulator-lacZ translational fusions and assayed as described in the Materials and Methods section. The results are plotted as an ‘RNA landscape’ in which values were compared to the vector control, set to 1 (Figure 1). Regulation by >2x was considered significant (black bars in Figure 1). Both positive and negative regulators should be detected in these screens, based on previous results (7,10).
For crp and fnr, no clear evidence of an sRNA regulator was found (Figure 1A and B), although a few sRNAs were close to the two-fold cutoff for repression of crp. For hns, the RNA landscape approach clearly confirmed the previous observation of negative regulation by DsrA (Figure 1C) (17,42,43). This result supports the likelihood of an Hfq-binding site and other critical sites for regulation within the hns 5′ end. None of the other known sRNAs were found to regulate hns (Figure 1C).
We found that MicF, GcvB and DsrA downregulated expression of the lrp-lacZ translational fusion (Figure 1D). These results confirm previous reports of negative regulation of lrp by MicF (24) and GcvB (23). The effect of DsrA on lrp has not previously been reported. Therefore, this approach was effective in identifying both known and new regulators of lrp.
soxS has not previously been demonstrated to be regulated by sRNAs. We found apparent strong negative regulation of soxS by Spot 42 sRNA (Figure 1E); however, this was later found to be an artifact of using the pBAD promoter with low levels of arabinose (J. Chen and S. Gottesman, in preparation). Other sRNAs (SdsR, ChiX and MgrR) were close to our two-fold cutoff.
The sRNA regulators of Lrp and SoxS were confirmed in independent transformations, using a host deleted for all three of the predicted sRNA regulators for Lrp (Supplementary Figure S1A) and using a wild-type strain for SoxS (Supplementary Figure S1B). These results demonstrate that the sRNA effects for Lrp are independent of the other sRNA regulators. For SoxS, MgrR showed the most significant regulation and was further examined (see below).
Lrp regulation by multiple sRNAs
While it is clear that high level of expression of three sRNAs can regulate the lrp-lacZ fusion, the contribution of each sRNA was assayed by measuring the lrp-lacZ fusion in strains deleted, individually, for each of the relevant sRNAs or hfq, both in minimal and rich media (Supplementary Figure S2). Only deletion of gcvB significantly increased expression of lrp-lacZ, in LB medium (Supplementary Figure S2A and C), but not in minimal MOPS or M9 Glycerol media (Supplementary Figure S2B and D). The lack of regulation in minimal medium may well reflect poor expression of GcvB under these growth conditions (44). Deletion of hfq increased fusion expression in LB, throughout the growth curve (Supplementary Figure S2C and E) and in minimal medium (Supplementary Figure S2D). The greater effect in minimal could reflect additive effects of loss of all of the sRNAs, could suggest that sRNAs not in the library are involved in lrp repression or could reflect other more indirect effects of Hfq.
lrp is transcribed by a single sigma 70-dependent promoter, with a 267-nt long leader (45). Transcription was reported to be negatively regulated by Lrp itself (45,46)), by H-NS and by NsrR (47) and positively regulated by GadE (48). Some of the Lrp binding sites are early in the 5′ UTR (45,46,49) and are present in our fusion. The leader is well conserved, particularly for the 34 nt upstream of the AUG, as well as for the region immediately downstream of the AUG (Supplementary Figure S3).
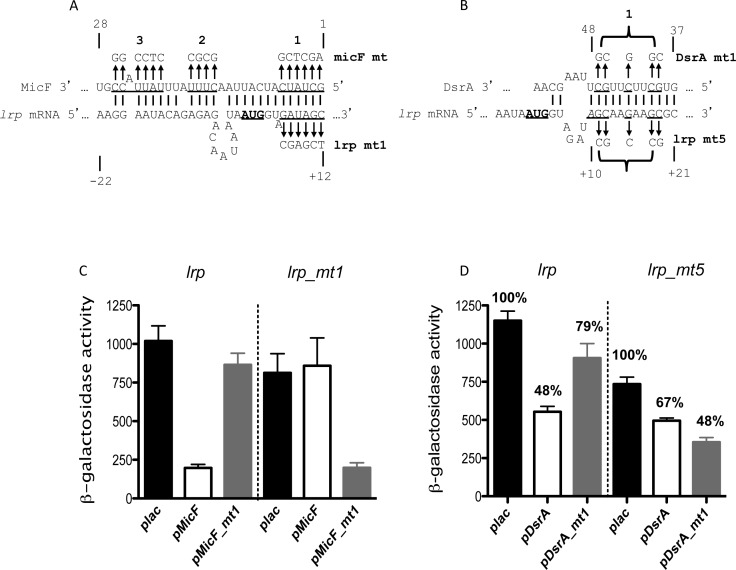
The MFold program (50) was used to predict base-pairing between each of the sRNAs and the lrp mRNA leader and initial translated region. For MicF, the predicted pairing overlaps that demonstrated by Holmqvist et al. (24) (region 1 in Figure 2A), overlapping the AUG of lrp and beginning at the 5′ end of MicF. Consistent with their results, mutations in region 1 of MicF abolished regulation of lrp; compensating mutations in region 1 of the lrp leader abolished regulation by wild-type MicF but allowed regulation by the complementary MicF mutant (lrp mt1) (Figure 2C and Supplementary Table S3). Mutations in region 2 or 3 (Supplementary Figure S4A) were less efficient in repressing lrp expression, but still retained significant negative regulation, compared to the vector control (Supplementary Figure S4B). Region 1 of MicF was also necessary for full repression of ompF, while regions 2 and 3 were not (Supplementary Figure S4C).
Figure 2.
MicF and DsrA direct pairing with the lrp 5′ UTR region. (A) Predicted base-pairing regions of sRNA MicF with the lrp mRNA are shown. Mutated nucleotides are underlined and the changes made to them are shown with arrows above or below the sequence. Numbering of lrp is relative to the AUG. The region of MicF shown extends from nt 1 on the right to nt 30. (B) Predicted base-pairing region of sRNA DsrA with the lrp mRNA is shown, annotated as for (A). The pairing region within DsrA is from nt 37 to nt 48. (C) β-galactosidase activity from cells with lrp-lacZ (HL1044) and lrp_mt1-lacZ (HL1079) translational fusions, in the presence of plasmids that overexpress either wild-type pMicF or pMicF_mt1. Cells were grown in LB Ampicillin with IPTG at 37°C to an OD600 of 2.5–3. (D) β-galactosidase activity from cells with lrp-lacZ (HL1100; ΔdsrA ΔmicF ΔgcvB) and lrp_mt5-lacZ (HL1154; ΔdsrA ΔmicF ΔgcvB) translational fusions, in the presence of plasmids that overexpress either wild-type DsrA or mutant DsrA variants. Cells were grown in LB Ampicillin with IPTG at 37°C to an OD600 of 3. Error bars indicate standard deviation.
Mfold predicted pairing by DsrA with lrp downstream from the AUG (Figure 2B), within the region of DsrA that is known to pair with hns (43). Wild-type DsrA reduced the expression of the wild-type fusion two-fold, compared to the vector control; mutating the lrp fusion (mt5; Figure 2B) reduced regulation modestly (Figure 2D). A mutant in DsrA, mt1 (Figure 2B), poorly repressed wild-type lrp expression but was able to repress the lrp mt5 fusion (Figure 2D), demonstrating direct interaction of DsrA with lrp mRNA. The pairing region for DsrA overlaps at positions +10 to +12 (AGC at 5′ end of DsrA predicted pairing region) with the region of MicF pairing (underlined and italics in Figure 2B). lrp mt1, regulated by the MicF mt1, has reduced regulation by DsrA and lrp mt5 cannot be regulated by wild-type MicF (Supplementary Table S3). This region early in the lrp ORF is thus important for regulation by both of these two sRNAs and presumably they could not act on the same lrp mRNA at the same time.
