Abstract
DNA circuit is a versatile and highly-programmable toolbox which can potentially be used for the autonomous sensing of dynamic events, such as biomolecular interactions. However, the experimental implementation of in silico circuit designs has been hindered by the problem of circuit leakage. Here, we systematically analyzed the sources and characteristics of various types of leakage in a split proximity circuit which was engineered to spatially probe for target sites held within close proximity. Direct evidence that 3′-truncated oligonucleotides were the major impurity contributing to circuit leakage was presented. More importantly, a unique strategy of translocating a single nucleotide between domains, termed ‘inter-domain bridging’, was introduced to eliminate toehold-independent leakages while enhancing the strand displacement kinetics across a three-way junction. We also analyzed the dynamics of intermediate complexes involved in the circuit computation in order to define the working range of domain lengths for the reporter toehold and association region respectively. The final circuit design was successfully implemented on a model streptavidin-biotin system and demonstrated to be robust against both circuit leakage and biological interferences. We anticipate that this simple signal transduction strategy can be used to probe for diverse biomolecular interactions when used in conjunction with specific target recognition moieties.
INTRODUCTION
Biomolecular interactions are involved in numerous biological processes, for example, cell receptor clustering for signal transduction, host–cell interaction during infections and within protein complexes (1,2). Despite the biological significance, a simple molecular toolbox for the direct detection of such multi-target activities is lacking (3). Conventional analysis methods often require artificial counterparts to fish for the interacting partner or extensive sample processing to isolate the biomolecules from their original environment, e.g. pull-down assay and gel retardation assay (4).
Simple yet effective techniques have emerged recently to bridge the methodology gap. For example, Wong's group has reported a novel DNA-only tool called DNA nanoswitches which is a simple, low-cost and label-free method for interrogating biomolecular interactions based on conformation changes in the DNA backbone (5). Though a versatile and powerful tool requiring only gel electrophoresis for readout, it cannot be used to perform in situ analysis and visualization. For such applications, the leading method in recent years is the proximity ligation assay (PLA) (6). An enzyme-free variant was recently developed based on the proximity-dependent initiation of hybridization chain reaction (HCR) (7). However, signal activation can only be achieved via a series of probe addition and washing steps.
We have previously reported a self-contained DNA circuit framework for evaluating bi-molecular recognition events within close proximity (8). The crux of the turn-on signal lies in the re-configuration of the aptamer sequence upon target binding to expose the sequestered toehold. However, the limited availability of well-validated aptamer sequences demands for the need to develop a more general signal transduction strategy to detect a wider pool of biomolecules (9). This motivates the design of a DNA-only split proximity circuit which draws inspirations from split-protein reporter systems to achieve real-time turn-on detection (10,11). The fundamental principle is based on the simple reassembly of readout signals mediated by the binding of interacting biomolecules. This principle can readily be extended to synthetic, cell-free systems as demonstrated by Ghosh's group using protein reporters synthesized from cell-free translation (12).
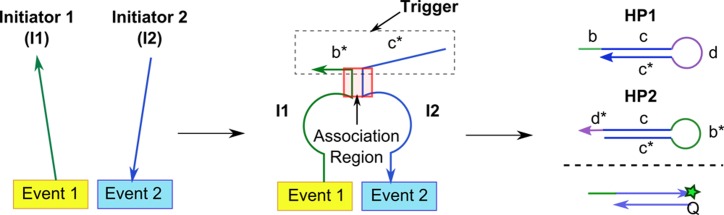
The central idea here is to split a single reporter trigger strand (c* b*) and attach the respective domains on separate initiator strands (I1 and I2) (Figure 1). The initiator strands bind to the respective targets and bring the split domains together to form the complete trigger strand. Previous study on remote toehold suggests that strand displacement can still take place across non-adjacent domains (13). The co-localization of the initiator strands within close proximity effectively increases their local concentration for enhanced reaction kinetics (14,15). An association region was also incorporated to thermodynamically stabilize the intermediate I1–I2 complex (16). The application of such DNA three-way junctions for protein detection was recently demonstrated by Le's group (17–19).
Figure 1.
The split proximity circuit involved three steps: (1) binding of initiator 1 (I1) and initiator 2 (I2) strands to the respective recognition sites. (2) The close proximity of I1 and I2 kinetically favored the formation of a complete trigger strand (c* b*) which was stabilized by a short DNA association region to (3) initiate readout signals, e.g. hybridization chain reaction (HCR) or fluorophore–quencher (F-Q) system.
The performance of any DNA circuit is closely related to its signal-to-background (S/B) ratio. While the signal can be enhanced by amplification methods, the issue of circuit leakage contributing to non-negligible amount of background noise still poses a great challenge in the community (20–22). There are two types of leakages: (i) initial leakage due to oligonucleotide synthesis defects and misfolded DNA, and (ii) asymptotic leakage due to spurious hybridization, conformational fluctuations or cross-talk between domains (23). The former may be minimized, though not entirely eliminated, by using purified (HPLC- or PAGE-grade) oligonucleotides. The latter is inherent at the point of sequence design and requires deliberate design effort to systematically minimize background noise for a particular circuit system. For example, Ellington's group has devised design principles and purification methods to minimize leakages in the catalytic hairpin assembly (CHA) system (23). As such, multiple CHA layers can be stacked to achieve thousands-fold signal gain without excessive background.
