Abstract
Splice-switching oligonucleotides (SSOs) are short, synthetic, antisense, modified nucleic acids that base-pair with a pre-mRNA and disrupt the normal splicing repertoire of the transcript by blocking the RNA–RNA base-pairing or protein–RNA binding interactions that occur between components of the splicing machinery and the pre-mRNA. Splicing of pre-mRNA is required for the proper expression of the vast majority of protein-coding genes, and thus, targeting the process offers a means to manipulate protein production from a gene. Splicing modulation is particularly valuable in cases of disease caused by mutations that lead to disruption of normal splicing or when interfering with the normal splicing process of a gene transcript may be therapeutic. SSOs offer an effective and specific way to target and alter splicing in a therapeutic manner. Here, we discuss the different approaches used to target and alter pre-mRNA splicing with SSOs. We detail the modifications to the nucleic acids that make them promising therapeutics and discuss the challenges to creating effective SSO drugs. We highlight the development of SSOs designed to treat Duchenne muscular dystrophy and spinal muscular atrophy, which are currently being tested in clinical trials.
OVERVIEW
Pre-mRNA splicing
Most protein-coding genes are comprised of coding sequences that are interspersed with non-coding sequences. Following gene transcription, these intervening, non-coding RNA sequences, called introns, are removed and the coding RNA sequences, called exons, are ligated together in a process called pre-mRNA splicing. This splicing gives rise to the final mRNA that is translated into a protein (1). Splicing of each intron involves two sequential trans-esterification reactions. The first reaction releases the 5′ exon from the downstream intronic sequence, which forms a lariat structure through an interaction with the branchpoint sequence at the 3′ end of the intron (1). The second reaction releases the lariat from the downstream, 3′ exon and ligates together the 5′ and 3′ exons. Pre-mRNA splicing requires precision and accuracy in order to ensure that the proper open reading frame is maintained for efficacious protein production during translation. This high fidelity is achieved, in large part, by sequences and structures within the RNA transcript that direct the binding of splicing proteins that aid in positioning the RNA in a manner that facilitates the correct cleavage and ligation reactions of splicing (2,3). These cleavage reactions occur at conserved sequences called the 5′ splice site at the 5′ end of an intron and the 3′ splice site at the 3′ end of an intron. The splice sites are recognized through interactions with a multi-megadalton ribonucleoprotein complex called the spliceosome (4,5). The spliceosome consists of small nuclear RNAs (snRNAs), which form specific RNA–RNA base-pairs with the splice sites, and proteins, all of which function together to direct the spliceosome to the splice sites and position the RNA for the catalytic steps of splicing (1).
In principle, a 5′ splice site can splice to any 3′ splice site, and the rules that determine the pairing of sites are not entirely clear. It is clear, however, that there can be variability in this process, giving rise to alternative splicing events (6). Indeed, transcriptome sequencing has revealed that splicing of pre-mRNA from most protein-coding genes can occur in a variety of different patterns, giving rise to multiple alternatively spliced isoforms from a single gene (7,8). The regulation of alternative splicing is directed in large part by differential protein binding to cis-acting sequences in the pre-mRNA transcript (6,9). These splicing factor proteins, depending on their function and the location of their binding sites relative to other splicing signals, can either promote or inhibit splicing at a particular site (Figure 1). In the most general terms, a splicing enhancer is defined as a sequence element that, when bound by its cognate protein, promotes the splicing of a nearby exon. In contrast, a splicing silencer is a sequence element that, when bound by its cognate protein, blocks or inhibits splicing at a particular site (Figure 1) (6,10). Splicing silencer and enhancer sequences are further defined by their location in either an exon (e.g. exonic splicing silencer or enhancer) or intron (e.g. intronic splicing silencer or enhancer). The proteins that bind splicing enhancer and silencer elements typically bind in a sequence-specific manner to single-stranded RNA. RNA secondary structures and chromatin structures can also act to influence alternative splicing (11). Much work has been devoted to understanding the splicing code, which aims to explain alternative splicing patterns by the location of cis-acting splicing element sequences, their trans-acting binding proteins, the interactions of these mRNA:protein complexes (mRNP) with surrounding mRNPs and their activity in repressing or enhancing splicing (12,13). An important component of the splicing code is the ability to predict alternative splicing regulation in a tissue-, cell-, condition- and developmental-specific manner and also to predict mutations and other sequence variations that disrupt normal splicing and potentially cause human pathological conditions (14–16). Understanding the splicing code is an important step toward designing strategies for manipulating and switching splicing in a predictable and potentially therapeutic manner.
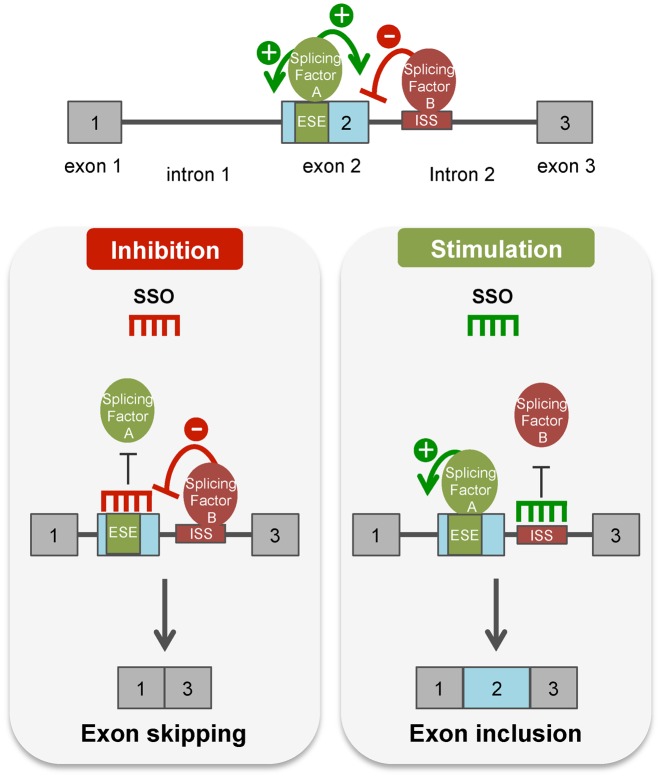
Figure 1.
Splice-switching oligonucleotides (SSOs) modulate alternative splicing. (top) Diagram of a pre-mRNA transcript with exons depicted as gray boxes and introns as lines. An intronic splicing silencer (ISS, red) and exonic splicing enhancer (ESE, green) are shown bound by a trans-acting inhibitory splicing factor protein (red oval) or stimulatory splicing factor (green oval). These SF proteins either block (−) or promote (+) splicing at splice sites bordering the surrounding exons. (left panel) An SSO that base-pairs to a splicing enhancer sequence creates a steric block to the binding of the stimulatory splicing factor to its cognate enhancer binding site. This block thereby disrupts splicing and results in exon skipping. (right panel) In contrast, an SSO that base-pairs to a splicing silencer sequence element blocks splicing silencer activity by preventing binding of a negatively acting splicing factor. Disruption of the binding of splicing inhibitory proteins to its cognate binding sequence activates splicing at the splice site that is negatively regulated by the silencer element, resulting in exon inclusion.
Alternative splicing expands the diversity of the human proteome and thereby has been hypothesized to contribute to organismal complexity (17,18). For example, though the mouse and human genomes have a similar number of genes, alternative splicing has been estimated to occur in 95–100% of human genes but only 63% of mouse genes (7,8). The prominence of alternative splicing likely explains a certain amount of the functional differences between cell types and suggests that cells can tolerate different isoforms of mRNA and proteins. At the same time, many genetic diseases result from mutations that either cause splicing abnormalities, other errors that alter canonical splicing or reading frameshifts (19–21). Indeed, a recent computational study predicted that the number of mutations that cause disease due to the disruption of splicing is far greater than previously appreciated (14). Given the centrality of splicing in gene expression and its prevalent deregulation in disease, there has been interest in identifying drugs that can specifically modulate splicing in ways that may work to treat disease symptoms. To this end, small molecule discovery and other approaches are being pursued as splice-targeting therapeutics. One particularly promising approach to specifically manipulate splicing at any given site involves the use of short antisense oligonucleotides (ASOs) that base-pair in an antisense orientation to a specific pre-mRNA sequence and, in so doing, modulate splicing by interfering with the normal protein:RNA or RNA:RNA interactions that direct splicing. ASOs that specifically target splicing are referred to here as splice-switching antisense oligonucleotides (SSOs).
Splice-switching antisense oligonucleotides (SSOs)
ASOs are synthetic molecules comprised of nucleotides or nucleotide analogues that bind to a complementary sequence through Watson–Crick base-pairing. Although all ASO approaches make use of short nucleic acids that specifically base-pair to a targeted sequence, the outcome of such base-pairing depends on the chemistry of the oligonucleotide and the binding location. SSOs are ASOs that are typically 15–30 nucleotides long and designed to base-pair and create a steric block to the binding of splicing factors to the pre-mRNA. In this way, SSO base-pairing to a target RNA alters the recognition of splice sites by the spliceosome, which leads to an alteration of normal splicing of the targeted transcript (Figure 1). Importantly, nucleotides of an SSO are chemically modified so that the RNA-cleaving enzyme RNase H is not recruited to degrade the pre-mRNA-SSO complex (22,23). Thus, SSOs modify splicing without necessarily altering the abundance of the mRNA transcript. The RNAse H-resistant features of SSOs are critical because the goal of SSOs is to alter splicing and not to cause the degradation of the bound pre-mRNA. Modifications to the SSO have also been crucial to stabilize the SSO in vivo and improve cellular uptake and release as well as binding affinity.
Common chemical modifications of splice-switching oligonucleotides
Key breakthroughs in the chemical design of antisense oligonucleotides have been instrumental in making SSOs a viable therapeutic approach. Natural, unmodified DNA and RNA oligonucleotides are generally unfavorable as therapeutics because they are vulnerable to nuclease degradation in serum and cells and thereby are unstable in vivo. Chemical modifications have improved oligonucleotide binding affinity, stability and pharmacodynamic properties. Modifications that improve these qualities involve changes to the phosphate backbone and/or sugar component of the oligonucleotide. These medicinal chemistry efforts have been comprehensively reviewed recently (24–26) and thus, here, we focus our discussion on the specific modifications that have been utilized in the development of SSOs that have shown promise in vivo in the treatment of disease/pathological conditions (Table 1).