The lrp leader has complex effects on translation and two regions of GcvB pairing
GcvB regulation of lrp has previously been described (23,51), but direct pairing was not demonstrated. Modi et al. predicted pairing of GcvB to lrp near the ribosome binding site (23). In our hands, a mutant at that site in lrp decreased basal activity of the fusion to less than 1% of the wild-type (Supplementary Figure S5, lrp mt7), consistent with disruption of the ribosome binding site; we have no evidence of pairing of GcvB at this site. Mutations in regions of GcvB implicated in pairing with other targets were tested by Sharma et al. but retained regulation of lrp (51).
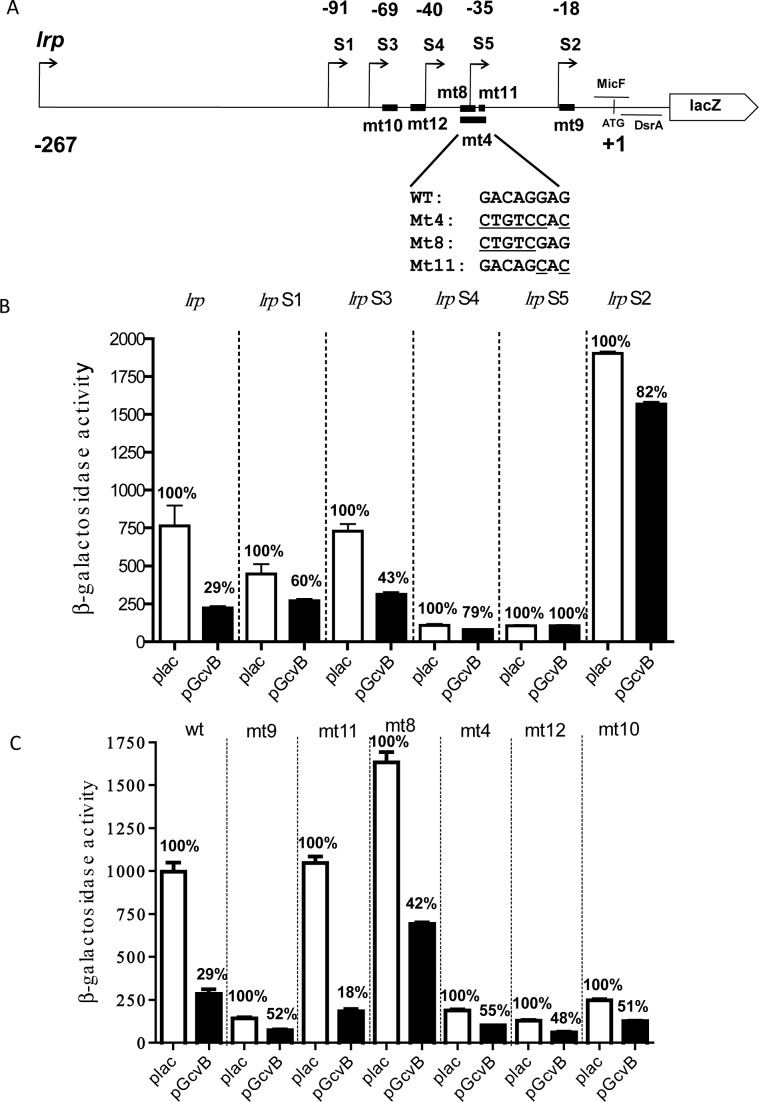
We revisited the question of how GcvB pairs with lrp, and began by deletion analysis of the long and well-conserved lrp leader. Deletions are shown schematically in Figure 3A; the sequence of the leader and the precise end-point of the truncations is in Supplementary Figure S3. The expression of the resulting fusions was measured in the presence of a vector or multicopy GcvB (Figure 3B), or in the absence of a plasmid (Supplementary Figure S5B). The first striking observation was that deletions showed very different basal levels of translation of the lrp-lacZ fusion, presumably reflecting changes in mRNA structure or stability that affect translation. Thus, deletions to −40 or −35 relative to the AUG start (lrp S4 and S5) gave extremely low levels of translation (8–10% compared to the full-length fusion), while deletion to −18 (lrp S2) increased expression more than two-fold compared to the full length fusion. Intermediate deletions (to −91 or −69; lrp S1 and S3) had more modest effects on translation. These results suggest that there are likely both negative and positive elements upstream of −18 that contribute to translation or mRNA stability, with the positive elements upstream of or overlapping −40 (but downstream of −69) and negative elements downstream of −35 but upstream of −18. Consistent with the importance of structure and/or sequence in this region, some point mutants between −69 and −30 (lrp_mt4, mt10, mt12) also significantly decreased translation, although lrp_mt8 and mt11, overlapping the region of mt4, did not decrease expression of the fusion (Figure 3A and C and Supplementary Figure S5B).
Figure 3.
Complex regulation of lrp translation by GcvB. (A) Schematic representation of various lrp-lacZ deletions and mutant fusions. The regions of pairing of MicF and DsrA are shown with horizontal lines. The positions of the point mutations are shown with heavy lines, with the sequence changes for mt4, mt8 and mt11 shown below the line. The precise end-point of each deletion is shown in Supplementary Figure S3. (B) Cells were grown and assayed as in Figure 2A (OD600 of between 2.5 and 3). β-galactosidase activity was determined from cells with lrp (HL1071), lrp S1 (HL1517), lrpS3 (HL1701), lrpS4 (HL1702), lrpS5 (HL1703) and lrp S2 (HL1518)-lacZ translational fusions in the presence of plasmids overexpressing GcvB (black bars) or with the pBRplac empty vector (white bars); the chromosomal copy of gcvB was deleted in all of these strains. Percentage regulation by GcvB was compared to the vector control for each deletion. Error bars indicate standard deviation. (C) Cells were grown and assayed as in Figure 2B. β-galactosidase activity was determined from cells with the following translational fusions: lrp (HL1071), lrp mt9 (HL1694), lrp mt11 (HL1731), lrp mt8 (HL1693), lrp mt4 (HL1149), lrp mt12 (HL1732) and lrp mt10 (HL1717).
Next we asked if any of these deletions abrogated regulation by the sRNAs. The shortest leader (S2, −18) lost regulation by GcvB (Figure 3B), but was still well regulated by DsrA and MicF (Supplementary Figure S5C). This is consistent with the regions of pairing identified for DsrA and MicF (Figure 2), and also suggests that any required Hfq binding sites within lrp are also downstream of −18. A possible Hfq binding region (ARN motif) is located between −7 and +2 (blue type in Supplementary Figure S3). None of the truncated leaders were regulated by GcvB to the same extent as the full-length leader (29% of the vector value; Figure 3B), but the results suggest a region downstream of −69 relative to AUG (lrp_S3) is sufficient for at least some GcvB regulation. Similarly, none of the point mutations fully abrogated regulation by GcvB (Figure 3C), although lrp_mt8 modestly decreased the ability of GcvB to regulate and lrp_mt11 improved regulation.
