Abstract
Cotranslational degradation of polypeptide nascent chains plays a critical role in quality control of protein synthesis and the rescue of stalled ribosomes. In eukaryotes, ribosome stalling triggers release of 60S subunits with attached nascent polypeptides, which undergo ubiquitination by the E3 ligase Ltn1 and proteasomal degradation facilitated by the ATPase Cdc48. However, the identity of factors acting upstream in this process is less clear. Here, we examined how the canonical release factors Sup45–Sup35 (eRF1–eRF3) and their paralogs Dom34-Hbs1 affect the total population of ubiquitinated nascent chains associated with yeast ribosomes. We found that the availability of the functional release factor complex Sup45–Sup35 strongly influences the amount of ubiquitinated polypeptides associated with 60S ribosomal subunits, while Dom34-Hbs1 generate 60S-associated peptidyl-tRNAs that constitute a relatively minor fraction of Ltn1 substrates. These results uncover two separate pathways that target nascent polypeptides for Ltn1-Cdc48-mediated degradation and suggest that in addition to canonical termination on stop codons, eukaryotic release factors contribute to cotranslational protein quality control.
INTRODUCTION
Protein quality control (QC) is essential for the production of a functional proteome. Inefficient or dysfunctional QC may cause accumulation of aberrant and aggregation-prone polypeptides, a condition implicated in a wide range of human disorders including neurodegenerative diseases and cancer (reviewed in (1)). Recent studies in eukaryotic cells have demonstrated that aberrant and misfolded polypeptide nascent chains (NCs) can undergo modification with ubiquitin while still attached to the ribosome, thereby initiating the degradation of the aberrant polypeptides by the proteasome (2–5). Given that as much as 6–15% of all newly generated polypeptides in eukaryotic cells fail to fold or assemble properly (3,5), the systems responsible for cotranslational QC are clearly important for maintaining protein homeostasis.
Currently, cotranslational degradation of NCs is best understood in the situations when ribosomes stall during elongation. Previous studies indicated that a complex of the release-like factors Dom34–Hbs1 (also known as Pelota–Hbs1 in mammalian cells) acts in concert with the recycling factor Rli1 (ABCE1 in mammals) to split stalled ribosomes into subunits (6–9). After dissociation from 40S, 60S subunits with associated NCs (60S·NC complexes) are recognized by the ribosome quality control (RQC) complex consisting of Ltn1, Rqc1 and Rqc2 (also known as Tae2) (2,10). Rqc2 binds to the intersubunit surface, prevents reassociation of the ribosomal subunits (11–13), signals translation stress to the heat shock factor Hsf1 (10,13), and promotes activity of the E3 ligase Ltn1 (12). Ltn1 (also known as Rkr1 in yeast and Listerin in mammals) modifies NCs in 60S·NC complexes with polyubiquitin chains, leading to the recruitment of the ubiquitin-selective chaperone complex Cdc48–Npl4–Ufd1 (2,4,10,14,15). It is currently believed that because of a steric clash with the 40S subunit, Ltn1 cannot bind to translating ribosomes, which likely serves to prevent ubiquitination of NCs by Ltn1 during elongation (11,12). The AAA family ATPase Cdc48 assists the clearance of the 60S-associated polypeptides, presumably by supplying mechanical force for the extraction of NCs and their presentation to the proteasome.
The RQC system is conserved in all eukaryotes and defects in its components have been linked to neurodegeneration in mice (16,17). The nature of the RQC substrates in the cell, however, remains an open question (18). The pioneering work by the Inada laboratory (19,20) demonstrated ubiquitin-proteasome-mediated degradation of NCs utilizing poly(Lys)- and poly(Arg)-containing model substrates, which were later investigated in conjunction with the RQC system (2,4,10,14). However, ribosomal stalling can occur in many other situations, including impaired cotranslational protein folding, the presence of rare codons and stable secondary structures within mRNA (3,5,21,22). Recent evidence indeed points to additional types of translational stalling contributing endogenous substrates to the RQC pathway (23,24). Additionally, the translation factors that generate RQC substrates are not completely understood. Dom34-Hbs1 play a pivotal role in splitting non-translating 80S ribosomes containing Stm1 in place of the mRNA (25) and have been shown to operate efficiently on ribosomal complexes with a short mRNA length relative to the ribosome's P-site, such as ribosomes stalled at the 3′ end of truncated mRNA and some nonstop messages (7,8,26). Moreover, aberrant nonstop polypeptides were observed to be stabilized by an ltn1Δ deletion in the absence of Dom34 (27). Thus, the contribution of Dom34-Hbs1 to cotranslational QC on a global scale remains unclear. Intriguingly, canonical release factors were recently reported to mediate release of ribosomes on polylysine-encoding segments of mRNA (28), although whether this contributes to degradation of other aberrant translation products is currently unknown. Here, we addressed the role of factors that operate upstream of Ltn1 ubiquitination in ribosome-associated degradation by investigating the global pattern of modification of distinct ribosomal species with ubiquitin.