One obvious strategy is to use low probe concentrations to kinetically suppress undesirable leakages (8,17). While it may be effective within the scope of a specific study, this approach lacks generality and may require arbitrary re-optimization for individual reaction conditions. Alternatively, a design-driven approach based on careful thermodynamics considerations can guarantee a more robust outcome and should be the method of choice wherever possible (24,25).
In this study, we identified the sources of leakages involved in the proposed split proximity circuit and systematically formulated design guidelines to minimize the circuit leakages. First, we presented direct experimental evidence that oligonucleotide defects due to 3′-truncation during solid-state synthesis was the main culprit for initial leakage. Next, we introduced a unique strategy of translocating a single nucleotide across split domains, which we termed ‘inter-domain bridging’, to eliminate toehold-independent side reaction while improving the strand displacement kinetics across the three-way junction by ca. 13-fold. Finally, we found that asymptotic leakage was inevitable given the inherent dynamics of a three-way junction circuit, and the best counter-strategy was to use the shortest possible association length, which we recommend to be less than 6 nt. These three key design guidelines culminated in a final optimized circuit design which was successfully implemented on a model streptavidin-biotin system and found to be robust against both circuit leakages and interferences from non-specific proteins or a biological environment of 10% fetal bovine serum.
MATERIALS AND METHODS
Materials
All DNA oligonucleotides used in this study were purchased from Integrated DNA (IDT), and HPLC purified by IDT. The sequences are provided in Supplementary Table S1 and the individual domain sequences are presented in Supplementary Table S2. The lyophilized DNA was reconstituted in 1× Tris–EDTA buffer (1× TE, pH 8.0) to give 100 μM stock and stored at 4°C, except for fluorescein (F)- and Dabcyl (Q)-modified DNAs which were stored at −20°C and protected from light. The following chemicals were used as received: sodium chloride (NaCl, ≥99.5%) and magnesium chloride (MgCl2, ≥98%) were purchased from Sigma Aldrich. SYBR gold nucleic acid stain (10 000× in DMSO) was purchased from Invitrogen. Agarose (molecular biology grade) and 10× Tris–borate–EDTA (TBE, pH 8.3) were purchased from Vivantis. 1× TE (pH 8.0) was purchased from 1st BASE. Milli-Q water with resistance >18.2 MP/cm was used throughout the experiment.
Gel electrophoresis for hybridization chain reaction (HCR) readout
The reaction buffer used in this study was 10 mM Tris (pH 7.0), 480 mM NaCl and 5 mM MgCl2. This was optimized using hairpin trigger (HT) on the HCR readout system (data not shown). Stock DNA (100 μM in 1× TE, pH 8.0) was diluted to 2.5 μM working concentration for the 5-components split trigger system in the reaction buffer. The respective DNA components (H1, H2, I1, I2 and ST) were heated to 95°C for 5 min and allowed to cool to room temperature for 30 min. Equal volume of the relevant components were mixed to obtain a final reaction concentration of 500 nM, except for the targets. The final sequence of adding the respective DNA components was: (i) Incubate target molecule (HT, DT, ST or streptavidin) with the initiator strands (I1 and I2) for 20 min; followed by (ii) adding the HCR readout strands (HP1 and HP2) for 10 min. Other addition sequences and incubation times were used when establishing the design guidelines which were stated within the main text.
Analysis was carried out on 3% agarose gel which was pre-stained with 1× SYBR gold nucleic acid stain. The gel was run at 90 V (6 V/cm) for 30–40 min in 0.5× TBE running buffer at 4°C. Distinct qualitative conclusions could be drawn due to the signal amplification of HCR and no further quantitative analysis was performed.
Real-time fluorescence measurement for fluorophore–quencher (F–Q) readout
All reactions proceeded at 24°C in 100 μl volume of the same reaction buffer: 10 mM Tris (pH 7.0), 480 mM NaCl and 5 mM MgCl2, unless otherwise stated. Stock DNA (100 μM in 1× TE, pH 8.0) was first diluted to 1.0 μM working concentration in the reaction buffer. Note that the F–Q complex was prepared in a 1:2 molar ratio (on the basis of 1.0 μM F) to ensure complete quenching of the fluorescence. The respective DNA components (F–Q, I1, I2 and trigger strands) were heated separately to 95°C for 5 min and allowed to cool to room temperature for 30 min. The final reaction concentration was 100 nM for all DNA components. The reactions for characterizing the various types of leakages were conducted in a one-pot format. The final reaction addition sequence (from the discussion of intermediate complexes onwards) was: (i) incubate the target (ST, streptavidin and thrombin) with initiator strands (I1 and I2) for 20 min; followed by (ii) adding the F–Q readout strand for an hour.
All fluorescence measurements were carried out using 96-well black Nunc microwell plate on a Tecan M200 Pro plate reader. The excitation and emission wavelength used for fluorescence measurement was 487 and 518 nm respectively. The kinetics of strand displacement was monitored by measuring the fluorescence of the reaction mixture every minute for a total of one hour.