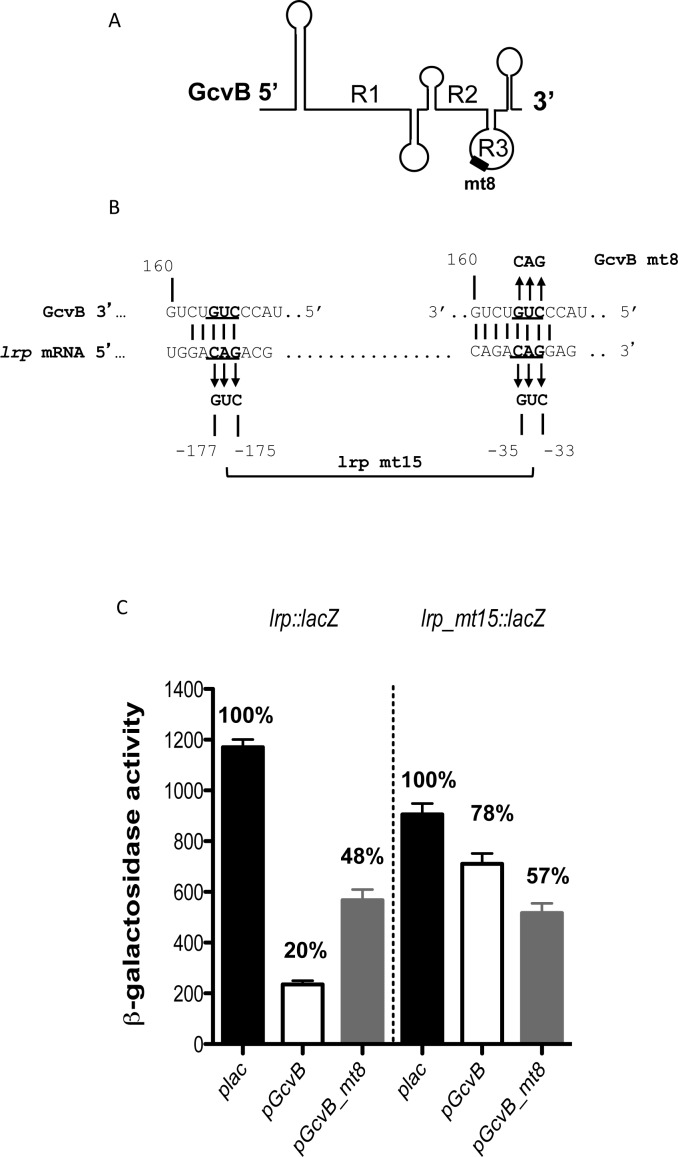
Multiple regions (R1, R2 and R3; Figure 4A) within GcvB have been implicated in pairing with targets (40,52). Regions R1 and R2 were shown not to be necessary for regulation of lrp (51). Coornaert et al. identified region R3 of GcvB as responsible for regulation of phoP, encoding a transcriptional regulator (53). We find that mutations in R3 abolish lrp regulation.
Figure 4.
GcvB regulation of lrp mRNA through redundant pairing. (A) Schematic drawing of the secondary structure of GcvB. Regions R1, R2 and R3 have been shown to be involved in pairing with various mRNA targets (51–53,83). (B) Predicted base-pairing region of sRNA GcvB with the lrp sequences is shown. Mutated nucleotides are underlined and changes made to them are shown. (C) β-galactosidase activity from cells with lrp-lacZ (HL1071) and lrp_mt15-lacZ (HL1810) translational fusion plasmids, in the presence of plasmids that overexpress either wild-type pGcvB or pGcvB_mt8. Assayed as in Figure 2. Error bars indicate standard deviation.
GcvB_mt4 changed seven nucleotides in the R3 region (Supplementary Figure S6A); this mutation fully abrogated GcvB regulation of lrp (Supplementary Figure S6B), but was functional for regulation of dppA, a GcvB target that does not use the R3 pairing region (Supplementary Figure S6C). Therefore, the levels of the GcvB_mt4 sRNA are sufficient for regulation. Two other mutations in the R3 region of GcvB, a 3-nt change to create GcvB_mt8 (Figure 4B) and a 5-nt change to create GcvB_mt6 (Supplementary Figure S6A), significantly decreased regulation (Figure 4C and Supplementary Figure S6D). GcvB_mt6 is the same as that used by Coornaert et al. (53) and was fully active for regulation of their compensating mutation in phoP (Supplementary Figure S6E), again suggesting that levels of this mutant sRNA are not significantly reduced.
While all of our results pointed to GcvB region R3 as necessary for regulation, and predicted pairing to this region could be found, compensating mutations lrp_mt4, lrp_mt8 and lrp_mt13 in lrp were still regulated by wild-type GcvB and were not better regulated by the GcvB derivatives that should have paired with them (Supplementary Figure S6B and D). Therefore, in vitro analysis was used to ask if GcvB regulated lrp via a direct pairing interaction, and then that information was used to design further in vivo tests.
Because lrp has a long 5′ UTR, we made two different versions of the lrp transcript. lrp 1 carries the full leader, between −267 and +60 relative to the ATG; lrp 2 is shorter, carrying from −69 to + 60 relative to the ATG (Supplementary Figure S7A). We initially used a gel mobility shift assay to see complexes between sRNAs and lrp mRNA. Both mRNAs formed complexes with sRNAs GcvB, MicF and DsrA, as measured by a shift in the lrp 1 or lrp 2 band (Supplementary Figure S7B and C); addition of Hfq led to modestly better interactions (right three lanes, Supplementary Figure S7B and C). GcvB gave the most significant shift, consistent with direct interaction of GcvB with lrp, with sites sufficient for this interaction present between −69 and +60.
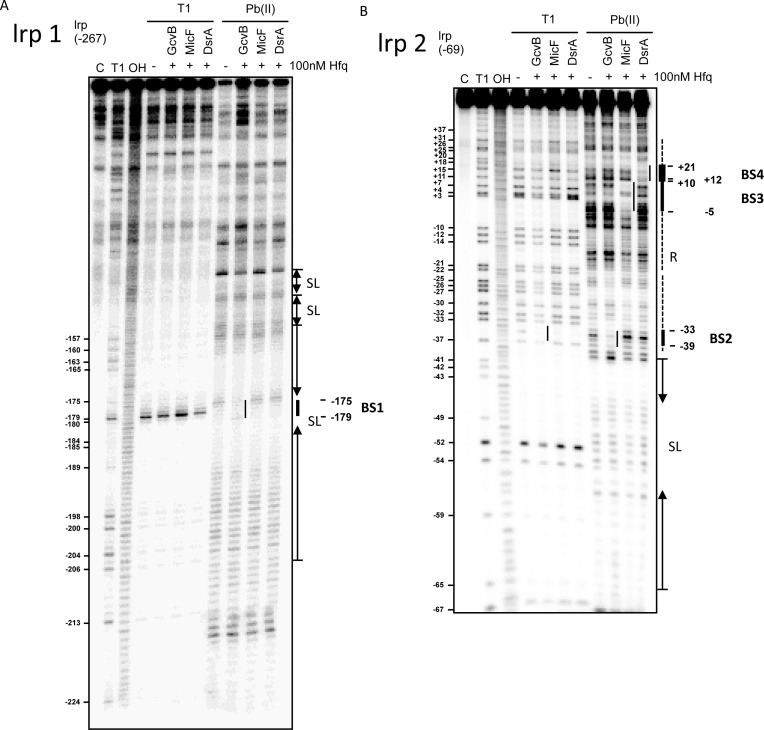
To identify the base-pairing regions, we probed the in vitro structures of lrp 1 and lrp 2 in the presence and absence of the sRNAs with lead(II) and RNase T1 (Figure 5A and B). Protection by the sRNAs were most visible for cleavage by lead(II) acetate (Pb(II) lanes in Figure 5). On lrp 2, the region closer to the ATG is resolved (Figure 5B), while the region further upstream in the leader is visible in lrp 1 (Figure 5A). Regions of T1 cleavage as well as protected regions were also modeled onto a secondary structure model predicted using the NUPACK software (54) for the full leader and first 24 nt of the coding region (Supplementary Figure S7D) and found to correlate. For instance, BS2, BS3 and BS4 are predicted to be unpaired in Supplementary Figure S7D and were subject to Pb(II) cleavage in Figure 5.
Figure 5.