Table 1. Splice-switching antisense oligonucleotides with activity in vivo. Examples of the most advanced SSO for each target are represented.
| Condition | Target gene | Stage/Model | SSO | Target (Action) | Route | Ref |
|---|---|---|---|---|---|---|
| Block cryptic/Aberrant splicing caused by mutations | ||||||
| β-Thalassemia | HBB | mouse | PPMO | intron 2 aberrant 5'ss (correct splicing) | IV | (144) |
| Fukuyama congenital muscular dystrophy | FKTN | mouse | VPMO | exon 10 aberrant 3'ss; alternative 5'ss; ESE (correct splicing) | IM | (145) |
| Hutchinson–Gilford progeria | LMNA | mouse | VPMO; 2′-MOE /PS | exon 10 5'ss; exon 11 cryptic 5'ss; exon 11 ESE (block exon 11 splicing) | IV/IP | (146,147) |
| Leber congenital amaurosis | CEP290 | mouse | 2′-OMe /PS; AAV | Intron 26 cryptic exon (correct splicing) | IVI | (56) |
| Myotonic dystrophy | CLCN1 | mouse | PMO | exon 7a 3'ss (exon 7a skipping) | IM | (53,148) |
| Usher syndrome | USH1C | mouse | 2′-MOE /PS | exon 3 cryptic 5'ss (correct splicing) | IP | (40) |
| X-linked agammaglobulinemia | BTK | mouse | PPMO | pseudoexon 4A ESS (pseudoexon skipping) | IV/SC | (149) |
| Switch alternative splicing | ||||||
| Alzheimer's disease | LRP8 | mouse | 2′-MOE /PS | intron 19 ISS (exon 19 inclusion) | ICV | (42) |
| Autoimmune diabetes susceptibility | CTLA4 | mouse | PPMO | exon 2 3'ss (exon skipping) | IP | (150) |
| Cancer | BCL2L1 | mouse | 2′-MOE /PS | exon 2 5'ss (alternative 5'ss) | IV/NP | (151) |
| Cancer | ERBB4 | mouse | LNA | exon 26 5'ss (exon skipping) | IP | (152) |
| Cancer | MDM4 | mouse | PMO | exon 6 5'ss (exon skipping) | ITM | (153) |
| Cancer | STAT3 | mouse | VPMO | exon 23 α 3'ss (β 3'ss use) | ITM | (154) |
| Inflammation | IL1RAP | mouse | 2-OMe /PS;LNA | exon 9 ESE (exon skipping) | IV/NP | (155) |
| Inflammation | TNFRSF1B | mouse | LNA /PS | exon 7 5'ss (exon skipping) | IP | (156) |
| Neovascularization | FLT1 | mouse | PMO | exon 13 5'ss (alternative pA site) | IVI / ITM | (157) |
| Neovascularization | KDR | mouse | PMO | exon 13 5'ss (alternative pA site) | IVI / SCJ | (158) |
| Spinal muscular atrophy | SMN2 | clinical trials | 2′-MOE /PS | intron 7 ISS (exon 7 inclusion) | IT | (43,142) |
| Correct open reading frame | ||||||
| cardiomyopathy | MYBPC3 | mouse | AAV | Exon 5 and 6 ESEs (exon 5, 6 skipping) | IV | (159) |
| Cardiomyopathy | TTN | mouse | VPMO | exon 326 ESE (exon skipping) | IP | (160) |
| Duchenne muscular dystrophy (DMD) | DMD | clinical trials | 2′-OMe / PMO | exon 51 ESE (exon skipping) | IV/SC | (46,98) |
| Nijmegen breakage syndrome | NBN | mouse | VPMO | exon 6/7 ESEs (exon skipping) | IV | (161) |
| Disrupt open reading frame/Protein function | ||||||
| Ebola | IL10 | mouse | PPMO | exon 4 3'ss (exon skipping) | IP | (162) |
| Huntington disease | HTT | mouse | 2′-OMe /PS | exon 12 skipping | IS | (163) |
| Hypercholesterolemia | APOB | mouse | 2′-OMe /PS | exon 27 3'ss (exon skipping) | IV | (164) |
| Muscle-Wasting/DMD | MSTN | mouse | PPMO/VPMO/ 2′-OMe | exon 2 ESE (exon skipping) | IV/ IM/ IP | (165,166) |
| Pompe disease | GYS2 | mouse | PPMO | exon 6 5'ss (exon skipping) | IM/IV | (167) |
| Spinocerebellar ataxia type 3 | ATXN3 | mouse | 2′-OMe /PS | exon 9, 10 skipping | ICV | (168) |
AAV (Adeno-associated viral expression of SSO); NP (nano-particle); PPMO (peptide-conjugated phosphorodiamidate morpholino); VPMO (Vivo-PMO).
ICV (intracerebroventricular); IM (intramuscular); IP (intraperitoneal); IS (intrastriatal); IT (intrathecal); ITM (intratumoral); IV (intravenous); IVI (intravitreal); SC (subcutaneous); SCJ (subconjunctival); ISE (Intronic splicing enhancer); ESE (Exonic splicing enhancer); ISS (intronic splicing silencer); pA (polyadenylation).
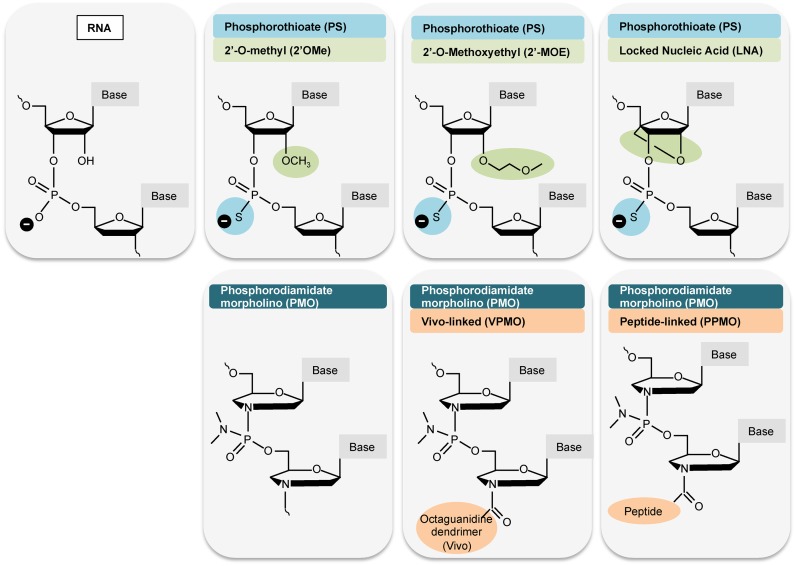
The phosphorothiate (PS) backbone modification was the first analog to be used in clinical applications and has been incorporated into many SSO designs that are currently being developed as potential therapeutics (Table 1, Figure 2) (27,28). SSOs with a PS backbone modification have modestly reduced binding affinities but have improved stability in vivo, with greater nuclease resistance (28). PS ASOs also bind to proteins in plasma, which reduces renal clearance and improves retention, allowing for broad biodistribution, but also increasing the risk of toxicity (28,29).
Figure 2.
Structures of oligonucleotide analogs commonly used in splice switching applications in vivo. Modifications that are used in the SSOs presented in Table 1 are depicted. Unmodified RNA is shown for reference. Base refers to unmodified adenine, cytosine, guanine or uracil.
Oligonucleotides with PS backbone modifications are not resistant to RNAse H and thus, to create a steric blocking SSO for splice-switching applications, additional modifications to the molecule are required. ASOs that are fully modified at the 2′ sugar position confer RNAse H-resistance and are commonly used as SSOs. The most widely used alterations at the 2′ position are 2′-O-methyl (2′-OMe) and 2′-O-methoxyethyl (2′-MOE) (Figure 2). Locked nucleic acid (LNA) chemistry, is another modification of the sugar, which involves bridging of the furanose ring (Figure 2) (30). A major benefit of LNA modified SSOs is the elevated binding affinity, which is an important consideration as high binding affinity can allow for the use of shorter SSO sequences. A shorter sequence can reduce the likelihood of binding to an incorrect site as a result of partial sequence complementarity to another sequence and thus lower the risk of unwanted off-target effects. Each of these modifications have been used together (2′-OMe/PS, 2′-MOE/PS; LNA/PS) to successfully target splicing in ways predicted to be therapeutic for a number of different pathological conditions (Table 1).
Phosphorodiamidate morpholinos (PMOs) are another type of modified oligonucleotide that has been used extensively to modify splicing. PMOs have a morpholine ring in place of the furanose ring found in natural nucleic acids and a neutral phosphorodiamidate backbone in place of the negatively charged phosphodiester backbone (Figure 2) (22). The neutral charge of PMOs results in their low binding of plasma proteins, which improves tolerability in vivo. However, they are also rapidly cleared by the kidney and for this reason exhibit lower accumulation in tissues compared to a charged PS backbone (29). As a result, high doses of PMOs may be necessary to elicit a pharmacological response (31,32). A number of approaches have been developed to improve PMO efficacy in vivo, as discussed below and shown in Figure 2.
Delivery routes and mechanisms
As the ultimate aim of most of the SSO drug designs is to treat a human condition, efficient delivery to cells in the body is imperative. Recent reviews have provided extensive details on the biological basis of ASO access to cells and tissues as well as approaches that are being used to enhance the delivery of ASOs in vivo (26,33,34). Thus, here, we highlight some of the approaches that have been used successfully for the delivery of SSOs in vivo to alleviate disease phenotypes (Table 1).
A number of different delivery paradigms have been utilized to deliver SSOs to cells in vivo, including intraperitoneal (IP), subcutaneous (SC) or intravenous (IV) administration. These methods result in exposure of many peripheral tissues to the oligonucleotide (35–37). Other approaches, such as intramuscular (IM), intratumoral (ITM), subconjunctival (SCJ) or intravitreal (IVI) injection of ASOs have been used to achieve more tissue-specific delivery (Table 1). ASOs do not readily cross the blood brain barrier when administered peripherally (29). However, for therapeutics intended for CNS applications and targets, direct delivery to the cerebrospinal fluid (CSF) by either intracerebroventricular (ICV) or intrathecal (IT) administration has been shown to result in therapeutic doses of SSOs throughout the CNS, though deeper brain regions are more challenging to access (32,38).
The pharmacokinetics, pharmacodynamics and other considerations of central and peripheral administration of ASOs have been expertly reviewed recently (26,39) and we highlight here only a few key points. The effects of a single injection of SSOs on splicing and/or disease have been found to last for up to a year in some tissues when delivered peripherally (40,41) or centrally (32,41,42). This persistent effect of SSOs suggests injections could be minimized. Nonetheless, depending on the condition, SSO therapeutics will likely require repeated dosing. While a repeat dosing regimen may not be a major drawback for the treatment of peripheral tissue, CNS delivery is more invasive and holds greater risk, though repeated IT injections of SSOs in pediatric patients have been shown to be well-tolerated (43). Because of the relative ease of peripheral delivery compared to direct delivery to the CNS, there have been efforts to develop SSO-conjugates that can cross the blood brain barrier (26). However, direct central application has some advantages over peripheral, systemic delivery in that it may allow for lower doses due to the tissue-specific delivery to the CNS and could minimize side effects associated with systemic delivery such as hepatotoxicity (44). Overall, although undoubtedly more invasive and technically challenging than peripheral dosing, a lower dose requirement and less frequent dosing compared to peripheral treatments could lower the amount of drug required for treatment and consequently drug-associated costs. The benefits of limiting systemic drug exposure by utilizing tissue-specific ASO delivery approaches apply to other tissues as well, including diseases of the eye, where intravitreal, and subconjunctival delivery have been efficacious in model systems (Table 1).