Data analysis and Interpretation
In each set of experiment, a negative control was established using 100 nM F–Q, I1 and I2 (without ST), which we termed as the background noise. A positive control was established using 100 nM F-Q and 150 nM direct trigger (DT) which completely displaced all quencher (Q) strands. The fluorescence intensity obtained here was taken to correspond to 100 nM of released fluorescence. All raw fluorescence readings were normalized against this value to calculate the relative fluorescence signals (RFU), i.e. 1 RFU was equivalent to 1 nM of released fluorescence. This minimized the random fluctuation of signals due to, e.g. instrument noise and environment.
The equilibrium concentrations of DNA complexes reported in our main text were generated from Nupack simulations (26). 100 nM of the required DNA components were inputted for each reaction at a temperature of 24°C under the salt conditions of [Na+] = 0.48 M and [Mg2+] = 0.005 M. The maximum complex size was set to be equal to the number of strand species for each scenario analyzed.
For the experimental analysis, all concentrations of DNA and DNA complexes were reported at the 1 h mark unless otherwise stated. Quantification of the initial signal burst was based on the fitted RFU at t = 0. We defined signal-to-background (S/B) ratio as (RFUsignal − RFUbackground)/RFUbackground, where signal was generated in presence of ST and background was generated in absence of ST.
Graphs used for the detailed characterization of minute leakages, i.e. Figures 2B, 3B and 4B, were shown without error bars (highlighted in their respective legends) to avoid congesting the narrow RFU range represented. Instead, we used scatter plots to represent the mean value for triplicate experiments, which were then fitted with a solid linear trend line to guide the eyes in identifying time-dependent trends.
Figure 2.
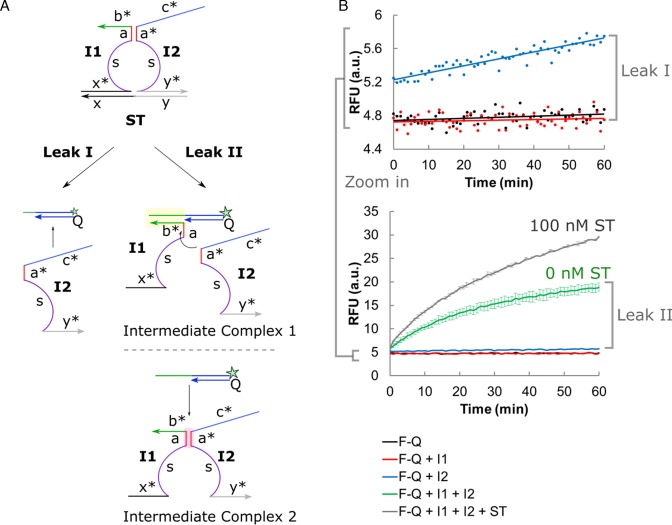
(A) The background noise could be attributed to two main sources of circuit leakage: Leak I - toehold-independent strand displacement by initiator 2 (I2), and Leak II - formation of intermediate complex 1 (I1–F–Q) or 2 (I1–I2) in presence of all DNA components except ST. (B) Different combinations of the DNA components (F–Q, I1, I2 and ST) revealed the types of leakages involved. Error bars shown indicate sample standard deviation of triplicate experiments. However, error bars were not shown for the narrow RFU range (top graph showing Leak I) to avoid congesting the plot.
Figure 3.
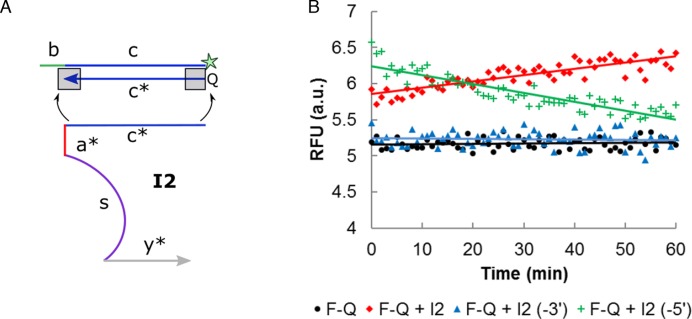
(A) The initial leakage by I2 was attributed to the synthesis defect of Q strand. The corresponding nucleotide on I2 was removed to investigate the end (3′- or 5′-) which contributed predominantly to the leakage. (B) The omission of a single nucleotide at the 3′-end of I2 domain c*, i.e.I2 (-3′), was found to be most effective in minimizing the initial leakage. The mean values of triplicate readings were presented as scatter plot, while error bars were not shown for the narrow RFU range to avoid congesting the plot.
Figure 4.