In vitro interaction of GcvB, MicF and DsrA with lrp mRNA. (A, B) 20 nM of 5′-end-labeled lrp 1 (A) and lrp 2 (B) were subjected to RNase T1 and lead(Pb(II)) cleavage in the absence and presence of 100-mM cold sRNAs, GcvB, MicF and DsrA, respectively. Lane C: untreated RNA control; lane T1: RNase T1 ladder of denatured RNA; lane OH: alkaline ladder. The position of G residues cleaved by RNase T1 is indicated at the left of the gels.
Consistent with the pairing defined in vivo (Figure 2), DsrA protected a region of the lrp mRNA between +10 and +21 relative to the ATG (BS4; Figure 5B). We also confirmed the interaction between lrp mRNA and MicF sRNA, from −5 to +12 relative to the ATG (BS3; Figure 5B). As suggested above, these regions overlap, and, as noted above, both regions appear to be unstructured, both by predicted folding and by RNase cleavage experiments.
GcvB protected two different five-nucleotide regions, one clearly visible in the full-length lrp 1 (BS1) and one most visible in the short form lrp 2 mRNA (BS2). The protected regions have the same sequence (GACAG; see Supplementary Figure S3). The BS1 region (Figure 5A) is located between −179 and −175 nucleotides; BS2 (Figure 5B) is located between −39 and −33 nucleotides from ATG. Furthermore, both GACAG protected sequences should be able to base-pair with the R3 portion of GcvB (CUGUC), mutated in the GcvB R3 mutant derivatives discussed above. Note that the BS1 region is only present in the full-length lrp-lacZ fusion, and not in the S1 and S3 truncated fusions. These shorter fusions were somewhat less repressed by wild-type GcvB (Figure 3), possibly reflecting loss of this upstream site. The shortest deletions, S4 and S2, delete BS1 and some or all of the BS2 region, and truncation S4 ends very close to the BS2 region, consistent with complete loss of GcvB regulation in these fusions (Figure 3). Both BS1 and BS2 are in unpaired loops (Supplementary Figure S7D). Note that the BS2 region shows some protection from T1 but not Pb in all three lanes in which Hfq is present (Figure 5B); this was not always seen. The sequence overlapping with and downstream from BS2 contains an (ARN)3 repeat, known to bind Hfq (see Supplementary Figure S3) (55). However, this site must not be essential for regulation by GcvB, since lrp_mt11 disrupts the middle ARN repeat, but has, if anything, better regulation by GcvB (Figure 3C). Possibly Hfq binding here limits the ability of GcvB to bind and repress.
Mutants in the in vitro-defined lrp pairing regions were tested. A double mutant that changed three nucleotides in each of the lrp regions (lrp mt15; Figure 4B) slightly reduced basal level expression of the fusion, and was poorly regulated by the wild-type GcvB (Figure 4C). The compensating mutant in GcvB, GcvB_mt8, lost some of its ability to regulate the wild-type fusion, and regulated the double mutant better than wild-type GcvB. The behavior of these mutants is consistent with direct pairing of GcvB at these two regions, but suggests that pairing in these compensating mutants may be suboptimal. Tests of the same three nucleotide change at BS2 in gel shifts or in the truncated lrpS1 or lrpS3 fusions were also consistent with direct pairing to this region (Supplementary Figure S7E and G).
Five-nucleotide changes in one or both sites in lrp were also tested, in the context of the full-length lrp-lacZ translational fusion; these were designed to be complementary to GcvB_mt6 (Supplementary Figure S6A). Wild-type GcvB was partially active on single mutants disrupting BS1 or BS2 (Supplementary Figure S6D; lrpmt8 and lrpmt13); activity was lowest on the double mutant lrpmt14 (Supplementary Figure S6D), suggesting that repression at either site is sufficient to partially regulate lrp. However, GcvB_mt6 was unable to regulate the wild-type fusion or either single mutant and had very weak activity on the double mutant, lrpmt14 (Supplementary Figure S6D). Consistent with the inability of GcvB_mt6 to regulate lrp in vivo, it was also unable to gel shift the lrp mRNA in vitro (Supplementary Figure S7H).
Overall, this change in folding and accessibility to regulation is consistent with our observations that deletions and many point mutants in the lrp leader disrupt translation and/or GcvB regulation (see Supplementary Table S4), suggesting that the structure of the leader is critical for efficient translation in ways that are not yet fully understood. For instance, two lrp point mutants, lrp_mt10 and lrp_mt12, reduce translation significantly (Supplementary Table S4), and both would disrupt the same stem-loop (SL4: between −65 and −39; Supplementary Figure S7D).
Physiological effects of sRNA regulation of lrp
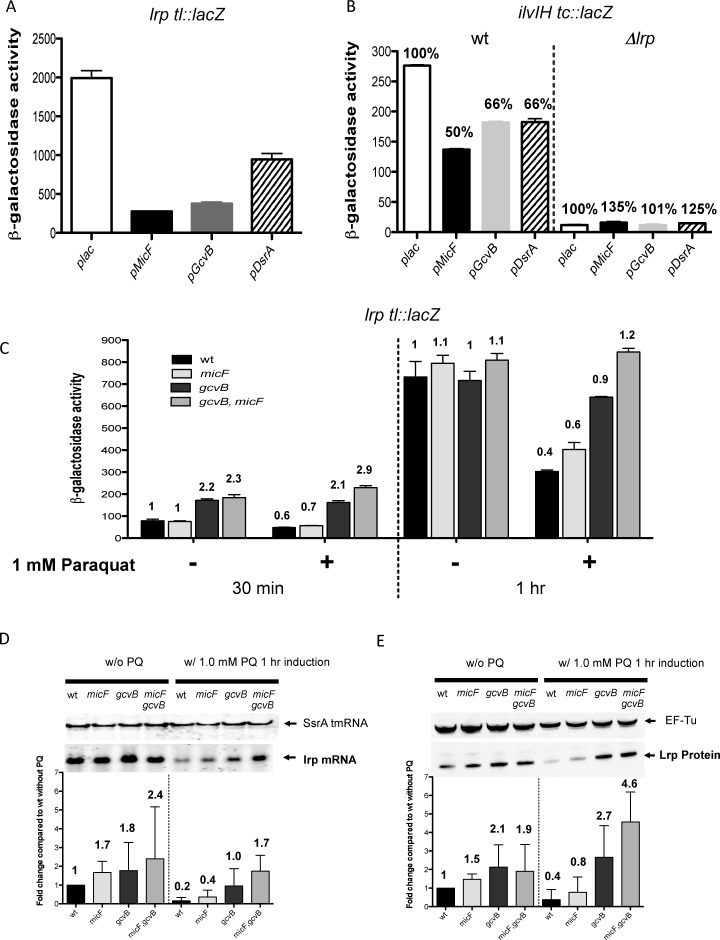
The experiments above demonstrate regulation of lrp translation by three sRNAs, at least when these sRNAs are overproduced. We asked whether, under these overproduction conditions, the effect of the sRNAs is sufficient to perturb Lrp regulation of one of its targets, ilvIH (Figure 6B). For this experiment, cells were grown in MOPS minimal, since the dependence of ilvIH expression on Lrp was seen in minimal media but not rich media. The sRNAs repressed the lrp translational fusion well in MOPS minimal (Figure 6A). The expression of an ilvIH-lacZ transcriptional fusion was strongly dependent upon Lrp (compare wild type (WT) and Δlrp plac bars; Figure 6B). Each of the sRNAs reduced expression of ilvIH about two-fold. The effects of the sRNAs on ilvIH were lost in an lrp mutant. Therefore, overproduced sRNAs are sufficient to reduce the activity of the abundant Lrp protein in the cell; this is consistent with and extends previous observations on the effect of multicopy MicF (24).
Figure 6.