MATERIALS AND METHODS
Yeast strains and media
We used standard recipes for YPDA (1% yeast extract, 2% peptone, 2% dextrose, 10 mg/l adenine), rich media without dextrose (1% yeast extract, 2% peptone, 10 mg/l adenine) and synthetic glucose or galactose-containing media. All strains used in this study are listed in Supplemental Table S1. To generate deletion strains, his5+ or LEU2 disruption cassettes were integrated via homologous recombination at the targeted genomic loci using standard PCR-based techniques (29). Genomic integration of the disruption cassettes was verified by PCR using two sets of primers. One set included a forward primer annealing ∼100 bp upstream of the integration site and a reverse primer annealing within the LEU2 or his5+ cassettes. The second set included primers annealing upstream and downstream of the integration site to discriminate between intact genes and the disruption cassettes. Sequences of all primers used in this study are available upon request.
All strains used in this study carry the prion form of the Rnq1 protein, [PIN+], which promotes formation of Sup35 aggregates. The [psi− PIN+] strain 74-D694 (30), also designated as OT60 (31), contains a UGA allele of the ADE1 gene, ade1-14. The strain GT181 was selected as a spontaneous Ade+ derivative of OT60 on –Ade medium and determined to bear a mutant allele of the SUP35 gene (designated sup35-R15) in a test for allelism using tester strains 66-8A-P3532 (MATα ade1-14 his7-1 met13-A1 sup45 [rho−]) and 68-8A-P3532 (MATα ade1-14 his7-1 met13-A1 sup35), kindly provided by S.G. Inge-Vechtomov (St. Petersburg State University, Russia). PCR amplification and sequencing of the sup35-R15 allele showed a single G-to-T substitution at nucleotide position 1615 (relative to the start of the Sup35 coding sequence), which created a premature stop codon instead of codon 535 in the SUP35 ORF. The sup35-R15 allele in the strain GT181 causes temperature sensitivity of growth on complete medium, with the permissive temperature of 25°C and non-permissive temperature of 30°C and above. Growth defects observed at 30°C were rescued by ectopic expression of the Sup35-ΔN protein, which lacks the QN-rich N-terminal prion domain but retains the translation termination activity.
Antibodies and chemicals
The polyclonal anti-FLAG antibodies were purchased from Sigma (F7425); the anti-ubiquitin antibodies (clone P4G7-H11) were from Stressgen (SPA-203); the anti-Rpl3 (ScRPL3) and anti-α-tubulin (12G10) antibodies were from the Developmental Studies Hybridoma Bank, University of Iowa; anti-Rps14 antibodies were kindly provided by John Woolford, Carnegie Mellon University, Pittsburgh; anti-Sup35C antibodies were kindly provided by D. Bedwell, University of Alabama, Birmingham. Doxycycline (Frontier Scientific, cat. # D10056) was used at a final concentration of 10 μg/ml. Cycloheximide (CHX) was purchased from Sigma (cat. # 01810) and used at 100 μg/ml, hygromycin B (Calbiochem cat. # 400053) was used at 100 μg/ml.
Plasmids
Full-length SUP35 was amplified by PCR from genomic yeast DNA with primers complementary to the 5′ and 3′ ends of the SUP35 coding sequence. To generate the ΔN truncation mutant (lacking the first 154 amino acids), the forward (5′) primer was chosen to anneal 462 bp downstream of the start codon of SUP35. To generate FLAG-tagged constructs of Sup35, we used a reverse (3′) primer containing a sequence that encoded the FLAG tag. PCR products were digested with BamHI and XbaI and cloned into BamHI and XbaI sites of the pLA1 vector (32).