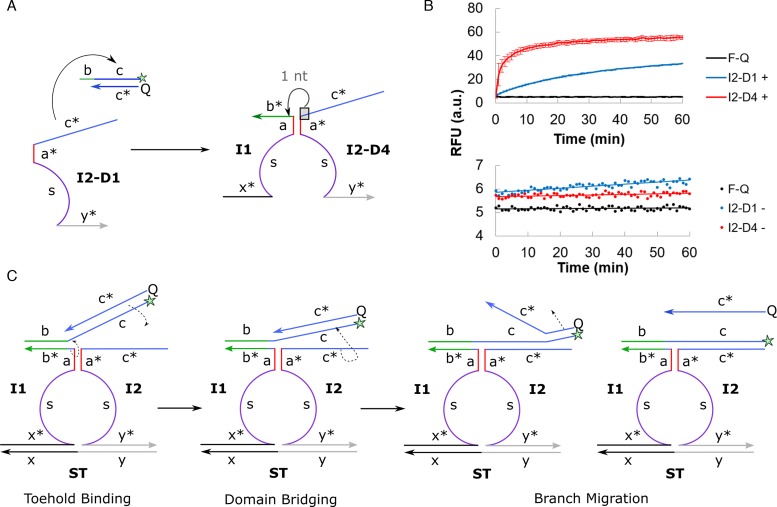
(A) A single nucleotide was translocated from domain c* to domain b* in I2–D4 design to minimize the asymptotic leakage in I2–D1. (B) The generation of positive signal (top) and Type I leakage (bottom) over time was evaluated in F–Q + I1 + I2 + ST and F–Q + I2 reaction mixture respectively. All reaction mixtures contained 100 nM of the relevant DNA components. Error bars shown for positive signal indicate sample standard deviation of triplicate experiments, while error bars were not shown for the narrow RFU range involved in circuit leakage to avoid congesting the plot. (C) The translocation of one nucleotide from domain c* to domain b* relieved the distinct separation of domains, in a strategy which we termed inter-domain bridging. First, toehold binding took place at domain b. Next, the single nucleotide brought over from domain c* initiated the displacement of the quencher (Q) strand and pulled the fluorophore–quencher (F–Q) complex towards domain c* of I2, effectively bridging the split domains spatially. Finally, strand displacement continued via the usual branch migration process.
RESULTS AND DISCUSSION
Sources of circuit leakage in split proximity circuit
We first demonstrated the circuit operations on an ideal system consisting of a single-stranded DNA split target (ST) strand with two recognition domains (domains x and y) to mimic bi-molecular binding events (Figure 2A). The split proximity circuit was evaluated at various design stages using the hybridization chain reaction (HCR) (27,28) and/or fluorescence-quencher (F–Q) readout (Figure 1). The former amplified the presence of circuit leakages and when used in conjunction with gel electrophoresis, allowed for the unambiguous confirmation of the presence of various DNA complexes. The latter was used to profile the kinetics evolution of various leakage types.
We attributed the leakages to two main sources (Figure 2A): Leak I - toehold-independent strand displacement by I2, and Leak II - formation of I1–F–Q and I1–I2 intermediate complexes when all DNA components, except ST, were present. Each source of leakage was systematically isolated and confirmed by the incremental addition of DNA components (Figure 2B).
I2 contributed to a small but non-negligible amount of background noise (Leak I) which was characterized by an initial burst of 8.9% increase in fluorescence signal followed by a gradual but steady growth over time. Surprisingly, the amount of background noise in absence of ST (Leak II) was overwhelmingly high and accounted for 56.5% of the positive signal. This was in spite of using one particular combination of association region length (6 nt) and toehold length (9 nt) previously reported by Le's group to give minimal background noise (nt = nucleotide) (17). This may be due to the significantly higher probe concentration (100 nM versus 10 nM) or different reaction buffer used in this study. This observation further reinforced our argument that kinetically-suppressed leakages lacked robustness.
3′-Truncated oligonucleotide as the source of initial leakage
Leak I was further decoupled into two types of leakages, i.e. initial leakage characterized by an initial signal burst and asymptotic leakage which steadily grew with time (Supplementary Figure S1). We first hypothesized that the synthesis defect on Q strand was the main culprit of the initial leakage (refer to Supplementary Figure S2 for the rationale of this hypothesis). To test this hypothesis, we eliminated one nucleotide respectively from the 5′- and 3′-end of the corresponding domain c* on I2 (Figure 3A). From Figure 3B, it is evident that the 3′-truncation of domain c* on I2 (I2 (-3′)) reduced leakages to the greatest extent, suggesting that the corresponding 3′-end on Q was most vulnerable to synthesis defects. We noted that the initial burst from the 5′-truncated I2 strand was higher than the full length I2. The reason for this unexpected observation is unknown at this point, however, the decreasing leakage over time can be explained by the subsequent strand displacement of I2 (-5′) by the full length Q strand.
This observation contradicted the commonly-held assumption that the 5′-end is more susceptible to truncation since synthesis proceeds in the 3′- to 5′-direction (25,29). We wish to clarify this misunderstanding by pointing out that in a solid-phase oligonucleotide synthesis, the first few nucleotides closer to the solid support, i.e. at the 3′-end, suffer from greater steric hindrance and hence product truncation. Temsamani et al. found that 55% of their n – 1 PAGE-purified products were truncated at the first four nucleotides from the 3′-end (30). Previously unexpected experimental results on the effect of DNA impurities and 5′/3′ orientation may be re-interpreted more intuitively in light of our alternative perspective and evidence presented (25). It can also explain why the presence of mismatches in the position corresponding to the 3′-end of the other CHA hairpin pair is most effective in suppressing the CHA leakage (31).
Oligonucleotides used in DNA nanotechnology works are often obtained via solid-phase synthesis, rendering this a common yet not often discussed issue across the field. We believe that the added knowledge of this non-trivial fact can guide future circuit designs or purification methods closer towards achieving an ideal leak-free system. Alternatively, other synthesis methods, e.g. using enzymes, may be considered to circumvent this technical issue (23).