Physiological effects of sRNA regulation of lrp. (A) Cells were grown in MOPS minimal medium containing ampicillin, IPTG, and 0.001% arabinose. Graph shows β-galactosidase activity of strain HL1100 (lrp-lacZ) transformed with plasmids overexpressing GcvB or MicF or DsrA, or with the pBRplac empty vector. (B) Cells were grown in MOPS minimal medium with ampicillin and IPTG. Graph shows β-galactosidase activity of strains HL1213 (wild-type) and HL1231 (Δlrp::kan) each carrying a chromosomal ilvIH transcriptional fusion and transformed with plasmids overexpressing GcvB or MicF or DsrA, or with the pBRplac empty vector. Percentages are relative to plac vector control for each strain. (C) Cells were grown on LB medium containing 0.001% arabinose to an OD600 of 0.5; one culture was treated with 1.0-mM paraquat and a control culture was not. Samples were collected 30 min and 1 h after paraquat addition and expression of the lrp-lacZ translational fusions in wild-type (HL1044), ΔgcvB (HL1071), ΔmicF (HL1078) or ΔmicF,gcvB (HL1083) backgrounds was determined. (D) Paraquat effects on lrp mRNA. Northern blot analysis of lrp and SsrA in wild-type (HL1434), ΔmicF (HL1437), ΔgcvB (HL1438) and ΔmicF,gcvB (HL1440) strains. Cells were grown in LB (to OD600 of 0.5) and treated with or without 1.0-mM paraquat and allowed to grow for the time indicated. RNA was extracted as described in the Materials and Methods section. (E) Paraquat effects on LRP-SPA protein. Western blot analysis of LRP-SPA and EF-Tu in the same strains as in panel (D). Protein was extracted as described in the Materials and Methods section. lrp mRNA bands and LRP-SPA protein bands were analyzed using ImageQuant software (GE Healthcare). Normalization of lrp mRNA and LRP-SPA protein was with SsrA or EF-Tu, respectively. The quantification is based on experiments performed in triplicate. Error bars in all panels indicate standard deviation.
We next investigated the extent of the effect of the sRNAs on lrp when present in single copy (expressed from their native promoters in the chromosome). In Supplementary Figure S2A and C, we found that, for cells grown in LB, only deletion of gcvB had a significant effect on expression of the lrp fusion (under the transcriptional control of pBAD). Even deletion of all three of the sRNAs had no detectable effect on the expression of either the lrp fusion or the ilvIH fusion in MOPS minimal media (Supplementary Table S5). These results, coupled with the modest effects of deletion of gcvB on Lrp expression in LB, suggest that the role of the sRNAs is likely important only under specific growth conditions. One of these, oxidative stress, was investigated further here.
Because MicF has been shown to increase its expression level when cells are under oxidative stress (56), we asked if the sRNAs had a more significant effect on lrp after oxidative stress. For these experiments, we focused on MicF and GcvB, since they were generally more effective than DsrA in regulating lrp. Isogenic strains carrying the lrp-lacZ fusion and deletions in gcvB, micF or both gcvB and micF were grown in LB to an OD600 of 0.5 and exposed to paraquat (PQ); samples were taken after 30′ and 1 h and assayed for the expression of the fusion (Figure 6C). After 30′ of PQ treatment, there was relatively little difference between the PQ treated cells and the control cells; in both cases, deletion of gcvB led to higher expression, as seen previously (Supplementary Figure S2C), but no significant effect of deletion of micF was seen. After 1 h, control cells were in stationary phase, and were unaffected by the sRNA deletions. However, expression was significantly lower for WT cells after PQ treatment (Figure 6C). While we had expected the effect of MicF to be particularly significant under these conditions, this was not the case. Deletion of micF led to a modest increase in expression of the fusion after PQ treatment. However, deletion of gcvB increased expression close to two-fold (Figure 6C). There was an additive effect of deleting both micF and gcvB, eliminating entirely the downregulation by PQ. Thus, it would appear that there is a significant GcvB-dependent repression of the lrp fusion after oxidative stress, and a modest repression by MicF (Figure 6C).
Parallel experiments were done to assess the effect of PQ treatment on both lrp mRNA (Figure 6D) and Lrp protein, tagged with SPA (Figure 6E). Note that for these two sets of assays, the native gene and its promoter activity are being assayed rather than the pBAD-lrp-lacZ fusion used in Figure 6C. In the absence of PQ, deletion of gcvB led to a significant increase in both lrp mRNA (1.8x) and protein (2.1x), consistent with the effect seen on the translational fusion in exponential phase (Supplementary Figure S2). We also detected an effect of MicF under these growth conditions (1.7x increase in lrp mRNA, 1.5x increase in Lrp protein in cells deleted for micF), and an additive effect of deleting both sRNAs, at least on lrp mRNA levels.
After treatment with PQ in the wild-type strain, both lrp mRNA and protein were significantly reduced, comparable to the reduced level of expression of the translational fusion. Deletion of gcvB had the strongest effect, on both lrp mRNA and protein. For lrp mRNA, deletion of gcvB reduced the effect of PQ from 5x (WT cells) to 1.8x; the double mutant reduced the effect of PQ treatment to 1.4x (Figure 6D). By comparison, deletion of micF alone reduced the effect of PQ from 5x to 4x. At the protein level, the general pattern was similar, although deletion of gcvB and deletion of both sRNAs led to an increase of Lrp protein above that found without PQ (Figure 6E). In spite of these observations implicating GcvB in significant lrp repression under oxidative stress, the levels of GcvB sRNA were not changed by this treatment (Supplementary Figure S8A).
Regulation of soxS by MgrR sRNA
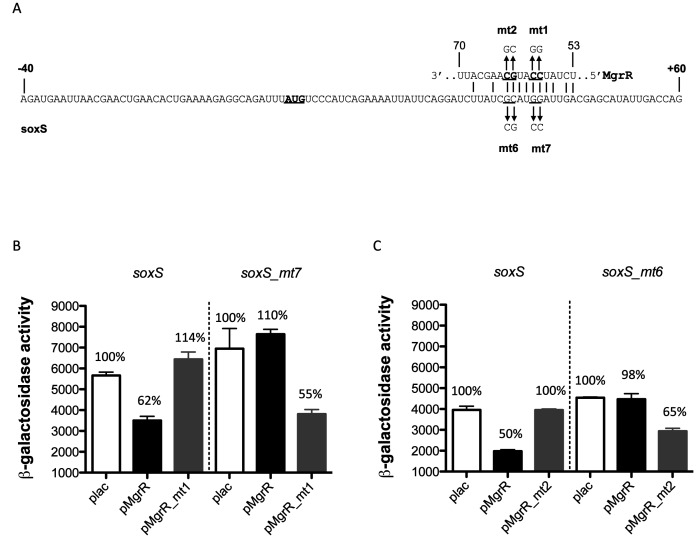
As described above, soxS was also found to be subject to regulation by sRNAs. The regulation by the MgrR sRNA was further investigated here. MgrR is synthesized in response to low Mg2+ or antimicrobials, dependent upon the PhoQ/PhoP two-component system (38). Relatively few targets of MgrR have been identified; one is eptB, encoding a protein that modifies LPS (57). The likely region of MgrR pairing within soxS was first identified by deletions of either the upstream 40 nt leader to 15 nt (soxS_3) or by shortening the soxS translated region from the 20 codons in the original fusion to nine codons (soxS_1) (Supplementary Figure S9A). MgrR was still able to repress soxS_3, suggesting very little of the 5′ UTR is necessary for regulation. However, regulation was lost in soxS_1, the fusion truncated to nine codons, and the basal level of expression was increased (Supplementary Figure S9B). Consistent with this, a region of pairing with the region downstream of +27 could be identified (Supplementary Figure S9A). Pairing with this region was tested using mutations mt1 and mt2 within MgrR and compensating mutations in the soxS ORF (Figure 7). Mutations in the predicted pairing region within MgrR led to complete loss of regulation of the wild-type fusion; the compensating mutation in soxS (soxS_mt7 or soxS_mt6) lost all regulation by wild-type MgrR (Figure 7B and C). When pairing was restored, regulation was restored. Therefore, MgrR directly pairs with soxS, well within the coding region.