Sucrose gradient analysis
Polysomes were prepared essentially as described previously (33). Cells were treated with CHX prior to harvesting, pelleted by centrifugation, washed twice in ice-cold lysis buffer A (100 mM NaCl, 3 mM MgCl2, 10 mM Tris–HCl pH 7.4, 100 μg/ml CHX, 200 μg/ml heparin and 100 μM PMSF). Cells were lysed in the lysis buffer by 8–10 cycles of 30 s vortexing followed by 30 sec incubation on ice in the presence of glass beads at 4°C. An aliquot corresponding to 50 OD260 units of the clarified lysate was loaded onto 15–45% (w/v) or 15–42% (w/v) sucrose gradients prepared in 70 mM NH4Cl, 4 mM MgCl2 and 10 mM Tris–HCl (pH 7.4). The volume of each gradient was 11 ml. Gradients were centrifuged at 188 000 × g at 4°C for 4 h 15 min (Beckman SW41Ti rotor, 36,000 rpm) and fractionated into 14 fractions (∼0.78 ml each) for 15–45% gradients or into 28 fractions (∼0.39 ml each) for 15–42% gradients using a Beckman fraction recovery system connected to an EM-1 UV monitor (Bio-Rad).
Protein analysis
To analyze proteins co-sedimented with various fractions of the sucrose gradients, each fraction was incubated with 0.1 mg/ml BSA and 10% TCA on ice for 30 min. Proteins were pelleted by centrifugation at 4°C for 20 min, pellets were washed once with ethanol and once with acetone, air-dried and resuspended in 1× SDS-PAGE loading buffer. Proteins were resolved in 10% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes, and stained with Ponceau S prior to immunodetection. Each membrane used for detection of ubiquitinated species was cut at ∼60 kDa and the lower part incubated with anti-Rpl3 antibodies to monitor 60S subunit distribution on the gradients. All gradient analyses shown in this paper were repeated a minimum of three times.
RNA analysis
To analyze RNA present in sucrose gradient fractions, each fraction was treated with 100 μg/ml proteinase K (Roche) in the presence of 1% SDS and 10 mM EDTA for 30 min at 42°C, followed by phenol/chloroform extraction and ethanol precipitation. RNA pellets were resuspended in formamide and separated on 1.5% agarose gels containing 1.3% formaldehyde (34). RNA was transferred to nylon membranes (Hybond N, GE Biosciences) and RNA was visualized by methylene blue staining. Individual RNA species were detected by Northern hybridizations using 32P-labeled oligonucleotide probes as described (35). Sequences of hybridization probes used in this study are listed in Supplemental Table S2. Hybridizations were analyzed using a Typhoon 9200 PhosphorImager and ImageQuant software (GE Biosciences).
Peptidyl-tRNA analysis
The presence of peptidyl-tRNA on 60S subunits was analyzed by acid-urea gel electrophoresis as described previously (36). First, the 60S fraction separated on a 15–45% sucrose gradient was subjected to ultracentrifugation at 67 000 × g for 90 min at 4°C (Beckman TLA-55 rotor, 55 000 rpm). The pelleted subunits were resuspended in 8 M urea, 10 mM sodium acetate (pH 5.0), 1 mM EDTA, 0.02% bromophenol blue and 0.03% xylene cyanol and loaded onto 0.75 mm, 10 × 10 cm 8% polyacrylamide gels containing 8 M urea. Gels were run at 4°C for 5 h at 100 V in 100 mM sodium acetate (pH 5.0) and 1 mM EDTA. RNA was transferred to nylon membranes in 0.5× TBE for 20 min at 10 V using a Trans-blot SD semi-dry apparatus (Bio-Rad), with subsequent staining and hybridizations done as described above.
SDD-AGE (semi-denaturating detergent agarose gel electrophoresis)
The distribution of the Sup35 prion polymers by size was determined by using SDD-AGE, as described previously (37) with some modifications (38). Crude cell extracts were prepared by vortexing cells with glass beads. Total lysates were cleared from cell debris by centrifugation at 3000 rpm for 2 min, 50 μg of total protein was incubated in the sample buffer (0.5× TAE, 2% SDS, 5% glycerol, 0.025% bromophenol blue) for 5 min at 37°C, and resolved on a 1.8% agarose gel in 1× TAE, 0.1% SDS, followed by wet transfer to a nitrocellulose membrane.