‘Inter-domain bridging’ for improving strand displacement across split domains
Since the strand displacement reaction is in dynamic equilibrium, the exposed domain c* of I2 can displace its identical counterpart, Q, over time and contribute to asymptotic leakage in the original design (Figure 4A). We conceptualized three designs to thermodynamically disfavor such toehold-independent strand displacement (refer to Section S3 for an extended discussion). Out of these, the modified I2–D4 exhibited the best performance in terms of both suppressing the asymptotic leakage, as seen from the relatively stable background noise over time, and in boosting the generation of positive signal by 65.9% as compared to I2–D1 (Figure 4B). This was in spite of the unconventional mixing of nucleotides between neighboring domains.
We propose the concept of inter-domain bridging arising from the translocation of nucleotide from domain c* to b* (Figure 4C). This anchored the F–Q complex towards domain c* on I2–D4 strand after toehold binding. The initial rate constant of the overall strand displacement process increased from kI2–D1 = 1.44 min−1 to kI2–D4 = 18.8 min−1. This suggests that the closer spatial proximity between F–Q and I2 strands promoted the kinetics of the remaining branch migration process. Overall, a 2-fold improvement in the signal-to-background (S/B) was achieved simply by shifting a single nucleotide across the split domains (S/B of I2–D4 = 9.5 versus S/B of I2–D1 = 5.2).
Analysis of intermediate complexes contributing to leak II
Two types of intermediate complexes involved in the formation of the three-way junction were responsible for Leak II (Figure 2A). We assumed that two out of the three strands involved (I1, I2 and F–Q) had to first form a stable intermediate complex for the last incoming strand to bind to and trigger the reporter signal since a simultaneous tri-molecular collision event is rare (32).
I1 and F could hybridize at domain b* while the association region (domain a*) doubled as the reporter toehold to trigger the formation of the reporter signals (intermediate complex 1). Also, I1 and I2 could associate at domain a* in the absence of the target (intermediate complex 2). It was difficult to isolate individual complexes experimentally because both domains a* and b* served dual roles of complex stabilization (for the formation of intermediate complex) and reporter triggering (branch migration to release the reporter signal) (Figure 2A). Regardless, it was possible to deduce the relative contribution by each intermediate complexes from their respective thermodynamics stability, which could be determined by the length of domains a* and b* respectively.
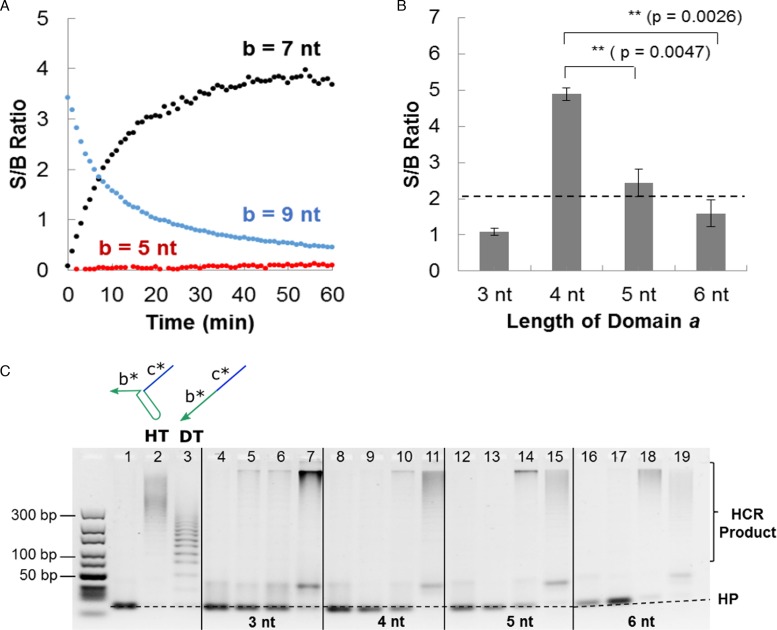
In our original configuration (a* = 6 nt and b* = 9 nt), the S/B ratio decreased over time (Figure 5A). This was due to the extensive amount of Leak II whereby the intermediate complexes consumed 39.7 nM of reporter strands. Based on Nupack simulation, I1-F-Q (intermediate complex 1) was found to be the predominant species at equilibrium (45.0 nM) compared to I1–I2 (intermediate complex 2, 0.61 nM). When the length of domain b* was reduced to 7 nt, the S/B ratio reversed to an increasing trend (Figure 5A). This was due to the significantly lower equilibrium concentration of I1–F–Q complex (2.6 nM) which reduced the Leak II consumption of reporter strands to 3.1 nM. Further reduction of the length of domain b* to 5 nt greatly penalized the generation of positive signal, leading to a negligibly low S/B ratio (Figure 5A).
Figure 5.
(A) The length of domain b* was varied (5 nt, 7 nt and 9 nt) to understand the contribution to signal and background by intermediate complex 1. Refer to Supplementary Figure S4 for the decoupled signal and background time profiles. (B) An optimal association length (domain a*) of 4 nt resulted in maximum signal-to-background (S/B) ratio at t = 10 min using the fluorophore–quencher (F–Q) readout. Refer to Supplementary Figure S5 for the time evolution of S/B ratio using different lengths of domain a*. The error bars correspond to sample standard deviation (n = 3). ** indicates that P < 0.005 for Student's t-test (one-sided). The actual p-values are shown in bracket. (C) Gel electrophoresis was used to visualize the extent of signal and background formation. Lane 1 corresponds to hairpins (HP1 and HP2) only, lanes 2 and 3 correspond to positive controls using hairpin trigger (HT) and direct trigger (DT) respectively. Lanes representing each association region length (domain a*) are demarcated by solid lines and are shown in the following sequence (from left to right): HP1 + HP2 + I1, HP1 + HP2 + I2, HP1 + HP2 + I1 + I2, HP1 + HP2 + I1 + I2 + ST. 500 nM of individual DNA components was used. A 10–300 bp DNA ladder is shown on the left-hand side of the gel.