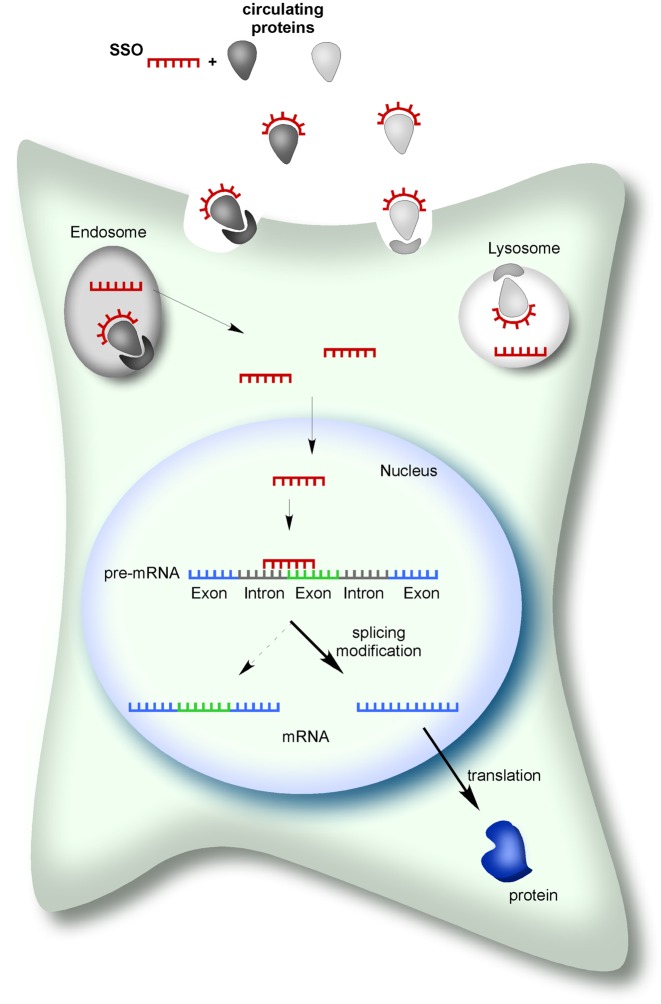
Once injected, ASOs can gain access to cells in vivo as naked/unformulated oligonucleotides. ASOs with a charged PS backbone are bound by high and low affinity circulating proteins in the plasma (Figure 3). Protein-binding is thought to be mediated, in part, the up-take of ASOs into cells via vesicular pathways that can either deliver ASOs to lysosomes or release them directly into the cytoplasm through a mechanism that is not well-understood (45) (Figure 3). In contrast, charge-neutral SSOs such as PMOs have reduced protein-binding properties compared to PS backbone-modified ASOs. Nonetheless, studies have demonstrated efficient, non-toxic in vivo delivery of both PS-modified SSOs and PMOs by direct injection of the naked oligomer (32,46,47). Once in the cytoplasm, ASOs can move into the nucleus to affect pre-mRNA splicing (48) (Figure 3). Upon entry into cells in vivo, ASOs have a long duration of action. SSOs have been shown to affect splicing and disease symptoms for up to a year after a single ICV (32,36) or IP (40) administration in mice.
Figure 3.
Splice-switching antisense oligonucleotides (SSOs) mechanism of action. SSOs can gain entry into cells in vivo following injection of a naked/unformulated ASO into the blood or cerebrospinal fluid. SSOs can be bound by circulating proteins and have been proposed to enter into cells by binding to receptors for these proteins on the cell surface. Subsequently, SSOs undergo compartmentalization followed by vesicle release at which point they are free to move into the nucleus, bind pre-mRNA and induce a splicing switch that results in an mRNA that is translated into a protein isoform in the cytoplasm.
Although SSOs can be delivered as naked oligonucleotides, modifications, carriers and other approaches offer opportunities to increase efficiency, lower doses and increase tissue-specific delivery, all of which can help to limit toxicity and off-target effects. For example, a number of approaches have been developed to improve PMO efficacy in vivo, including modifications of the 2′ position of the sugar with cell-penetrating peptides, or octaguanidine dendrimers (Vivo-morpholinos), which enhance cellular uptake and endosomal release (Table 1, Figure 2) (49–51). Co-administration of PMOs with different agents such as hexose (52), bubble liposomes (53,54) and F127 copolymer (55) have also shown some promise in improving PMO activity in vivo. Adeno-associated virus (AAV)-packaged SSO has also been explored for the treatment of disease (Table 1). Though AAV-mediated gene therapy itself is a major therapeutic platform under development for the treatment of a number of diseases, the approach is constrained by cargo size limitations, which makes them less valuable as a therapeutic option for genes with coding sequences greater than 5 kb. A recent study has shown similar efficacy of naked SSOs and AAV-packaged SSOs in vivo (56). However, AAV-mediated expression of SSOs may offer advantages of improved delivery to specific cell types and the potential for a longer lasting effect.
SSO STRATEGIES TO THERAPEUTICALLY MANIPULATE GENE EXPRESSION
SSOs have many of the key attributes that make an ideal drug. They are relatively easy to synthesize and deliver, as they do not strictly require packaging or other delivery intermediates. They are highly target-specific due to their base-pairing requirements, and they exhibit widespread entry into most cell types in the body (29,50,57). SSOs are also well tolerated, particularly in the CNS, and they have a long-lasting effect in vivo (23). Another important feature of SSOs is that they can be easily designed to have any number of different effects on the expression of a gene by either inhibiting or enhancing the use of a specific splice site (20).
Inhibition of splicing using SSOs can be achieved by targeting the molecule to base-pair at a splice site, which will interfere with splicing protein interactions at the location, which is a key step for splicing catalysis. SSO basepairing at a splicing enhancer sequence can also cause splicing inhibition at a particular splice site (Figure 1). Inhibition of a splice site by an SSO offers a way to block a cryptic splice site that is created by a genetic mutation or to switch or modulate alternative splicing patterns in a manner that are predicted to be therapeutic. Inhibition of splicing at a particular site can also be utilized to restore the reading frame of an mRNA by skipping out an exon that either has a premature termination codon created by a mutation or a deletion resulting in a frameshift. In these cases, the exclusion of an exon from the mRNA restores the mRNA reading frame, albeit producing a shortened version of the protein with an internal deletion. SSO-induced exon skipping can also be used to create a frame-shift in an mRNA in order to down-regulate protein production from a gene or to eliminate unwanted or pathological sequences from a protein. Each of these strategies is being pursued for the treatment of disease and has been shown to be efficacious in vivo for a number of different disease models and we present examples of some of the more advanced studies with SSOs for specific genes in Table 1.
SSOs can also be designed to activate or enhance splicing at a particular site. For this, SSOs are often targeted to basepair at a cis-acting splicing silencer sequence, thereby blocking the binding of the associated trans-acting inhibitory protein factor (Figure 1). In this way, SSOs can be used to promote splicing at a splice site that has been weakened by a mutation in the region of the splice site sequence itself or by a mutation in a splicing enhancer sequence that promotes the normal use of the site. SSOs can also be designed to disrupt RNA secondary structures, which can function to either enhance or inhibit splicing (58). Though not a focus of this review, ASOs can also be designed to target other RNA processing events in the pathway to mRNA maturation such as polyadenylation (59).
The first demonstration that ASOs could be used to target splicing came from studies of a thalassemia-associated defect in splicing caused by a mutation in the human β-globin gene that creates a cryptic 5′ splice site, which is used preferentially over the natural site (60). In this study, Dominski and Kole demonstrated that a 2′-OMe SSO, designed to base-pair to the region encompassing the cryptic splice site, blocks splicing at the site and redirects splicing to the correct splice site. Since these early studies, targeting splicing with SSOs has been used as a tool to identify cis-acting splicing elements and to modify splicing in ways that are designed to be therapeutic in disease (20). Advances in SSO-based therapeutics and their comparisons to other therapeutic platforms have been extensively reviewed (23,26,39,61–67). Many SSO strategies have now been demonstrated to be effective in modulating splicing in animal models of human disease and some have entered clinical trials (Table 1). Results from tests of splice-switching ASOs in humans were first reported in 2007 for the treatment of Duchenne Muscular Dystrophy (DMD) (65). The most advanced SSOs are now in Phase 3 clinical trials for the treatment of DMD and another pediatric genetic disorder, Spinal Muscular Atrophy (SMA) (Tables 1 and 2).
Table 2. Clinical trials for Eteplirsen, Kyndrisa and Nusinersen.
| Trial number | Start | End | Age | n | Status | Additional information | Design* | Phase |
|---|---|---|---|---|---|---|---|---|
| Eteplirsen™- Duchenne Muscular Dystrophy | ||||||||
| NCT00159250 | Oct ‘07 | Mar ‘09 | 10–17 yr | 7 | Completed | non-ambulatory | single blind | 1/2 |
| NCT00844597 | Jan ‘09 | Dec ‘10 | 5–15 yr | 19 | Completed | 25 m unaided walk | open label | 1/2 |
| NCT01396239 | Jul ‘11 | Jun ‘12 | 7–13 yr | 12 | Completed | 200–400 m 6MWD | placebo control | 2 |
| NCT01540409 | Feb ‘12 | Sep ‘16 | 7–13 yr | 12 | Active | 01396239 extension | open label | 2 |
| NCT02255552 | Sep ‘14 | May ‘19 | 7–16 yr | 160 | Recruiting | >300 m 6MWD | open label, untreated control | 3 |
| NCT02286947 | Oct ‘14 | Sep ‘17 | 7–21 yr | 20 | Active | non-ambulatory or ≤300m 6MWD | open label | 2 |
| NCT02420379 | Mar ‘15 | Feb ‘18 | 4–6 yr | 40 | Recruiting | open label | 2 | |
| Kyndrisa™- Duchenne Muscular Dystrophy | ||||||||
| NCT01910649 | Mar ‘08 | Dec ‘16 | 5–16 yr | 12 | Terminated | open label | 1/2 | |
| NCT01128855 | Jul ‘10 | Oct ‘11 | ≥9 yr | 20 | Completed | non-ambulatory | placebo control | 1 |
| NCT01153932 | Sep ‘10 | Sep ‘12 | ≥5 yr | 53 | Completed | >75 m 6MWD | placebo control | 2 |
| NCT01254019 | Dec ‘10 | Jun ‘13 | ≥5 yr | 186 | Completed | >75 m 6MWD | placebo control | 3 |
| NCT01480245 | Sep ‘11 | Mar ‘14 | ≥5 yr | 233 | Terminated | 01254019, 01153932 extension | open label | 3 |
| NCT01462292 | Oct ‘11 | Nov ‘13 | ≥5 yr | 51 | Completed | >75 m 6MWD | placebo control | 2 |
| NCT01803412 | May ‘13 | Jun ‘17 | ≥5 yr | 67 | Terminated | 01480245, 01462292, 01254019 extension | open label | 3 |
| NCT02636686 | Dec ‘15 | Jan ‘18 | 5–80 yr | 220 | Terminated | extension, ineligible for other trials | open label | 3 |
| Nusinersen™- Spinal Muscular Atrophy | ||||||||
| NCT01494701 (CS1) | Nov ‘11 | Jan ‘13 | 2–14 yr | 28 | Completed | Type 2/3 SMA | open label | 1 |
| NCT01703988 (CS2) | Oct ‘12 | Jan ‘15 | 2–15 yr | 34 | Completed | Type 2/3 SMA | open label | 1/2 |
| NCT01780246 (CS10) | Jan ‘13 | Feb ‘14 | 2–15 yr | 18 | Completed | CS1 extension | open label | 1 |
| NCT01839656 | May ‘13 | Nov ‘16 | 0–210 d | 20 | Active | Type 1 SMA | open label | 2 |
| NCT02052791 (CS12) | Jan ‘14 | Jan ‘17 | Any | 52 | Active | CS2, CS10 extension | open label | 1 |
| NCT02193074 (ENDEAR) | Jul ‘14 | Jul ‘17 | 0–210 d | 111 | Recruiting | Type 1 SMA | sham control | 3 |
| NCT02292537 (CHERISH) | Nov ‘14 | Jun ‘17 | 2–12 yr | 117 | Active | Type 2 SMA, non-ambulatory | sham control | 3 |
| NCT02386553 (NURTURE) | May ‘15 | Apr ‘20 | 0–6 wk | 25 | Recruiting | Pre-symptomatic, Type I SMA | open label | 2 |
| NCT02462759 (EMBRACE) | Jun ‘15 | Oct ‘17 | Any | 21 | Active | ineligible for ENDEAR or CHERISH | sham control | 2 |
| NCT02594124 (SHINE) | Nov ‘15 | Feb ‘20 | 13 mo–21 yr | 274 | Active | ENDEAR, CHERISH, CS12 extension | open label | 3 |
*Unless otherwise noted, open label trials were non-randomized and non-controlled and placebo and sham-controlled studies were double-blind and randomized.