Figure 7.
MgrR regulation of soxS. (A) Sequence of soxS 5′ UTR and first 60 nt of the translated gene, with predicted pairing of MgrR. Mutant changes tested are shown. Predicted base-pairing region of sRNA MgrR with the soxS mRNA are shown, annotated as for (A). (B, C) All strains are deleted for mgrR and were transformed with either the empty vector (plac), the plac-MgrR plasmid (pMgrR), the plac-MgrR mutant 1 plasmid (pMgrR_mt1) or the plac-MgrR mutant 2 plasmid (pMgrR_mt2). Cells were grown on LB medium containing 100-μg/ml ampicillin, 100-μM IPTG and 0.0002% arabinose. Samples were collected at stationary phase (OD600 of between 2.5 and 3) and assayed for β-galactosidase. Strains used: soxS (HL1755), soxS_ mt7 (HL1775) and soxS_mt6 (HL1791)-lacZ translational fusions. Error bars indicate standard deviation.
SoxS transcription is activated upon oxidative stress (58); its regulon overlaps with that of two other global regulators, Mar and Rob. To understand if the repression of soxS by MgrR was also found for these other two regulators, translational fusions to both proteins were made and screened with the RNA library. There was no significant regulation by sRNAs of Mar and Rob expression (Supplementary Figure S10). Thus, MgrR provides a unique input to this complex regulon. The conditions under which this is important remain to be determined. When cells were grown in LB, deletion of mgrR led to a modest (30%) increase in expression of the translational reporter, while deletion of hfq increased expression two-fold (Supplementary Figure S9C), suggesting that other sRNAs or Hfq itself may also regulate SoxS translation.
DISCUSSION
Transcriptional regulators are central players in the response of bacteria to changes in growth conditions. sRNAs can positively or negatively modulate the translation of the transcriptional regulator. The approach used here allowed us to examine transcriptional regulators of interest for regulation by Hfq-dependent sRNAs. The library of sRNAs used here includes 26 different Hfq-dependent sRNAs, the majority of this family of sRNAs.
Not everything is regulated by sRNAs
Of the seven transcriptional regulators tested here, four of them, Crp, Fnr, Mar and Rob, did not show significant sRNA regulation (Figure 1A and B and Supplementary Figure S10). However, we note that our translational fusions will only detect regulation in the 5′ UTR and the first 20 codons of the targets; while this is the most common location for regulation, some examples of pairing deeper within ORFs have been found (59). A second caveat is that regulation could be via sRNAs not currently in our library. We tentatively suggest that while Fnr and Crp control the synthesis of sRNAs (Spot 42, CyaR and McaS are regulated by Crp and cAMP, and FnrS is regulated by Fnr; (9,60–62)), they are not themselves regulated by the sRNAs.
HNS, an abundant silencer of transcription in E. coli and related bacteria, has been implicated in silencing of foreign DNA (63). It has previously been shown to be negatively regulated by DsrA (43); we confirmed this with our hns-lacZ translational fusion (Figure 1C), but no other sRNA regulators were found. The physiological significance of DsrA repression of HNS is not fully understood, but it is worth noting that HNS silencing and RpoS stimulation are often found as alternatives at some promoters (64), and thus DsrA may modulate this switch, reducing HNS and inducing RpoS under conditions of its expression. This modulation may be important specifically under expression conditions for DsrA, and not those leading to increased RpoS due to RprA and ArcZ sRNAs. The robust regulation by DsrA of the hns::lacZ translational fusion (Figure 1C) suggests that pairing of DsrA with the 3′ end of hns mRNA, suggested by Lease and Belfort (17), is not necessary for regulation.
SoxS, Mar and Rob bind to similar target promoters to help the cell respond to antibiotics and other stresses (65); each is transcriptionally induced under a different condition. SoxS induction depends on SoxR. Under superoxide or nitric oxide stress, SoxR activates SoxS, which in turn activates expression of genes that are related to amino acid biosynthesis, cell wall synthesis and divalent metal ion transport (66). Levels of SoxS, rather than any modification of SoxS, determine whether or not it is an effective regulator (67), and consistent with that, SoxS is subject to post-transcriptional regulation of protein levels. Degradation of SoxS by the Lon protease helps in returning protein levels to basal levels once induction stops (68–70). We found here that soxS was also regulated by MgrR, by pairing within the ORF (Figures 1 and 7). Because MgrR is itself positively regulated by PhoQ/PhoP (38), this observation links these two global regulatory systems and suggests that inhibition of the SoxS response may be useful under the low Mg++ conditions that activate the PhoQ/P system.
Lrp is a global regulator with many targets, most of them involved with amino acid transport and use (20,71–73). In addition, synthesis of Lrp is negatively autoregulated, and is also reported to be regulated by GadE (positively) and HNS (negatively), suggesting that the levels of Lrp, as well as its activity, are important for proper regulation. Three sRNAs, MicF, GcvB and DsrA, negatively regulate lrp expression. MicF and GcvB have previously been shown to regulate lrp expression (23,24), demonstrating the ability of our approach to identify regulators found by other means. In addition, DsrA, identified here, had not previously been shown to be a regulator of lrp. Further analysis of the regulation of lrp is discussed below.
Overall, we find that while not all transcription factors are subject to sRNA regulation, many are. Those in which levels of the protein have a significant effect on regulatory activity are, not surprisingly, more likely to be subject to post-transcriptional regulation by sRNAs. The ‘RNA landscape’ approach is an easy and accurate way to screen genes of interest for sRNA regulation, and allows appreciation of the contributions of multiple sRNAs in the cases of complex regulation.
Pairing interactions by sRNAs: complex regulation by GcvB
Many sRNAs carry out negative regulation by pairing close to the ribosome binding region or initiation codon. MicF and DsrA pair early in the lrp ORF, at overlapping sites (Figure 2). MicF pairs at its 5′ end, consistent with previous results (24); DsrA pairs in a region that is also used for negative regulation of hns (17) (Figure 2B). A possible Hfq binding site is found just upstream of the ATG (highlighted in Supplementary Figure S3). A fusion carrying the region from −18 to +60 is sufficient for efficient regulation by both DsrA and MicF (Supplementary Figure S5B), consistent with an Hfq site within this region. MicF has been shown to block ribosome binding to the lrp translation initiation region (24), and the expectation is that DsrA would as well.
Pairing by GcvB with lrp was more difficult to define. GcvB has multiple regions that have been shown to interact with targets (51–53); mutations in R3 (SL4) blocked the ability of GcvB to regulate lrp (Figure 4 and Supplementary Figure S6). In vitro protection experiments, confirmed with mutagenesis (Figures 4 and 5) identified a site, BS1, far upstream in the leader (−175 to −179) and a second site, BS2 (−33 to −37), for GcvB interaction. These sites contain a conserved GACAG sequence which pairs to R3 (Figure 4). Pairing with either site was sufficient for significant repression of lrp expression, although pairing at both sites gave the best repression (compare repression of the full-length and truncated fusions in Figure 3, and effects of mutations in single or double sites in Supplementary Figure S6D).