From the above result, we propose that domain b* is the de facto reporter toehold in the proper ST–I1–I2 assembly and should be kept effectively long to significantly enhance the triggering of reporter signals in the presence of ST (refer to Section S5 for an extended discussion) (33). As such, a better strategy would be to temporally separate the addition of initiator strands (I1 and I2) and reporter strands (F–Q or HP1 and HP2). Under this format, the main design criterion was to reduce the formation of intermediate complex 2 for which the stability was controlled by the length of domain a. 4 nt was found to be the optimal association region length giving the highest S/B ratio (Figure 5B) within as quickly as 10 min without forming I1–I2 complex at equilibrium.
Unfortunately, we were unable to completely eliminate the background noise due to Leak II (Supplementary Figure S7). This was not surprising as the thermodynamically-favored I1–F–Q complex could form once the reporter strands were introduced. Nonetheless, the involvement of a short domain a* (4 nt) as the reporter toehold implied exponentially slower reporter triggering as compared to the 9 nt toehold (domain b*) triggering in the proper ST–I1–I2 assembly (34). Overall, we observed that the S/B ratio was not negatively affected even when HCR amplification was involved, except when domain a* was longer at 6 nt (Figure 5C).
Evaluation of performance on model biomolecular systems
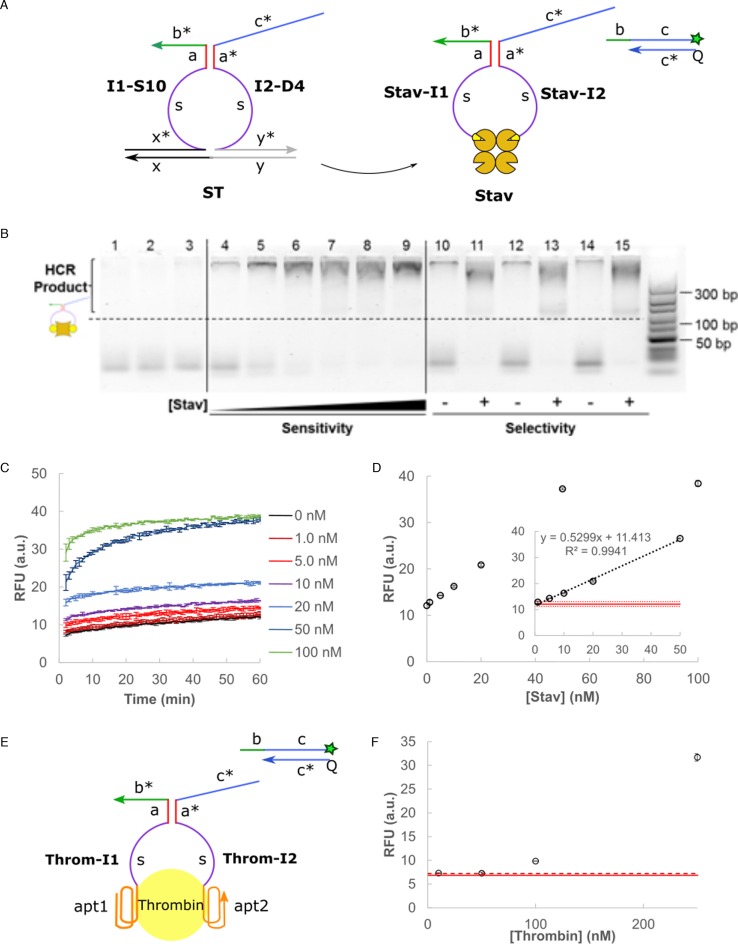
Having successfully developed a DNA split proximity circuit with minimized circuit leakages (Supplementary Figure S7), we tested its applicability first on a model streptavidin-biotin system. Streptavidin was chosen for its high binding affinity (KD ≈ 10−14 to 10−15 nM) to biotin which can be modified onto commercially-available oligonucleotides (Figure 6A). This allowed us to validate the inherent DNA circuit properties without further complications from the target recognition step which was not the focus of this study. The HCR product bands were visibly more intense than the negative control (lane 4) for the range of streptavidin concentrations tested (10–500 nM, lanes 5–9) (Figure 6B). The circuit remained selective under the interferences by non-specific bovine serum albumin (BSA) protein (lanes 10–13) and a biological environment of 10% fetal bovine serum (FBS) (lanes 14 and 15). The detection result was further quantified using F–Q readout signal (Figure 6C). Clear dosage dependence was observed for 0–50 nM streptavidin target with a limit of detection at 2.95 nM (mean of background noise ±3 standard deviation) (Figure 6D). These results demonstrated the good sensitivity, selectivity and fast detection speed (10 min) of our split proximity circuit. To further demonstrate the versatility of this method, we replaced the recognition moiety for thrombin binding aptamers which have known binding affinities (KD ≈ 100 nM and 0.5 nM) to their thrombin target (Figure 6E) (35). Dosage dependence of the fluorescence signal on thrombin concentration was similarly observed (Figure 6F).