Extension indicates patients participated in a previous study. 6MWD, 6-min walk distance; Yr, years; Mo, months; wk, weeks; d, days.
SPLICE-MODIFYING ANTISENSE APPROACHES IN HUMANS
SSOs for the treatment of Duchenne Muscular Dystrophy (DMD)
DMD is an X-linked neuromuscular disorder that affects 1:5000-10 000 male births (68–70), and is caused by mutations in the DMD gene, which codes for dystrophin protein. Dystrophin is an important structural protein in muscle cells that anchors proteins from the internal cytoskeleton to those in the fiber membrane (71). Approximately 70% of DMD mutations are deletions of exons that disrupt the mRNA reading frame and create premature termination codons that produce truncated and usually non-functional dystrophin protein. This lack of functional dystrophin results in progressive muscle weakness beginning typically before the age of 6, followed by loss of ambulation by the age of 12. Death usually occurs in the second decade of life and results from complications related to failure of the respiratory muscles, though most patients also develop cardiomyopathy, which is the primary cause of death in up to 30% of patients (72).
Becker Muscular Dystrophy (BMD) is also caused by mutations in the DMD gene and has similar symptoms to DMD but with later onset and slower progression. This difference in phenotypes is related to the type of DMD mutation. DMD mutations resulting in BMD do not disrupt the reading frame and thus produce an altered dystrophin protein with sufficient functionality to ameliorate the severity of disease symptoms (73,74). This spectrum of disease severity, which is driven by the nature of the mutations, gave rise to the idea that inducing exon skipping to correct the reading frame of mRNA from mutated DMD could be an effective way to produce a BMD-type dystrophin protein that would partially compensate for the loss of full-length protein and ameliorate the symptoms of DMD (Figure 3). SSOs targeting different exons would allow reading frame correction of over 50% of deletions and 22% of duplications reported in the Leiden DMD-mutation Database (http://www.dmd.nl/). In order for an SSO drug for DMD to treat a greater number of patients with different DMD mutations, the use of cocktails containing multiple different SSOs targeting different exons is also a possibility, and could allow for the correction of more than 90% of patient mutations by a frame-correcting exon skipping therapy (75,76).
One of the first demonstrations that SSOs could be used to modulate splicing of a DMD exon involved the use of an SSO to inhibit splicing of exon 19 from a minigene-derived pre-mRNA in an in vitro splicing assay (77). This study was followed by reports that SSO-induced exon skipping could be achieved in cells from a mouse model of DMD and in DMD patient-derived cells in culture (78,79). Subsequently, numerous groups tested 2′-OMe and PMO SSOs in mouse models of DMD, and found that exon skipping could be effectively induced by the different SSOs administered by intramuscular, subcutaneous or intravenous injection (55,80–82). Furthermore, weekly injections of SSO, for 7 weeks to 6 months, proved to be well tolerated and resulted in enough dystophin protein to improve muscle function in dystrophic mice (83,84). SSO cocktails aimed at skipping multiple exons for reading frame correction have also been tested and shown to be effective in mice (85) and dogs (31), an advancement that could benefit future clinical development in humans (76).
The promising results in animal models of DMD led to the initiation of clinical trials of SSOs in DMD patients. The first SSO to be tested in a clinical trial for the treatment of DMD was a 31-mer oligodeoxynucleotide phosphorothioate (DNA/PS), which was administered by intravenous injection (86). Though exon skipping and an increase in dystrophin was observed in muscle biopsy tissue, this DNA/PS SSO may have also made dystrophin RNA a substrate for RNAse H-targeted degradation decreasing its effectiveness. Today, SSOs targeting numerous DMD exons (8,35,43–45,50,52–55) are being developed as therapies. SSOs that induce skipping of exon 44 (PRO044, BMN 044), exon 45 (SRP-4045, BMN 045), exon 51 (Eteplirsen, Kyndrisa) and exon 53 (SRP-4053, BMN 053) have advanced to clinical trials http://investorrelations.sarepta.com/phoenix.zhtml?c=64231&p=irol-newsArticle&ID=2007537 (67,87,88) (Table 2). Skipping of exon 51 would benefit the most DMD patients, as ∼13% have a frame-shift that could be corrected by the exclusion of this exon from the mRNA (http://www.dmd.nl/) (89) (Figure 4). This exon was first tested as a target for SSO-mediated skipping in patient cells lines via retroviral expression of an exon 51 antisense sequence (90). Following a systematic study in human cells to evaluate the optimal chemistry and sequences for a DMD SSO (91), two SSO approaches were pursued. Both SSOs are designed to base-pair in the same DMD RNA target region but they have different lengths and chemical modifications. Both of these SSOs have been tested in clinical trials to treat DMD. Eteplirsen (AVI-4658) is being developed by Sarepta Therapeutics and Kyndrisa™ (Drisapersen/PRO051/GSK2402968), was pursued by Biomarin (formerly developed by Prosensa and GlaxoSmithKline) though has recently been withdrawn from development, as detailed below.
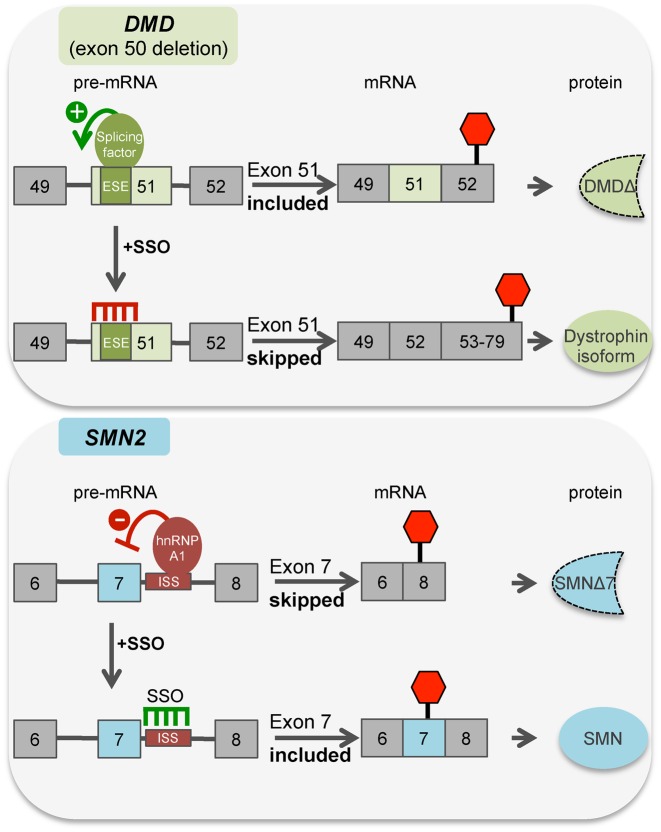
Figure 4.
Schematic representation of disease associated splicing in DMD (top panel) and SMA (bottom panel) and the SSO targeting strategy used to therapeutically switch splicing for the treatment of the disease. Boxes are exons and horizontal lines are introns. Splicing regulatory sequences and protein regulators are noted. ESE, exonic splicing enhancer; ISS, intronic splicing silencer N1. SMNΔ7 refers to a form of SMN lacking amino acids encoded by exon 7. DystrophinΔ refers to a form of dystrophin truncated after amino acids encoded by exon 52 before encountering a premature termination codon in exon 52. Dystrophin isoform refers to a form of the Dystrophin protein encoded by mRNA lacking exons 50 and 51. The position of the stop codon is indicated by a red hexagon.
Eteplirsen is a 30-nucleotide, PMO targeting DMD exon 51 for skipping. Studies in mice and non-human primates demonstrated the safety and tolerability of the drug (92–94). Treatment with the SSO was also concluded to be safe and capable of causing exon 51 skipping and increasing dystrophin protein expression in clinical trials in humans when delivered either by intramuscular or intravenous injection (Table 2) (47,95,96). A longitudinal study of 12 patients treated with Eteplirsen for four years concluded that patients receiving the drug had a statistically significant increase on a 6-min walk test (6MWT) compared to natural history data from historical control patients with similar mutations that had not been treated with the SSO. This clinical outcome is accompanied by a very modest 0.9% increase in dystrophin protein in Eteplirsen-treated patients compared to untreated DMD patients http://www.fda.gov/downloads/AdvisoryCommittees/ CommitteesMeetingMaterials/Drugs/PeripheralandCentralNervousSystemDrugsAdvisory Committee/UCM481913.pdf (46). A recent FDA review of the drug data acknowledged the safety of the drug treatments but questioned the evidence that Eteplirsen treatment benefited patients http://www.fda.gov/downloads/AdvisoryCommittees/ CommitteesMeetingMaterials/Drugs/PeripheralandCentralNervousSystemDrugsAdvisory Committee/UCM481912.pdf. Specifically, the FDA questioned the use of natural history as the control, as the patients in that group may have had more severe symptoms than those enrolled in the clinical trial as clinical trial patients were often required to be ambulatory (46)(Table 2). Additionally, the increases in the 6MWT in the treated patients were within the range of the normal progression of the disease. Indeed, independent statistical review and analysis of the data by the FDA concluded that the comparison of Eteplirsen with historical controls was not statistically interpretable. In light of this evidence, an independent FDA review panel declined to recommend Eteplirsen for approval. The FDA is expected to announce its decision on the drug in 2016. Many of the issues holding back the advancement of Eteplirsen are related to the small patient sample size available for clinical trials and the variable, progressive nature of the disease, which make analysis of statistical and clinical significance challenging.