Because the same region of GcvB appears to interact with two regions of the lrp mRNA, either one site at a time must be targeted in a given lrp mRNA, or, if both sites are targeted at once, two GcvB molecules would need to collaborate to regulate. If each GcvB requires its own Hfq, that would also suggest that two Hfq rings may bind to a single lrp mRNA simultaneously. One Hfq binding site is likely to be the AAT repeat near the start of translation; a second possible site is near the BS2 pairing region, but is not essential for GcvB regulation (see above and Supplementary Figure S3). Further dissection of the long lrp leader will be necessary to clarify how these two sites cooperate. Other cases where sRNAs interact with multiple sites within an mRNA have been described. Rice et al. (74) found that the complex manXYZ operon is regulated by SgrS binding to two different sites, each capable of translation repression of different genes, but both needed for RNaseE-dependent degradation of the mRNA. In this case, as with our GcvB experiments, the same region of SgrS pairs with both targets. RyhB represses msrB, a methionine oxidase, by similar interactions at two sites in the mRNA, one upstream and one close to the translational start, and mutation of one site does not abrogate regulation (75). In another example of redundant sites within a target, RybB pairs with ompD using two overlapping sites within the target; either one is sufficient for full regulation (76). Spot 42 interacts with a number of targets via two different sites, but in that case, targets were specifically identified that interacted with two different regions within Spot 42 (77), so that one can imagine a single Spot 42 carrying out the interaction.
The finding of these two sites of GcvB interaction does not immediately provide an explanation for how pairing to these sites leads to regulation. Direct interference with ribosome entry is unlikely, given the distance of these sites from the ribosome binding site. As shown in Figure 3 and Supplementary Figure S5, truncations and some point mutants had dramatic effects on the basal level of translation. Our results suggest the existence of a translation or mRNA stability enhancer between −69 and −40; this enhancer acts to overcome a translation inhibitory region between −40 and −18. These effects are independent of GcvB, but are consistent with a model in which GcvB negatively regulates lrp by interfering with the translational enhancer. This is similar to the previous demonstration that GcvB region R1 pairs with and blocks the action of CA-rich translational enhancers (40,51). Our results now suggest that other regions of GcvB have evolved to deal with yet other translational enhancers. How regulation at the far upstream site occurs also remains unexplained; we have no evidence that this pairing region acts as an enhancer. Certainly pairing near the 5′ mRNA end and recruitment of a ribonuclease could have an effect far downstream, for instance by providing a 5′ OH for the downstream mRNA (78). A possible short ORF was also found near this upstream site (underlined in Supplementary Figure S3) and translation of this could play a role in the regulation by GcvB. We find it intriguing that the two targets for GcvB region R3 binding are both transcriptional regulators (lrp, here, and phoP (53)), while the majority of targets of GcvB interact with the R1 region and are components of amino acid transport and metabolism pathways. We also note that GcvB repression of cycA involved in glycine transport and thus possibly providing feedback control of GcvB synthesis is also complex, possibly involving pairing within a GACAG sequence (51). Thus, region R3 of GcvB may have evolved to specifically regulate regulators.
Lrp regulated by sRNAs under stress conditions
Each of the sRNAs regulating lrp is expressed under different conditions, suggesting that they will regulate lrp under different growth conditions. Deletions of micF and dsrA gave little or no change in translational expression of lrp in rich or minimal growth conditions (Supplementary Figure S2). DsrA is known to be expressed at low temperatures (79), and possibly DsrA repression becomes important to decrease Lrp levels under the slow growth/translation conditions at low temperature. Holmqvist et al. saw an increase in expression of an lrp-GFP translational fusion in rich medium in the absence of MicF (24); their experiments differed from ours in that their fusion contained the native lrp promoter, while ours is driven by a PBAD promoter. When the native lrp gene was measured, deletion of micF led to a modest increase in both the lrp mRNA (1.7x) and Lrp protein (1.5x) (Figure 6D and E; -PQ samples), consistent with the Holmqvist observation. Thus, it seems likely that MicF regulation of Lrp, under rich medium growth conditions, may have an indirect effect on the lrp promoter as well as directly repressing Lrp translation; possibly the promoter effect is more significant under lab growth conditions. Thus, our results reinforce the idea that MicF and Lrp regulation are entangled at multiple levels.
Deletion of the gene encoding GcvB, an sRNA that is highly abundant when bacteria grow in rich medium, increased lrp expression in rich medium, both at the level of translation (Supplementary Figure S2C and Figure 6C) and at the level of mRNA and protein (Figure 6D and E). This increase is very similar to the increase in lrp mRNA measured by Sharma et al. in Salmonella when gcvB was deleted (51).
Oxidative stress is known to lead to increased MicF levels (56,80); therefore, we examined the effects of GcvB and MicF on lrp after oxidative stress, expecting MicF to play a more significant role under these conditions. Instead, we saw an increased role for GcvB (Figure 6C–E). The level of expression of the PBAD-lrp::lacZ translational fusion, levels of native lrp mRNA and Lrp protein were all significantly decreased after 1 h of paraquat treatment and this was relieved by deletion of gcvB, with some additional effect of deleting micF (Figure 6C–E). Therefore, GcvB repression is apparently effective under this oxidative stress condition, even though its levels do not change (Supplementary Figure S8).
A number of mechanisms might explain why GcvB is apparently better able to repress lrp (and possibly other targets) under oxidative stress, via changes in the availability or activity of the sRNA. GcvB has been shown to be subject to N-terminal modification by Nicotinamide adenine dinucleotide (NAD) (81), although the effect of this modification on GcvB activity is not yet known. It is easy to imagine, however, that oxidative stress may change the nature and/or extent of the GcvB NAD cap, affecting sRNA activity. It was recently reported that GcvB is negatively regulated by an RNA sponge, SroC sRNA (82). SroC pairs with the same region of GcvB as the lrp mRNA. Thus, SroC, in addition to leading to degradation of GcvB, may directly compete with other targets pairing to this region of GcvB. If the level of this RNA sponge (and possibly others, if they exist) decreases after oxidative stress, this may leave GcvB more able to repress lrp. Alternatively, if GcvB acts by blocking a translational enhancer, trans-acting factors active at that enhancer may be less available after oxidative stress, leading to increased activity of any GcvB that is present.
Our results have expanded our understanding of the complex network of sRNAs that regulate lrp. Our results also suggest the existence of translational signals throughout the lrp leader, which may themselves respond to yet other signals. GcvB plays a central role in this regulation, repressing lrp via two separate but redundant sites within the lrp mRNA. After PQ treatment, lrp becomes unavailable, and this is in large part dependent on repression by GcvB, providing a previously unknown role for GcvB in the response to oxidative stress.
Supplementary Material
Acknowledgments
We thank G. Storz and members of the S. Gottesman lab for comments on the manuscript. We thank T. Updegrove for advice on the structure probing assay and the S. Adhya lab for providing facilities. We also thank M. Guillier for providing strains and the Sarah Woodson lab for providing Hfq protein, and a referee for pointing out a possible ORF in the lrp leader.