Figure 6.
(A) The DNA split proximity circuit was tested on a model streptavidin (Stav)-biotin system. (B) The circuit performance was evaluated using gel electrophoresis and HCR readout. Lane 1 corresponds to hairpins (HP1 and HP2) only, lanes 2 and 3 correspond to controls with HP1 + HP2 + I1 and HP1 + HP2 + I2 respectively. Lanes 4–9 represent the signal developed in presence of 0 nM, 10 nM, 50 nM, 100 nM, 250 nM and 500 nM of Stav. Lanes 10– 15 represent the circuit selectivity in presence of interference from 500 nM BSA (lanes 10 and 11), 5000 nM BSA (lanes 12 and 13) and 10% FBS (lanes 14 and 15). 500 nM of individual DNA circuit components was used. A 10 – 300 bp DNA ladder is shown on the right-hand side of the gel. (C) Signal trace over time in 1 min intervals when varying concentration of Stav (0–100 nM) was added to the split proximity circuit. Error bars correspond to sample standard deviation (n = 3). (D) The equilibrium RFU signal at t = 1 h was plotted for the range of Stav concentrations tested. A linear trend was observed between 0 and 50 nM (inset). The solid red line corresponds to the mean background noise while the dotted red line corresponds to 3 standard deviations from the mean noise level. (E) The recognition moiety was replaced by thrombin binding aptamer with known binding affinities to thrombin target to demonstrate the versatility of this method. (F) Dosage dependence of the fluorescence signal on thrombin concentration was observed for the range tested (10–250 nM). The solid red line corresponds to the mean background noise while the dotted red line corresponds to 3 standard deviations from the mean noise level. The reaction buffer was modified as 10 mM Tris (pH 7.0), 140 mM NaCl, 10 mM KCl and 5 mM MgCl2 to maintain good aptamer-thrombin binding.
The final sequences designed function independently of the specific target(s) and can be used directly as split proximity probes upon conjugation to desired recognition moieties, e.g. antibody, peptide and aptamers. The key concepts introduced in this work, i.e. inter-domain bridging and 3′-truncation as the main source of defect in solid-state oligonucleotide synthesis, are applicable to any DNA circuits in general. In particular, the novel inter-domain bridging concept improved both the migration kinetics and equilibrium signal while suppressing asymptotic leakage, which was essentially a triple-win design strategy. At the same time, we acknowledge that most biomolecular interactions are weaker than the streptavidin-biotin system presented in this work and are currently working towards implementing the final circuit design on actual biological systems. The use of autonomous DNA circuits is gaining momentum for performing complex sensing applications, e.g. cell surface profiling (36,37), detection of receptor interactions (7) and protein activity regulation (38). We are hopeful that our optimized split proximity design can contribute along this direction as a simple yet robust one-pot signal transduction strategy for modular adaptation in other related DNA circuits.
CONCLUSION
In conclusion, a simple DNA split proximity circuit was engineered through a systematic approach aimed primarily at minimizing circuit leakages. A novel concept of inter-domain bridging was introduced which enhanced strand displacement across the split domains. The final design with minimized circuit leakage was applied on a model streptavidin-biotin system which returned decent performance in terms of sensitivity and selectivity even in a biological environment. Other target recognition moieties, e.g. antibodies, peptides and aptamers, can be incorporated through chemical modifications to probe for diverse biomolecular interactions, as demonstrated using thrombin target. We are working towards using this general signal transduction strategy for evaluating actual biological proximity events, e.g. protein complexes and host–cell interactions.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Millennium Foundation and the Singapore Ministry of Education Academic Research Fund Tier 1; National University of Singapore and Ministry of Education for the President Graduate Fellowship (Y.S.A.). Funding for open access charge: Singapore Ministry of Education Academic Research Fund Tier 1.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fukuyama S., Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr. Opin. Immunol. 2011;23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legrain P., Rain J.-C. Twenty years of protein interaction studies for biological function deciphering. J. Proteomics. 2014;107:93–97. doi: 10.1016/j.jprot.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 4.Berggård T., Linse S., James P. Methods for the detection and analysis of protein–protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 5.Koussa M.A., Halvorsen K., Ward A., Wong W.P. DNA nanoswitches: a quantitative platform for gel-based biomolecular interaction analysis. Nat. Methods. 2015;12:123–126. doi: 10.1038/nmeth.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 7.Koos B., Cane G., Grannas K., Lof L., Arngarden L., Heldin J., Clausson C.-M., Klaesson A., Hirvonen M.K., de Oliveira F.M.S. Proximity-dependent initiation of hybridization chain reaction. Nat. Commun. 2015;6:7294. doi: 10.1038/ncomms8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang Y.S., Yung L.Y.L. Engineering self-contained DNA circuit for proximity recognition and localized signal amplification of target biomolecules. Nucleic Acids Res. 2014;42:9523–9530. doi: 10.1093/nar/gku655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou W., Jimmy Huang P.