Kyndrisa is a 20-nucleotide, 2′-O-methyl-phosphorothioate oligomer (2′OMePS) (97). The sequence of Kyndrisa is similar to Eteplirsen, though Eteplirsen is longer, with an additional eight and two nucleotides on its 5′ and 3′ ends, respectively. Initial clinical trials found that intramuscular injection resulted in exon 51 skipping and a modest increase in dystrophin protein (97). However, repeated, systemic administration of the drug did not result in dystrophin protein above the range found in untreated patients. Following 25 weeks in a Phase 2 clinical trails, during which DMD patients were treated by subcutaneous injection with Kyndrisa, patients receiving the SSO showed consistent improvements in the 6MWT when compared with patients receiving placebo or intermittent treatment (98,99). However, after 49 weeks of treatment, the difference between Kyndrisa-treated and placebo groups was not statistically significant. Follow-up studies in a larger patient population also did not show a statistically significant improvement in the 6MWT after 48 weeks of treatment. In addition, unlike Eteplirsen, which was well-tolerated by patients, Kyndrisa treatment resulted in adverse events in several organ systems including life-threatening thrombocytopenia. The FDA recently denied an application to approve the drug for DMD, concluding that the drug was not ready for approval and recommended additional placebo-controlled clinical trials, some of which are on-going (Table 2) (http://www.fda.gov/downloads/AdvisoryCommittees/ CommitteesMeetingMaterials/Drugs/PeripheralandCentralNervousSystemDrugsAdvisory Committee/UCM473737.pdf). An application for marketing approval to the the European Medicines Agency was recently withdrawn and Biomarin programs developing similar SSOs to target exon 44, 45 and 53 skipping have been discontinued (http://investors.bmrn.com/releasedetail.cfm?ReleaseID=973536). Biomarin has announced plans to invest in the development of next generation oligonucleotides that will overcome the safety issues associated with Kyndrisa.
The development of an SSO for the treatment of DMD has been challenging and several caveats must be considered when interpreting the clinical results for the SSO DMD trials to date and evaluating the potential of SSO therapies for DMD (100,101). First, the two SSO drugs being tested in DMD patients differ in their chemistries which likely influences the pharmacodynamics and clinical outcomes and may present unique challenges to delivery and dosing. Second, the variable nature of the disease makes it difficult to assess cohort results (102). Third, rare diseases such as DMD have a small patient population available for clinical trials, which make statistical analysis problematic. Fourth, restored dystrophin protein is truncated and semi-functional and therefore, at best, the clinical outcome is conversion to the BMD phenotype. Finally, the timing of treatment and cellular targeting in the clinical trials to date are likely not optimal. Many human muscle fibers are formed embryonically starting at approximately week 11 of gestation and most muscle cells express membrane-associated dystrophin by gestational week 22 (103). Therefore, improving dystrophin expression may be more effective if initiated at an earlier developmental stage. Indeed, studies in mice suggest that SSO-based treatment of DMD symptoms must occur early in pathology, as treatment later in life was not effective at ameliorated disease phenotype (104,105). In addition, skeletal muscles are not the only tissues impacted by DMD. As mentioned previously, most DMD patients die from a combination of respiratory and cardiac failure. Both 2′-OMe and PMO modifications on ASOs have been found to result in low efficiency in cardiac muscles (57,83). Therefore, additional modifications to the SSOs may be required to target cardiac tissue. To this end, peptide-conjugated PMOs have been developed that improve DMD exon skipping in cardiac tissue (49,57). Although more development is clearly necessary, the progress to date with SSO therapeutics for DMD is substantial and demonstrates that a disease treatment that redirects splicing can be safe and may be therapeutically beneficial to some patients.
An SSO therapeutic for spinal muscular atrophy
Splice-switching ASO therapeutics are also in clinical trails for the treatment of SMA, an autosomal-recessive disease characterized by motor neuron degeneration that leads to progressive muscle weakness and, in severe cases, respiratory failure and death (106). SMA is the most common genetic cause of infant mortality and affects ∼1:10 000 live births with ∼1:50 carrier frequency (107–109). Studies on the natural history of infants with the most severe forms of the disease, SMA Type 1, found that 50% had died or were on permanent ventilation by 6.1–13.5 months of age and 80% had these outcomes by 18 months of age (110–112). The severity of SMA warrants an aggressive approach to therapeutic development for a disease that so far has no FDA-approved treatment.
SMA is caused by insufficient production of the protein SMN, a highly conserved and ubiquitously expressed protein involved in pre-mRNA splicing. In humans, SMN protein is produced from two different genes, SMN1 and SMN2, which arose from a duplication at chromosome 5q where the genes are located. Most SMN protein in the cell is produced from the SMN1 gene. The lack of SMN protein production from SMN2, despite the fact that SMN1 and SMN2 are nearly identical, is due to the fact that SMN2 has a single nucleotide C>T difference in exon 7 compared to SMN1, which disrupts splicing and results in skipping of exon 7 in most SMN2 mRNA transcripts (Figure 4) (113–117). This SMN2 exon 7-skipped mRNA isoform (SMNΔ7) codes for an SMN protein isoform that is unstable and does not function in the same manner as the full-length SMN isoform (118,119). People with SMA do not have a functional version of SMN1 most often due to deletion of part of the gene (120) and the small amount of full-length SMN produced from the limited amount of fully-spliced SMN2 mRNA cannot fully compensate for the loss of SMN1 (113). However, the intrinsic instability and variability of the 5q chromosomal region, which is responsible for the high incidence of SMN1 deletion, can also give rise to genomes with multiple copies of SMN2, and a higher SMN2 copy number is inversely correlated with disease severity (110,121–123). Patients with the most severe form of SMA (Type 1), have the fewest copies of SMN2, and most often die in the first few months of life, whereas those with a high SMN2 copy-number have a less severe form of the disease (Type 2, 3, 4) (110,124,125). Thus, SMN2 is a clear genetic modifier of SMA, and consequently elevating SMN2 expression and full-length SMN protein production from the gene has been a major focus of SMA therapeutic strategies. Because skipping of exon 7 is the major cause of the low SMN production from SMN2, SSOs have been heavily studied as a means to target exon 7 splicing and promote its inclusion to improve SMN protein expression (Figure 4).
The first SSOs used to increase SMN2 exon 7 splicing were targeted to the 3′ splice site of exon 8 and resulted in an increase in the use of the 3′ splice site of exon 7 and, thereby, more exon 7 inclusion (126). This study was followed by reports that SSOs could be used to increase exon 7 splicing by blocking putative splicing silencer elements surrounding exon 7 (127–129). Since this early work, numerous ASO-based approaches have been shown to effectively increase inclusion of exon 7 in SMN2 mRNA (23,130). Much of the current work on SSOs for SMA is focused on optimizing the length and the oligonucleotide target sequence as well as testing different backbone modifications to improve the efficacy and pharmacokinetics of the SSO (131–135). Some of the most widely-studied SSOs to date base-pair and block recognition of a splicing silencer element called intronic splicing silencer-N1 (ISS-N1), which is located in intron 7 of the SMN2 gene (136). The ISS-N1 RNA sequence is recognized by hnRNPA1/A2, which, upon binding, repressed SMN2 exon 7 splicing (137). SSOs that base-pair with ISS-N1 block hnRNPA1/A2 and relieve the splicing inhibition, which increases exon 7 inclusion full length SMN production (41) (Figure 4). ISS-N1 targeting by SSOs has been shown to dramatically increase the survival of SMA mice (36,131,132,138–141).
The most advanced splice-modulating SSO for the treatment of SMA is Nusinersen (formerly ISIS-SMNRx, ASO-10-27, ISIS 396443), first designed and reported by Krainer and colleagues in 2008 and now in Phase 3 clinical trials (Table 2 and Figure 4) (137). Ionis Pharmaceuticals (formerly Isis Pharmaceuticals) is developing the ASO in conjunction with Biogen Idec. Nusinersen is an 18-nucleotide 2′-MOE SSO with a fully modified PS backbone. Pre-clinical studies in SMA mice and non-human primates validated Nusinersen functionality, deliverability and safety (32,36,139).
Nusinersen was first administered to humans in 2011 in a Phase 1 open-label safety, tolerability and dosing study in Type 2 and Type 3 SMA children aged 2–14 years of age (Table 2). The SSO was delivered directly to the CNS by IT injection via lumbar puncture. Though there were only 28 participants with 6 children in each of three different dose cohorts and 10 children in a fourth dose cohort, the results of this first-in-human study were promising in several aspects. First, the lumbar puncture procedure was found to be feasible, well-tolerated and safe method for repeated IT delivery of the SSO (43). Second, the half-life of the SSO in the cerebrospinal fluid (CSF) was 4–6 months (142). This long half-life is within the tolerated range of feasible repeat IT dosing, which was performed total of three times, once at initial dosing and again at eight days and 9–14 months later (43,142). Finally, the children treated with the highest dose (9 mg) of SSO had a significant increase in SMN protein levels in the CSF and an improvement in clinical assessment measurements (Hammersmith Functional Motor Scale Expanded and Pediatric Quality of Life Inventory) at 9–14 months after treatment (142). These results, though encouraging, are preliminary and will require confirmation by larger, controlled studies.
To date, 10 clinical trials with Nusinersen have been completed or are on-going and current trials are expected to continue through 2020 (Table 2). Together, these trials have treated infants, children and young adults between the ages of ≤6 weeks old up to 21 years of age. All SSO treatments have been administered by IT injection by lumbar puncture of either a single or a repeated dose three times over the course of three months. Maintenance doses every 4–6 months are now in place for individuals that have completed initial dosing regimens (http://Clinicaltrials.gov). Although no results from Phase 2 or 3 clinical trials have been published yet, recent press releases from Ionis Pharmaceuticals claim that there had been no serious adverse effects related to the drug treatment and some children have been on treatment for more than 46 months (http://ir.ionispharma.com/phoenix.zhtml?c=222170&p=irol-newsArticle&ID=2061208). The company further reports that Type 1 SMA children that had been on Nusinersen treatment for the longest had a median event-free age of more than 20 months and 73% of the infants still enrolled in the study were event free, older than 15 months of age, had achieved motor milestones and had increased muscle function including some three year old children from the studies that are not on permanent ventilation and some that are walking, which is unprecedented for Type 1 SMA children (http://ir.ionispharma.com/phoenix.zhtml?c=222170&p=irol-presentations) (http://ir.ionispharma.com/phoenix.zhtml?c=222170&p=irol-newsArticle&ID=2061208). Although two deaths were reported in children that received the full dosing regimens, this number is less than expected from natural history studies (110). The molecular effects of Nusinersen have also been promising. Ionis reports that analysis of autopsy tissue found that the concentration of Nusinersen in the CNS was greater than the concentration that showed biological activity in animal studies (http://ir.ionispharma.com/phoenix.zhtml?c=222170&p=irol-newsArticle&ID=2061208). In addition, there was a higher abundance of full-length SMN2 mRNA and SMN protein in infants treated with Nusinersen compared to untreated SMA infants.