Footnotes
Present address: Hyun-Jung Lee, Department of Biomedical Sciences, Ajou University School of Medicine, Suwon 443-380, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Funding for open access charge: Center for Cancer Research intramural funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 2.Hanamura A., Aiba H. A new aspect of transcriptional control of the Escherichia coli crp gene: positive autoregulation. Mol. Microbiol. 1992;6:2489–2497. doi: 10.1111/j.1365-2958.1992.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 3.Kobir A., Shi L., Boskovic A., Grangeasse C., Franjevic D., Mijakovic I. Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta. 2011;1810:989–994. doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Battesti A., Hoskins J.R., Tong S., Milanesio P., Mann J.M., Kravats A., Tsegaye Y.M., Bougdour A., Wickner S., Gottesman S. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27:2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majdalani N., Hernandez D., Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 7.Mandin P., Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomason M.K., Fontaine F., De Lay N., Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Lay N., Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehm A., Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol. Microbiol. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 12.Moon K., Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masse E., Vanderpool C.K., Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma C.M., Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Dorman C.J. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Li G.W., Chen C., Xie X.S., Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lease R.A., Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernsting B.R., Atkinson M.R., Ninfa A.J., Matthews R.G. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 1992;174:1109–1118. doi: 10.1128/jb.174.4.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brinkman A.B., Ettema T.J., de Vos W.M., van der Oost J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 20.Calvo J.M., Matthews R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew E., Zhi J., Freundlich M. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 1996;178:7234–7240. doi: 10.1128/jb.178.24.7234-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart B.R., Blumenthal R.M. Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 2011;193:1054–1064. doi: 10.1128/JB.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modi S.R., Camacho D.M., Kohanski M.A., Walker G.C., Collins J.J. Functional characterization of bacterial sRNAs using a network biology approach. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15522–15527. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmqvist E., Unoson C., Reimegard J., Wagner E.G. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- 25.Fic E., Bonarek P., Gorecki A., Kedracka-Krok S., Mikolajczak J., Polit A., Tworzydlo M., Dziedzicka-Wasylewska M., Wasylewski Z. cAMP receptor protein from escherichia coli as a model of signal transduction in proteins—a review. J. Mol. Microbiol. Biotechnol. 2009;17:1–11. doi: 10.1159/000178014. [DOI] [PubMed] [Google Scholar]
- 26.Matsui M., Tomita M., Kanai A. Comprehensive computational analysis of bacterial CRP/FNR superfamily and its target motifs reveals stepwise evolution of transcriptional networks. Genome Biol. Evol. 2013;5:267–282. doi: 10.1093/gbe/evt004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semchyshyn H., Bagnyukova T., Lushchak V. Involvement of soxRS regulon in response of Escherichia coli to oxidative stress induced by hydrogen peroxide. Biochemistry. 2005;70:1238–1244. doi: 10.1007/s10541-005-0253-6. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.H., Lee K.L., Yeo W.S., Park S.J., Roe J.H. SoxRS-mediated lipopolysaccharide modification enhances resistance against multiple drugs in Escherichia coli. J. Bacteriol. 2009;191:4441–4450. doi: 10.1128/JB.01474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunoshiba T., Hidalgo E., Amabile Cuevas C.F., Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung C.T., Niemela S.L., Miller R.H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandin P., Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol. Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battesti A., Tsegaye Y.M., Packer D.G., Majdalani N., Gottesman S. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J. Bacteriol. 2012;194:2470–2478. doi: 10.1128/JB.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bougdour A., Cunning C., Baptiste P.J., Elliott T., Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 35.Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 36.Miller J.H. A Short Course in Bacterial Genetics. NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 37.Masse E., Escorcia F.E., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon K., Gottesman S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009;74:1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J.C., Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma C.M., Darfeuille F., Plantinga T.H., Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sledjeski D., Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lease R.A., Cusick M., Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbanowski M.L., Stauffer L.T., Stauffer G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q., Wu J., Friedberg D., Plakto J., Calvo J.M. Regulation of the Escherichia coli lrp gene. J. Bacteriol. 1994;176:1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lintner R.E., Mishra P.K., Srivastava P., Martinez-Vaz B.M., Khodursky A.B., Blumenthal R.M. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 2008;8:60. doi: 10.1186/1471-2180-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima T., Ito K., Kabayama H., Nakamura Y. Regulation of lrp gene expression by H-NS and Lrp proteins in Escherichia coli: dominant negative mutations in lrp. Mol. Gen. Genet. 1995;247:521–528. doi: 10.1007/BF00290342. [DOI] [PubMed] [Google Scholar]
- 48.Hommais F., Krin E., Coppee J.Y., Lacroix C., Yeramian E., Danchin A., Bertin P. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004;150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 49.Partridge J.D., Bodenmiller D.M., Humphrys M.S., Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 50.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma C.M., Papenfort K., Pernitzsch S.R., Mollenkopf H.J., Hinton J.C., Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 52.Pulvermacher S.C., Stauffer L.T., Stauffer G.V. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol. Lett. 2008;281:42–50. doi: 10.1111/j.1574-6968.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- 53.Coornaert A., Chiaruttini C., Springer M., Guillier M. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013;9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 55.Schu D.J., Zhang A., Gottesman S., Storz G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015;34:2557–2573. doi: 10.15252/embj.201591569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou J.H., Greenberg J.T., Demple B. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J. Bacteriol. 1993;175:1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moon K., Six D.A., Lee H.J., Raetz C.R., Gottesman S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013;89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Touati D. Sensing and protecting against superoxide stress in Escherichia coli—how many ways are there to trigger soxRS response? Redox Rep. 2000;5:287–293. doi: 10.1179/135100000101535825. [DOI] [PubMed] [Google Scholar]
- 59.Gutierrez A., Laureti L., Crussard S., Abida H., Rodriguez-Rojas A., Blazquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J., et al. Beta-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013;4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durand S., Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polayes D.A., Rice P.W., Garner M.M., Dahlberg J.E. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J. Bacteriol. 1988;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Navarre W.W., McClelland M., Libby S.J., Fang F.C. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 64.Battesti A., Majdalani N., Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duval V., Lister I.M. MarA, SoxS and Rob of Escherichia coli—global regulators of multidrug resistance, virulence and stress response. Int. J. Biotechnol. Wellness Ind. 2013;2:101–124. doi: 10.6000/1927-3037.2013.02.03.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seo S.W., Kim D., Szubin R., Palsson B.O. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 2015;12:1289–1299. doi: 10.1016/j.celrep.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 67.Gaudu P., Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffith K.L., Shah I.M., Wolf R.E., Jr Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 2004;51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 69.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 70.Chubiz L.M., Glekas G.D., Rao C.V. Transcriptional cross talk within the mar-sox-rob regulon in Escherichia coli is limited to the rob and marRAB operons. J. Bacteriol. 2012;194:4867–4875. doi: 10.1128/JB.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho B.-K., Barrett C.L., Knight E.M., Park Y.S., Palsson B.O. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tani T.H., Khodursky A., Blumenthal R.M., Brown P.O., Matthews R.G. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung S.-P., Baldi P., Hatfield G.W. Global gene expression profiling in Escherichia coli K12: the effects of leucine-responsive regulatory protein. J. Biol. Chem. 2002;277:40309–40323. doi: 10.1074/jbc.M204044200. [DOI] [PubMed] [Google Scholar]
- 74.Rice J.B., Balasubramanian D., Vanderpool C.K. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2691–E2698. doi: 10.1073/pnas.1207927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bos J., Duverger Y., Thouvenot B., Chiaruttini C., Branlant C., Springer M., Charpentier B., Barras F. The sRNA RyhB regulates the synthesis of the Escherichia coli methionine sulfoxide reductase MsrB but not MsrA. PLoS ONE. 2013;8:e63647. doi: 10.1371/journal.pone.0063647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balbontin R., Fiorini F., Figueroa-Bossi N., Casadesus J., Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 2010;78:380–394. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- 77.Beisel C.L., Updegrove T.B., Janson B.J., Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bandyra K.J., Said N., Pfeiffer V., Gorna M.W., Vogel J., Luisi B.F. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell. 2012;47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sledjeski D.D., Gupta A., Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 80.Chen S., Zhang A., Blyn L.B., Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cahova H., Winz M.L., Hofer K., Nubel G., Jaschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 82.Miyakoshi M., Chao Y., Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015;34:1478–1492. doi: 10.15252/embj.201490546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stauffer L.T., Stauffer G.V. The Escherichia coli GcvB sRNA uses genetic redundancy to control cycA expression. ISRN Microbiol. 2012;2012:636273. doi: 10.5402/2012/636273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.