-J., Ding J., Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- 10.Morell M., Ventura S., Avilés F.X. Protein complementation assays: approaches for the in vivo analysis of protein interactions. FEBS Lett. 2009;583:1684–1691. doi: 10.1016/j.febslet.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Stynen B., Tournu H., Tavernier J., Van Dijck P. Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 2012;76:331–382. doi: 10.1128/MMBR.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter J.R., Stains C.I., Jester B.W., Ghosh I. A general and rapid cell-free approach for the interrogation of protein−protein, protein−DNA, and protein−RNA interactions and their antagonists utilizing split-protein reporters. J. Am. Chem. Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 13.Genot A.J., Zhang D.Y., Bath J., Turberfield A.J. Remote toehold: a mechanism for flexible control of DNA hybridization kinetics. J. Am. Chem. Soc. 2011;133:2177–2182. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- 14.Li F., Zhang H., Wang Z., Li X., Li X.-F., Le X.C. Dynamic DNA assemblies mediated by binding-induced DNA strand displacement. J. Am. Chem. Soc. 2013;135:2443–2446. doi: 10.1021/ja311990w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teichmann M., Kopperger E., Simmel F.C. Robustness of localized DNA strand displacement cascades. ACS Nano. 2014;8:8487–8496. doi: 10.1021/nn503073p. [DOI] [PubMed] [Google Scholar]
- 16.Chen X. Expanding the rule set of DNA circuitry with associative toehold activation. J. Am. Chem. Soc. 2012;134:263–271. doi: 10.1021/ja206690a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F., Lin Y., Le X.C. Binding-induced formation of DNA three-way junctions and its application to protein detection and DNA strand displacement. Anal. Chem. 2013;85:10835–10841. doi: 10.1021/ac402179a. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y., Lin Y., Yang X., Wang Z., Le X.C., Li F. Universal strategy to engineer catalytic DNA hairpin assemblies for protein analysis. Anal. Chem. 2015;87:8063–8066. doi: 10.1021/acs.analchem.5b02504. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y., Wang Z., Yang X., Chen J., Liu L., Zhao W., Le X.C., Li F. Constructing real-time, wash-free, and reiterative sensors for cell surface proteins using binding-induced dynamic DNA assembly. Chem. Sci. 2015;6:5729–5733. doi: 10.1039/c5sc01870f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi H.M.T., Beck V.A., Pierce N.A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B., Ellington A.D., Chen X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D.Y., Turberfield A.J., Yurke B., Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Briggs N., McLain J.R., Ellington A.D. Stacking nonenzymatic circuits for high signal gain. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5386–5391. doi: 10.1073/pnas.1222807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machinek R.R.F., Ouldridge T.E., Haley N.E.C., Bath J., Turberfield A.J. Programmable energy landscapes for kinetic control of DNA strand displacement. Nat. Commun. 2014;5:5324. doi: 10.1038/ncomms6324. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D.Y., Winfree E. Robustness and modularity properties of a non-covalent DNA catalytic reaction. Nucleic Acids Res. 2010;38:4182–4197. doi: 10.1093/nar/gkq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 27.Ang Y.S., Yung L.Y.L. Rational design of hybridization chain reaction monomers for robust signal amplification. Chem. Commun. 2016;52:4219–4222. doi: 10.1039/c5cc08907g. [DOI] [PubMed] [Google Scholar]
- 28.Dirks R.M., Pierce N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B., Jiang Y., Chen X., Ellington A.D. Probing spatial organization of DNA strands using enzyme-free hairpin assembly circuits. J. Am. Chem. Soc. 2012;134:13918–13921. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temsamani J., Kubert M., Agrawal S. Sequence identity of the n-1 product of a synthetic oligonucleotide. Nucleic Acids Res. 1995;23:1841–1844. doi: 10.1093/nar/23.11.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Y.S., Bhadra S., Li B., Ellington A.D. Mismatches improve the performance of strand-displacement nucleic acid circuits. Angew. Chem. Int. Ed. 2014;53:1845–1848. doi: 10.1002/anie.201307418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis M.E., Davis R.J. Fundamentals of Chemical Reaction Engineering. Boston: McGraw-Hill; 2003. [Google Scholar]
- 33.Zhang D.Y., Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 34.Zhang D.Y., Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 35.Deng B., Lin Y., Wang C., Li F., Wang Z., Zhang H., Li X.-F., Le X.C. Aptamer binding assays for proteins: the thrombin example—a review. Anal. Chim. Acta. 2014;837:1–15. doi: 10.1016/j.aca.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 36.Rudchenko M., Taylor S., Pallavi P., Dechkovskaia A., Khan S., Butler V.P., Jr, Rudchenko S., Stojanovic M.N. Autonomous molecular cascades for evaluation of cell surfaces. Nat. Nanotechnol. 2013;8:580–586. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu G., Zhang S., Song E., Zheng J., Hu R., Fang X., Tan W. Building fluorescent DNA nanodevices on target living cell surfaces. Angew. Chem. Int. Ed. 2013;52:5490–5496. doi: 10.1002/anie.201301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han D., Zhu Z., Wu C., Peng L., Zhou L., Gulbakan B., Zhu G., Williams K.R., Tan W. A logical molecular circuit for programmable and autonomous regulation of protein activity using DNA aptamer–protein interactions. J. Am. Chem. Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.