Although the early company reports from the Phase 2 clinical trials suggest drug efficacy, the results have not yet been published so critical evaluation is not possible. Furthermore, these reports are of results from open-label trials, which are not randomized, well-controlled studies (Table 2). Randomized, double-blind, sham-procedure controlled Phase 3 clinical trials are currently underway to help definitively assess the efficacy and safety of Nusinersen in infants and children with SMA. In addition, there is some question as to whether CNS delivery of Nusinersen will be sufficient as a disease therapeutic, considering the fact that in mouse models of SMA, it has become increasingly clear that restoring SMN in peripheral tissues is important for treatment of the disease in mice (36,140,141). In these studies, systemic ASO delivery to peripheral tissue resulted in greatly improved long-term rescue of SMA phenotypes. Therefore, it will be important that human clinical trials consider ASO delivery to both the CNS and the periphery. The coming months and years will reveal the full potential of Nusinersen as an SMA therapeutic as the effects of optimized dosing regimens and treatment windows become available from the current clinical trials.
CONCLUSIONS AND FUTURE PERSPECTIVES
The most developed therapeutic SSO programs are designed to treat two severe pediatric diseases. The SSO drugs for their treatment have different chemistries but all aim to alter splicing to increase functional protein expression from the targeted gene transcript. Eteplirsen and Nusinersen, have been well-tolerated by subjects, which is a major victory for the use of SSOs in the clinic. Outcome measurements of efficacy have been mixed for the DMD SSOs but early results from Nusinersen in SMA trials appear promising (142). It is important to consider the differences between the treatment paradigms for SMA and DMD when comparing their efficacy in the clinic. For example, the SSO for SMA, when effectively targeted, results in the production of the functional, full-length SMN protein. In contrast, the DMD SSO treatments are expected to reduce the severity of the disease to a condition similar to the less devastating BMD by inducing exon skipping and the production of a partially functional protein isoform. Thus, the DMD SSOs may not be expected to be as therapeutically beneficial as the SSO used to treat SMA. Additionally, the SSO drugs in clinical trials (Table 2) have different modifications and are delivered as naked/unformulated oligomers. It is possible and even likely that future drug optimization and delivery will lead to better efficacy. Finally, the DMD trials involve subcutaneous or intravenous administration to target skeletal muscles, whereas the SMA therapeutic is administered by intrathecal injection to specifically target the CNS. The pharmacokinetic profiles of ASOs in the CNS and peripheral organs are distinct, which likely affects the efficacy of an ASO (32,35,143). The fact that clinical trials with SSOs for DMD resulted in statistically significant changes in primary outcome measures suggest that the SSO may cause some exon skipping, but the lack of convincing clinical benefit to patients suggests that more work must be done to improve SSO efficacy by optimizing delivery, dosing or other features of the treatment and or SSO. A similar critical evaluation of Nusinersen awaits further reporting of results from current clinical trials with the drug.
The correction of aberrant gene expression has long been a focus of therapeutic development for the treatment of the human disease. ASOs, and in particular SSOs, offer a treatment approach that allows for specific and defined control of gene expression that can be easily tailored to correct or bypass the effects of a specific mutation. For this reason, the use of SSOs for the manipulation of splicing and gene expression is gaining favor as a drug platform for the treatment of disease. The fact that the mechanism of action of any ASO is known a priori by the nature of its design makes it amenable to rapid and systematic optimization. Methods and modifications that improve SSO and ASO drug profiles, in general, are being actively investigated as this drug platform gains traction and favor in the therapeutics arena. Efforts to increase cellular uptake in vivo, limit off-target effects and gain tissue and cell-specific entry are just some of the aims of creating better antisense drugs. Table 1 provides examples of SSOs that have been shown to be effective in vivo for the molecular and, in some cases, functional correction of disease defects, and provides insight into potential SSO-based therapeutics on the horizon.
Acknowledgments
The authors thank Jennifer Chang, Frederic Depreux, Dominik Duelli, Anthony Hinrich, Tiffany Chairuden and Francine Jodelka for comments on the manuscript, and Linda Schwartz for critical comments and editing.
FUNDING
National Institutes of Health (NIH) [NS069759 and DC012596 to M.L.H.]; Foundation Fighting Blindness; Batten Disease Support and Research Association; Batten Research Alliance; Noah's Hope; Hope for Bridget; and Charlotte and Gwenyth Gray Foundation. The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. M.L.H. receives funding from Ionis Pharmaceuticals.
REFERENCES
- 1.Papasaikas P., Valcarcel J. The Spliceosome: The ultimate RNA chaperone and sculptor. Trends Biochem. Sci. 2016;41:33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Hang J., Wan R., Yan C., Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 3.Wan R., Yan C., Bai R., Wang L., Huang M., Wong C.C., Shi Y. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 4.Brody E., Abelson J. The “spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- 5.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y., Rio D.C. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 10.Singh G., Pratt G., Yeo G.W., Moore M.J. The clothes make the mRNA: Past and present Trends in mRNP fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luco R.F., Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu X.D. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 13.Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 14.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:144–151. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 16.Leung M.K., Xiong H.Y., Lee L.J., Frey B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014;30:i121–i129. doi: 10.1093/bioinformatics/btu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueroussov S., Gonatopoulos-Pournatzis T., Irimia M., Raj B., Lin Z.Y., Gingras A.C., Blencowe B.J. An alternative splicing event amplifies evolutionary differences between vertebrates. Science. 2015;349:868–873. doi: 10.1126/science.aaa8381. [DOI] [PubMed] [Google Scholar]
- 18.Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R., et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 19.Chabot B., Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J. Cell Biol. 2016;212:13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havens M.A., Duelli D.M., Hastings M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterne-Weiler T., Sanford J.R. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol. 2014;15:201–208. doi: 10.1186/gb4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 23.Rigo F., Seth P.P., Bennett C.F. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol. 2014;825:303–352. doi: 10.1007/978-1-4939-1221-6_9. [DOI] [PubMed] [Google Scholar]
- 24.Sharma V.K., Watts J.K. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med. Chem. 2015;7:2221–2242. doi: 10.4155/fmc.15.144. [DOI] [PubMed] [Google Scholar]
- 25.Saleh A.F., Arzumanov A.A., Gait M.J. Overview of alternative oligonucleotide chemistries for exon skipping. Methods Mol. Biol. 2012;867:365–378. doi: 10.1007/978-1-61779-767-5_23. [DOI] [PubMed] [Google Scholar]
- 26.Evers M.M., Toonen L.J., van Roon-Mom W.M. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 29.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 30.Campbell M.A., Wengel J. Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011;40:5680–5689. doi: 10.1039/c1cs15048k. [DOI] [PubMed] [Google Scholar]
- 31.Yokota T., Lu Q.L., Partridge T., Kobayashi M., Nakamura A., Takeda S., Hoffman E. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann. Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigo F., Chun S.J., Norris D.A., Hung G., Lee S., Matson J., Fey R.A., Gaus H., Hua Y., Grundy J.S., et al. Pharmacology of a central nervous system delivered 2′-o-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther. 2014;350:46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw236. doi:10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Q.L., Wu B. Systemic delivery of antisense oligomer in animal models and its implications for treating DMD. Methods Mol. Biol. 2012;867:393–405. doi: 10.1007/978-1-61779-767-5_25. [DOI] [PubMed] [Google Scholar]
- 35.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung G., Xiao X., Peralta R., Bhattacharjee G., Murray S., Norris D., Guo S., Monia B.P. Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther. 2013;23:369–378. doi: 10.1089/nat.2013.0443. [DOI] [PubMed] [Google Scholar]
- 38.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H., et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southwell A.L., Skotte N.H., Bennett C.F., Hayden M.R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Lentz J.J., Jodelka F.M., Hinrich A.J., McCaffrey K.E., Farris H.E., Spalitta M.J., Bazan N.G., Duelli D.M., Rigo F., Hastings M.L. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 2013;19:345–350. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinrich A.J., Jodelka F.M., Chang J.L., Brutman D., Bruno A.M., Briggs C.A., James B.D., Stutzmann G.E., Bennett D.A., Miller S.A., et al. Therapeutic correction of ApoER2 splicing in Alzheimer's disease mice using antisense oligonucleotides. EMBO Mol. Med. 2016;8:328–345. doi: 10.15252/emmm.201505846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hache M., Swoboda K.J., Sethna N., Farrow-Gillespie A., Khandji A., Xia S., Bishop K.M. Intrathecal injections in children with spinal muscular atrophy: Nusinersen clinical trial experience. J. Child Neurol. 2016;31:899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calias P., Banks W.A., Begley D., Scarpa M., Dickson P. Intrathecal delivery of protein therapeutics to the brain: a critical reassessment. Pharmacol. Ther. 2014;144:114–122. doi: 10.1016/j.pharmthera.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study G., Network D.M.D.I. Longitudinal effect of eteplirsen vs. historical control on ambulation in DMD. Ann. Neurol. 2015;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenz P., Misteli T., Baker B.F., Bennett C.F., Spector D.L. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betts C., Saleh A.F., Arzumanov A.A., Hammond S.M., Godfrey C., Coursindel T., Gait M.J., Wood M.J. Pip6-PMO, A New Generation of Peptide-oligonucleotide Conjugates With Improved Cardiac Exon Skipping Activity for DMD Treatment. Mol. Ther. Nucleic Acids. 2012;1:e38. doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moulton J.D., Jiang S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules. 2009;14:1304–1323. doi: 10.3390/molecules14031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Andaloussi S.A., Hammond S.M., Mager I., Wood M.J. Use of cell-penetrating-peptides in oligonucleotide splice switching therapy. Curr. Gene Ther. 2012;12:161–178. doi: 10.2174/156652312800840612. [DOI] [PubMed] [Google Scholar]