RESULTS
Gradient analysis of ubiquitinated polypeptides associated with ribosomes in Cdc48-depleted cells
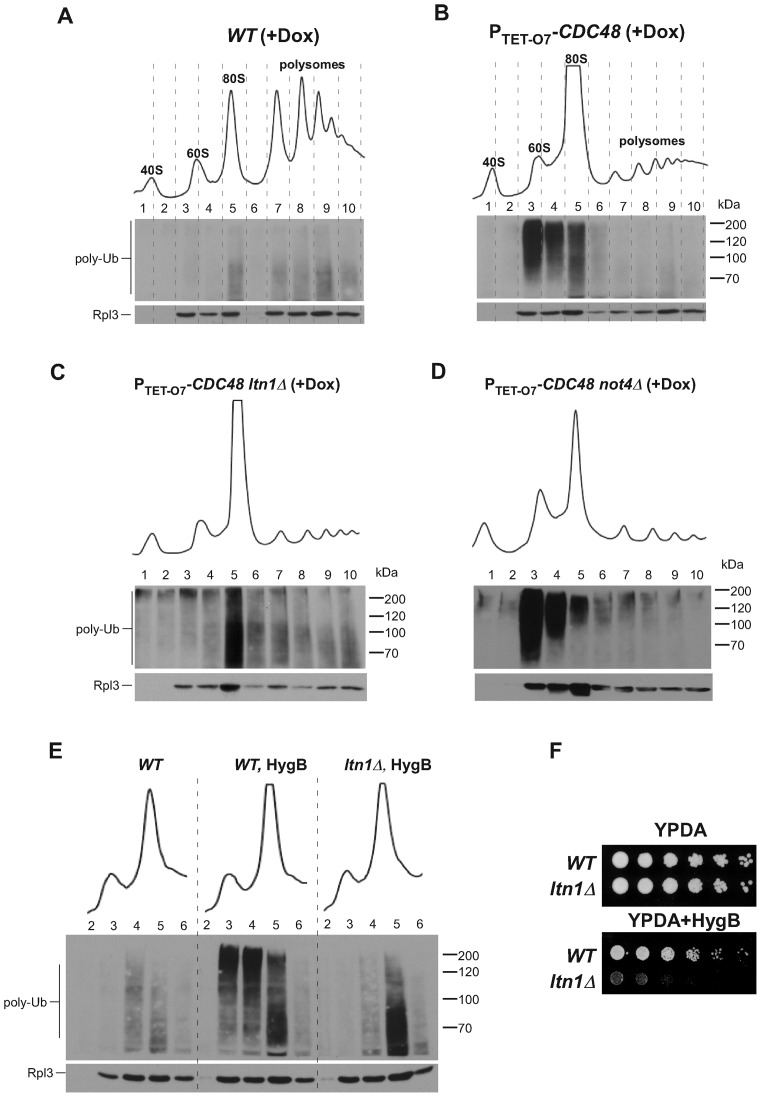
To examine how endogenous targets of cotranslational protein QC are distributed among different subpopulations of ribosomes in yeast cells, we fractionated cytoplasmic lysates by sedimentation through sucrose gradients. We observed a smear of ubiquitinated translation products in fractions containing polysomes and 80S monosomes in wild-type cells (Figure 1A, Supplementary Figure S1A), consistent with the previously described ubiquitination of ribosome-associated NCs (3,4). Previous studies have shown that loss of Cdc48 function increases accumulation of ubiquitinated NCs associated with ribosomes (2–4). To better define this effect, we analyzed yeast strains in which CDC48 is placed under the control of a tetracycline-regulatable promoter (PTET-O7-CDC48) (39). Depletion of Cdc48 by addition of doxycycline (Dox) to growing yeast cultures significantly increased the intensity of the ubiquitin signal in these cells (Figure 1B, Supplementary Figure S1A-D), as expected from stabilization of ubiquitinated NCs that normally undergo Cdc48-assisted degradation by the proteasome (4). The ubiquitin signal in Cdc48-depleted cells was mainly concentrated in fractions 3–5 (Figure 1B), corresponding to the peaks of 60S subunits (fractions 3 and 4) and 80S monosomes (fractions 4 and 5). In this and other experiments, the equivalent separation of ribosomal fractions on gradients was controlled by comparing alignment of A260 traces of the gradients, gel analysis of rRNA from the same fractions that were used for protein isolation, Ponceau S staining to control protein transfer, and probing with antibodies against ribosomal protein Rpl3 (Supplementary Figure S1A). When it was necessary to compare absolute levels of ubiquitinated species between different strains, we analyzed fractions side by side on the same gel to rule out variations between processing of individual membranes (Supplementary Figure S1D). Notably, the increase in ubiquitination of the 80S fraction was not due to redistribution of ribosomes from polysomes to the 80S monosome form in Cdc48-depleted cells, as shift of wild-type cells to medium lacking glucose (a condition known to inhibit translation initiation) resulted in elevated 80S levels comparable to those in Dox-treated PTET-O7-CDC48 cells, but did not cause accumulation of polyubiquitinated peptides in the 60S–80S fractions (Supplementary Figure S1E). Thus, Cdc48 depletion leads to increased accumulation of ubiquitinated polypeptides associated predominantly with 60S and 80S ribosomes.