- 52.Han G., Gu B., Cao L., Gao X., Wang Q., Seow Y., Zhang N., Wood M.J., Yin H. Hexose enhances oligonucleotide delivery and exon skipping in dystrophin-deficient mdx mice. Nat. Commun. 2016;7:10981. doi: 10.1038/ncomms10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koebis M., Kiyatake T., Yamaura H., Nagano K., Higashihara M., Sonoo M., Hayashi Y., Negishi Y., Endo-Takahashi Y., Yanagihara D., et al. Ultrasound-enhanced delivery of morpholino with Bubble liposomes ameliorates the myotonia of myotonic dystrophy model mice. Sci. Rep. 2013;3:2242. doi: 10.1038/srep02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Negishi Y., Ishii Y., Shiono H., Akiyama S., Sekine S., Kojima T., Mayama S., Kikuchi T., Hamano N., Endo-Takahashi Y., et al. Bubble liposomes and ultrasound exposure improve localized morpholino oligomer delivery into the skeletal muscles of dystrophic mdx mice. Mol. Pharm. 2014;11:1053–1061. doi: 10.1021/mp4004755. [DOI] [PubMed] [Google Scholar]
- 55.Lu Q.L., Rabinowitz A., Chen Y.C., Yokota T., Yin H., Alter J., Jadoon A., Bou-Gharios G., Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garanto A., Chung D.C., Duijkers L., Corral-Serrano J.C., Messchaert M., Xiao R., Bennett J., Vandenberghe L.H., Collin R.W. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum. Mol. Genet. 2016;2016:ddw118. doi: 10.1093/hmg/ddw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Peacey E., Rodriguez L., Liu Y., Wolfe M.S. Targeting a pre-mRNA structure with bipartite antisense molecules modulates tau alternative splicing. Nucleic Acids Res. 2012;40:9836–9849. doi: 10.1093/nar/gks710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vorlova S., Rocco G., Lefave C.V., Jodelka F.M., Hess K., Hastings M.L., Henke E., Cartegni L. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benchaouir R., Robin V., Goyenvalle A. Gene and splicing therapies for neuromuscular diseases. Front. Biosci. 2015;20:1190–1233. doi: 10.2741/4367. [DOI] [PubMed] [Google Scholar]
- 62.Siva K., Covello G., Denti M.A. Exon-skipping antisense oligonucleotides to correct missplicing in neurogenetic diseases. Nucleic Acid Ther. 2014;24:69–86. doi: 10.1089/nat.2013.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McClorey G., Wood M.J. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr. Opin. Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Beaudet A.L., Meng L. Gene-targeting pharmaceuticals for single-gene disorders. Hum. Mol. Genet. 2015;25:R18–R26. doi: 10.1093/hmg/ddv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Disterer P., Kryczka A., Liu Y., Badi Y.E., Wong J.J., Owen J.S., Khoo B. Development of therapeutic splice-switching oligonucleotides. Hum. Gene Ther. 2014;25:587–598. doi: 10.1089/hum.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arechavala-Gomeza V., Khoo B., Aartsma-Rus A. Splicing modulation therapy in the treatment of genetic diseases. Appl. Clin. Genet. 2014;7:245–252. doi: 10.2147/TACG.S71506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jirka S., Aartsma-Rus A. An update on RNA-targeting therapies for neuromuscular disorders. Curr. Opin. Neurol. 2015;28:515–521. doi: 10.1097/WCO.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 68.Ellis J.A., Vroom E., Muntoni F. 195th ENMC International Workshop: Newborn screening for Duchenne muscular dystrophy 14-16th December, 2012, Naarden, The Netherlands. Neuromuscul. Disord. 2013;23:682–689. doi: 10.1016/j.nmd.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 69.Romitti P.A., Zhu Y., Puzhankara S., James K.A., Nabukera S.K., Zamba G.K., Ciafaloni E., Cunniff C., Druschel C.M., Mathews K.D., et al. Prevalence of Duchenne and Becker muscular dystrophies in the United States. Pediatrics. 2015;135:513–521. doi: 10.1542/peds.2014-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mendell J.R., Lloyd-Puryear M. Report of MDA muscle disease symposium on newborn screening for Duchenne muscular dystrophy. Muscle Nerve. 2013;48:21–26. doi: 10.1002/mus.23810. [DOI] [PubMed] [Google Scholar]
- 71.Nowak K.J., Davies K.E. Duchenne muscular dystrophy and dystrophin: Pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox G.F., Kunkel L.M. Dystrophies and heart disease. Curr. Opin. Cardiol. 1997;12:329–343. [PubMed] [Google Scholar]
- 73.Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 74.Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Muller C.R., Lindlof M., Kaariainen H., et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 75.Aartsma-Rus A., Janson A.A., Kaman W.E., Bremmer-Bout M., van Ommen G.J., den Dunnen J.T., van Deutekom J.C. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am. J. Hum. Genet. 2004;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Echigoya Y., Yokota T. Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther. 2014;24:57–68. doi: 10.1089/nat.2013.0451. [DOI] [PubMed] [Google Scholar]
- 77.Takeshima Y., Nishio H., Sakamoto H., Nakamura H., Matsuo M. Modulation of in vitro splicing of the upstream intron by modifying an intra-exon sequence which is deleted from the dystrophin gene in dystrophin Kobe. J. Clin. Invest. 1995;95:515–520. doi: 10.1172/JCI117693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunckley M.G., Manoharan M., Villiet P., Eperon I.C., Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum. Mol. Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- 79.van Deutekom J.C., Bremmer-Bout M., Janson A.A., Ginjaar I.B., Baas F., den Dunnen J.T., van Ommen G.J. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 80.Mann C.J., Honeyman K., Cheng A.J., Ly T., Lloyd F., Fletcher S., Morgan J.E., Partridge T.A., Wilton S.D. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. U.S.A. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClorey G., Fletcher S., Wilton S. Splicing intervention for Duchenne muscular dystrophy. Curr. Opin. Pharmacol. 2005;5:529–534. doi: 10.1016/j.coph.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 82.Heemskerk H.A., de Winter C.L., de Kimpe S.J., van Kuik-Romeijn P., Heuvelmans N., Platenburg G.J., van Ommen G.J., van Deutekom J.C., Aartsma-Rus A. In vivo comparison of 2′-O-methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. J. Gene Med. 2009;11:257–266. doi: 10.1002/jgm.1288. [DOI] [PubMed] [Google Scholar]
- 83.Alter J., Lou F., Rabinowitz A., Yin H., Rosenfeld J., Wilton S.D., Partridge T.A., Lu Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 84.Tanganyika-de Winter C.L., Heemskerk H., Karnaoukh T.G., van Putten M., de Kimpe S.J., van Deutekom J., Aartsma-Rus A. Long-term Exon Skipping Studies With 2′-O-Methyl Phosphorothioate Antisense Oligonucleotides in Dystrophic Mouse Models. Mol. Ther. Nucleic Acids. 2012;1:e44. doi: 10.1038/mtna.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aoki Y., Yokota T., Nagata T., Nakamura A., Tanihata J., Saito T., Duguez S.M., Nagaraju K., Hoffman E.P., Partridge T., et al. Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13763–13768. doi: 10.1073/pnas.1204638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takeshima Y., Yagi M., Wada H., Ishibashi K., Nishiyama A., Kakumoto M., Sakaeda T., Saura R., Okumura K., Matsuo M. Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr. Res. 2006;59:690–694. doi: 10.1203/01.pdr.0000215047.51278.7c. [DOI] [PubMed] [Google Scholar]
- 87.Koo T., Wood M.J. Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum. Gene Ther. 2013;24:479–488. doi: 10.1089/hum.2012.234. [DOI] [PubMed] [Google Scholar]
- 88.A phase I/IIa clinical trial in duchenne muscular dystrophy using systemically delivered morpholino antisense oligomer to Skip Exon 53 (SKIP-NMD) Hum. Gene Ther. Clin. Dev. 2015;26:92–95. doi: 10.1089/humc.2015.2528. [DOI] [PubMed] [Google Scholar]
- 89.van Deutekom J.C., van Ommen G.J. Advances in Duchenne muscular dystrophy gene therapy. Nat. Rev. Genet. 2003;4:774–783. doi: 10.1038/nrg1180. [DOI] [PubMed] [Google Scholar]
- 90.De Angelis F.G., Sthandier O., Berarducci B., Toso S., Galluzzi G., Ricci E., Cossu G., Bozzoni I. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arechavala-Gomeza V., Graham I.R., Popplewell L.J., Adams A.M., Aartsma-Rus A., Kinali M., Morgan J.E., van Deutekom J.C., Wilton S.D., Dickson G., et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- 92.Sazani P., Weller D.L., Shrewsbury S.B. Safety pharmacology and genotoxicity evaluation of AVI-4658. Int. J. Toxicol. 2010;29:143–156. doi: 10.1177/1091581809359206. [DOI] [PubMed] [Google Scholar]
- 93.Sazani P., Ness K.P., Weller D.L., Poage D., Nelson K., Shrewsbury A.S. Chemical and mechanistic toxicology evaluation of exon skipping phosphorodiamidate morpholino oligomers in mdx mice. Int. J. Toxicol. 2011;30:322–333. doi: 10.1177/1091581811403504. [DOI] [PubMed] [Google Scholar]
- 94.Sazani P., Ness K.P., Weller D.L., Poage D.W., Palyada K., Shrewsbury S.B. Repeat-dose toxicology evaluation in cynomolgus monkeys of AVI-4658, a phosphorodiamidate morpholino oligomer (PMO) drug for the treatment of duchenne muscular dystrophy. Int. J. Toxicol. 2011;30:313–321. doi: 10.1177/1091581811403505. [DOI] [PubMed] [Google Scholar]
- 95.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P., et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 97.van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M., et al. Local dystrophin restoration with antisense oligonucleotide PRO051. New Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 98.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., De Kimpe S.J., Eagle M., Guglieri M., Hood S., et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 99.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A., et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. New Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 100.Lu Q.L., Cirak S., Partridge T. What can we learn from clinical trials of exon skipping for DMD? Mol. Ther. Nucleic Acids. 2014;3:e152. doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arechavala-Gomeza V., Anthony K., Morgan J., Muntoni F. Antisense oligonucleotide-mediated exon skipping for Duchenne muscular dystrophy: progress and challenges. Curr. Gene Ther. 2012;12:152–160. doi: 10.2174/156652312800840621. [DOI] [PubMed] [Google Scholar]
- 102.Ricotti V., Muntoni F., Voit T. Challenges of clinical trial design for DMD. Neuromuscul. Disord. 2015;25:932–935. doi: 10.1016/j.nmd.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 103.Prelle A., Chianese L., Moggio M., Gallanti A., Sciacco M., Checcarelli N., Comi G., Scarpini E., Bonilla E., Scarlato G. Appearance and localization of dystrophin in normal human fetal muscle. Int. J. Dev. Neurosci. 1991;9:607–612. doi: 10.1016/0736-5748(91)90022-e. [DOI] [PubMed] [Google Scholar]
- 104.Wu B., Cloer C., Lu P., Milazi S., Shaban M., Shah S.N., Marston-Poe L., Moulton H.M., Lu Q.L. Exon skipping restores dystrophin expression, but fails to prevent disease progression in later stage dystrophic dko mice. Gene Ther. 2014;21:785–793. doi: 10.1038/gt.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merrick D., Stadler L.K., Larner D., Smith J. Muscular dystrophy begins early in embryonic development deriving from stem cell loss and disrupted skeletal muscle formation. Dis. Models Mech. 2009;2:374–388. doi: 10.1242/dmm.001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kolb S.J., Kissel J.T. Spinal muscular atrophy. Neurol. Clin. 2015;33:831–846. doi: 10.1016/j.ncl.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prior T.W., Snyder P.J., Rink B.D., Pearl D.K., Pyatt R.E., Mihal D.C., Conlan T., Schmalz B., Montgomery L., Ziegler K., et al. Newborn and carrier screening for spinal muscular atrophy. Am. J. Med. Genet. A. 2010;152:1608–1616. doi: 10.1002/ajmg.a.33474. [DOI] [PubMed] [Google Scholar]
- 108.Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 2012;20:27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Larson J.L., Silver A.J., Chan D., Borroto C., Spurrier B., Silver L.M. Validation of a high resolution NGS method for detecting spinal muscular atrophy carriers among phase 3 participants in the 1000 Genomes Project. BMC Med. Genet. 2015;16:100–113. doi: 10.1186/s12881-015-0246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Finkel R.S., McDermott M.P., Kaufmann P., Darras B.T., Chung W.K., Sproule D.M., Kang P.B., Foley A.R., Yang M.L., Martens W.B., et al. Observational study of spinal muscular atrophy type I and implications for clinical trials. Neurology. 2014;83:810–817. doi: 10.1212/WNL.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Farrar M.A., Vucic S., Johnston H.M., du Sart D., Kiernan M.C. Pathophysiological insights derived by natural history and motor function of spinal muscular atrophy. J. Pediatr. 2013;162:155–159. doi: 10.1016/j.jpeds.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 112.Rudnik-Schoneborn S., Berg C., Zerres K., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Heller R. Genotype-phenotype studies in infantile spinal muscular atrophy (SMA) type I in Germany: implications for clinical trials and genetic counselling. Clin. Genet. 2009;76:168–178. doi: 10.1111/j.1399-0004.2009.01200.x. [DOI] [PubMed] [Google Scholar]
- 113.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 115.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 116.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 118.Cho S., Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 120.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 121.Gavrilov D.K., Shi X., Das K., Gilliam T.C., Wang C.H. Differential SMN2 expression associated with SMA severity. Nat. Genet. 1998;20:230–231. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 122.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 123.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wirth B., Brichta L., Schrank B., Lochmuller H., Blick S., Baasner A., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 125.Mercuri E., Bertini E., Iannaccone S.T. Childhood spinal muscular atrophy: controversies and challenges. Lancet Neurol. 2012;11:443–452. doi: 10.1016/S1474-4422(12)70061-3. [DOI] [PubMed] [Google Scholar]
- 126.Lim S.R., Hertel K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 127.Miyajima H., Miyaso H., Okumura M., Kurisu J., Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- 128.Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 129.Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tisdale S., Pellizzoni L. Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J. Neurosci. 2015;35:8691–8700. doi: 10.1523/JNEUROSCI.0417-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mitrpant C., Porensky P., Zhou H., Price L., Muntoni F., Fletcher S., Wilton S.D., Burghes A.H. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLoS One. 2013;8:e62114. doi: 10.1371/journal.pone.0062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou H., Janghra N., Mitrpant C., Dickinson R.L., Anthony K., Price L., Eperon I.C., Wilton S.D., Morgan J., Muntoni F. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum. Gene Ther. 2013;24:331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Osman E.Y., Miller M.R., Robbins K.L., Lombardi A.M., Atkinson A.K., Brehm A.J., Lorson C.L. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum. Mol. Genet. 2014;23:4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Keil J.M., Seo J., Howell M.D., Hsu W.H., Singh R.N., DiDonato C.J. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol. Ther. Nucleic Acids. 2014;3:e174. doi: 10.1038/mtna.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sahashi K., Ling K.K., Hua Y., Wilkinson J.E., Nomakuchi T., Rigo F., Hung G., Xu D., Jiang Y.P., Lin R.Z., et al. Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol. Med. 2013;5:1586–1601. doi: 10.1002/emmm.201302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hua Y., Liu Y.H., Sahashi K., Rigo F., Bennett C.F., Krainer A.R. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Verhaart I.E., Tanganyika-de Winter C.L., Karnaoukh T.G., Kolfschoten I.G., de Kimpe S.J., van Deutekom J.C., Aartsma-Rus A. Dose-dependent pharmacokinetic profiles of 2′-O-methyl phosphorothioate antisense oligonucleotidesin mdx mice. Nucleic Acid Ther. 2013;23:228–237. doi: 10.1089/nat.2012.0398. [DOI] [PubMed] [Google Scholar]
- 144.Svasti S., Suwanmanee T., Fucharoen S., Moulton H.M., Nelson M.H., Maeda N., Smithies O., Kole R. RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1205–1210. doi: 10.1073/pnas.0812436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taniguchi-Ikeda M., Kobayashi K., Kanagawa M., Yu C.C., Mori K., Oda T., Kuga A., Kurahashi H., Akman H.O., DiMauro S., et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–131. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Osorio F.G., Navarro C.L., Cadinanos J., Lopez-Mejia I.C., Quiros P.M., Bartoli C., Rivera J., Tazi J., Guzman G., Varela I., et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 2011;3:106ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 147.Lee J.M., Nobumori C., Tu Y., Choi C., Yang S.H., Jung H.J., Vickers T.A., Rigo F., Bennett C.F., Young S.G., et al. Modulation of LMNA splicing as a strategy to treat prelamin A diseases. J. Clin. Invest. 2016;126:1592–1602. doi: 10.1172/JCI85908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bestas B., Moreno P.M., Blomberg K.E., Mohammad D.K., Saleh A.F., Sutlu T., Nordin J.Z., Guterstam P., Gustafsson M.O., Kharazi S., et al. Splice-correcting oligonucleotides restore BTK function in X-linked agammaglobulinemia model. J. Clin. Invest. 2014;124:4067–4081. doi: 10.1172/JCI76175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mourich D.V., Oda S.K., Schnell F.J., Crumley S.L., Hauck L.L., Moentenich C.A., Marshall N.B., Hinrichs D.J., Iversen P.L. Alternative splice forms of CTLA-4 induced by antisense mediated splice-switching influences autoimmune diabetes susceptibility in NOD mice. Nucleic Acid Ther. 2014;24:114–126. doi: 10.1089/nat.2013.0449. [DOI] [PubMed] [Google Scholar]
- 151.Bauman J.A., Li S.D., Yang A., Huang L., Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38:8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nielsen T.O., Sorensen S., Dagnaes-Hansen F., Kjems J., Sorensen B.S. Directing HER4 mRNA expression towards the CYT2 isoform by antisense oligonucleotide decreases growth of breast cancer cells in vitro and in vivo. Br. J. Cancer. 2013;108:2291–2298. doi: 10.1038/bjc.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dewaele M., Tabaglio T., Willekens K., Bezzi M., Teo S.X., Low D.H., Koh C.M., Rambow F., Fiers M., Rogiers A., et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J. Clin. Invest. 2016;126:68–84. doi: 10.1172/JCI82534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zammarchi F., de Stanchina E., Bournazou E., Supakorndej T., Martires K., Riedel E., Corben A.D., Bromberg J.F., Cartegni L. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yilmaz-Elis A.S., Aartsma-Rus A., t Hoen P.A., Safdar H., Breukel C., van Vlijmen B.J., van Deutekom J., de Kimpe S., van Ommen G.J., Verbeek J.S. Inhibition of IL-1 signaling by antisense oligonucleotide-mediated exon skipping of IL-1 Receptor Accessory Protein (IL-1RAcP) Mol. Ther. Nucleic Acids. 2013;2:e66. doi: 10.1038/mtna.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Graziewicz M.A., Tarrant T.K., Buckley B., Roberts J., Fulton L., Hansen H., Orum H., Kole R., Sazani P. An endogenous TNF-alpha antagonist induced by splice-switching oligonucleotides reduces inflammation in hepatitis and arthritis mouse models. Mol. Ther. 2008;16:1316–1322. doi: 10.1038/mt.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Owen L.A., Uehara H., Cahoon J., Huang W., Simonis J., Ambati B.K. Morpholino-mediated increase in soluble Flt-1 expression results in decreased ocular and tumor neovascularization. PLoS One. 2012;7:e33576. doi: 10.1371/journal.pone.0033576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Uehara H., Cho Y., Simonis J., Cahoon J., Archer B., Luo L., Das S.K., Singh N., Ambati J., Ambati B.K. Dual suppression of hemangiogenesis and lymphangiogenesis by splice-shifting morpholinos targeting vascular endothelial growth factor receptor 2 (KDR) FASEB J. 2013;27:76–85. doi: 10.1096/fj.12-213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gedicke-Hornung C., Behrens-Gawlik V., Reischmann S., Geertz B., Stimpel D., Weinberger F., Schlossarek S., Precigout G., Braren I., Eschenhagen T., et al. Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO Mol. Med. 2013;5:1060–1077. doi: 10.1002/emmm.201202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gramlich M., Pane L.S., Zhou Q., Chen Z., Murgia M., Schotterl S., Goedel A., Metzger K., Brade T., Parrotta E., et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 2015;7:562–576. doi: 10.15252/emmm.201505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Salewsky B., Hildebrand G., Rothe S., Parplys A.C., Radszewski J., Kieslich M., Wessendorf P., Krenzlin H., Borgmann K., Nussenzweig A., et al. Directed alternative splicing in Nijmegen Breakage Syndrome: Proof of principle concerning its therapeutical application. Mol. Ther. 2016;24:117–124. doi: 10.1038/mt.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Panchal R.G., Mourich D.V., Bradfute S., Hauck L.L., Warfield K.L., Iversen P.L., Bavari S. Induced IL-10 splice altering approach to antiviral drug discovery. Nucleic Acid Ther. 2014;24:179–185. doi: 10.1089/nat.2013.0457. [DOI] [PubMed] [Google Scholar]
- 163.Evers M.M., Tran H.D., Zalachoras I., Meijer O.C., den Dunnen J.T., van Ommen G.J., Aartsma-Rus A., van Roon-Mom W.M. Preventing formation of toxic N-terminal huntingtin fragments through antisense oligonucleotide-mediated protein modification. Nucleic Acid Ther. 2014;24:4–12. doi: 10.1089/nat.2013.0452. [DOI] [PubMed] [Google Scholar]
- 164.Disterer P., Al-Shawi R., Ellmerich S., Waddington S.N., Owen J.S., Simons J.P., Khoo B. Exon skipping of hepatic APOB pre-mRNA with splice-switching oligonucleotides reduces LDL cholesterol in vivo. Mol. Ther. 2013;21:602–609. doi: 10.1038/mt.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kang J.K., Malerba A., Popplewell L., Foster K., Dickson G. Antisense-induced myostatin exon skipping leads to muscle hypertrophy in mice following octa-guanidine morpholino oligomer treatment. Mol. Ther. 2011;19:159–164. doi: 10.1038/mt.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu-Nguyen N.B., Jarmin S.A., Saleh A.F., Popplewell L., Gait M.J., Dickson G. Combination antisense treatment for destructive exon skipping of myostatin and open reading frame rescue of dystrophin in neonatal mdx mice. Mol. Ther. 2015;23:1341–1348. doi: 10.1038/mt.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Clayton N.P., Nelson C.A., Weeden T., Taylor K.M., Moreland R.J., Scheule R.K., Phillips L., Leger A.J., Cheng S.H., Wentworth B.M. Antisense oligonucleotide-mediated suppression of Muscle Glycogen Synthase 1 synthesis as an approach for substrate reduction therapy of pompe disease. Mol. Ther. 2014;3:e206. doi: 10.1038/mtna.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Evers M.M., Tran H.D., Zalachoras I., Pepers B.A., Meijer O.C., den Dunnen J.T., van Ommen G.J., Aartsma-Rus A., van Roon-Mom W.M. Ataxin-3 protein modification as a treatment strategy for spinocerebellar ataxia type 3: removal of the CAG containing exon. Neurobiol. Dis. 2013;58:49–56. doi: 10.1016/j.nbd.2013.04.019. [DOI] [PubMed] [Google Scholar]