Figure 1.
Cdc48 and ubiquitin ligases affect the distribution pattern of ribosome-associated ubiquitinated species. Sucrose gradient sedimentation analysis of ribosomes extracted from the wild-type RS1158 strain (A) and its derivative strain harboring a tetracycline-repressible PTET-O7-CDC48 allele (B). Cells were grown in the presence of doxycycline (Dox) for 20 h. Cell lysates were centrifuged through 15%–45% sucrose gradients and fractionated with the continuous measurement of absorbance at 254 nm to visualize ribosomal peaks. Proteins isolated from individual fractions were separated on 10% SDS-acrylamide gels and the same membrane was probed with anti-ubiquitin (top) and anti-Rpl3 (bottom) antibodies. (C) and (D) The same analysis performed in PTET-O7-CDC48 cells lacking either LTN1 or NOT4. All gradients were run at least five times, representative blots are shown; see Supplemental Figure S1 for additional controls. (E) Hygromycin B (HygB) treatment induces accumulation of Ltn1-ubiquitinated 60S products. Where indicated, cells were treated with 100 μg/mL hygromycin for 2 h. Gradient fractions 2–6 were separated on the same gel for a better comparison of total ubiqutination levels. (F) Lack of LTN1 makes cells hypersensitive to hygromycin. Five-fold dilutions of wild-type B4741 (WT) and ltn1Δ strains were grown on plates without antibiotic or with 100 μg/ml hygromycin for 3 days at 30°C.
We next examined the distribution of ubiquitin-conjugated species in the 60S–80S fractions of PTET-O7-CDC48 strains containing additional deletions in genes of E3 ligases implicated in cotranslational QC. Consistent with previous findings obtained with model substrates (4,14,15,40), an ltn1Δ deletion completely abrogated the accumulation of ubiquitin-modified NCs in fractions that contained free 60S subunits, but not in the 80S fraction (Figure 1C). In contrast, a Cdc48-depleted strain lacking Not4, part of the Ccr4-Not deadenylase complex reported to ubiquitinate polypeptides associated with stalled ribosomes (19,27), showed a diminished intensity of the ubiquitin signal in the 80S fraction, whereas accumulation of ubiquitinated products in the 60S fraction remained constitutively high (Figure 1D). These results support the notion (2,10,14,15) that Ltn1 is a primary, and possibly unique, E3 ligase responsible for ubiquitin tagging of the 60S-associated NCs, a process necessary for Cdc48 recruitment and polypeptide degradation.
To further investigate whether the observed accumulation of 60S-associated products ubiquitinated by Ltn1 is related to aberrant polypeptide synthesis during translation, we treated cells for 2 h with hygromycin B, an aminoglycoside that causes decoding errors, stop codon readthrough and ribosomal stalling (41–43). In wild-type cells, hygromycin treatment significantly increased the amount of polyubiquitinated products present in the 60S–80S fractions (Figure 1E). Similarly to PTET-O7-CDC48 ltn1Δ cells, deletion of LTN1 abrogated the accumulation of ubiquitinated products in the 60S, but not 80S fraction in the hygromycin-treated cells (Figure 1E). Consistent with a role of Ltn1 in resolving translation errors caused by hygromycin, ltn1Δ cells are hypersensitive to this antibiotic (Figure 1F). These results support the idea that the Ltn1-Cdc48-mediated QC mechanism can capture a variety of defective translational products that remain associated with the large ribosomal subunit after splitting of the ribosome.
Dom34 has little impact on the levels of ubiquitinated NCs
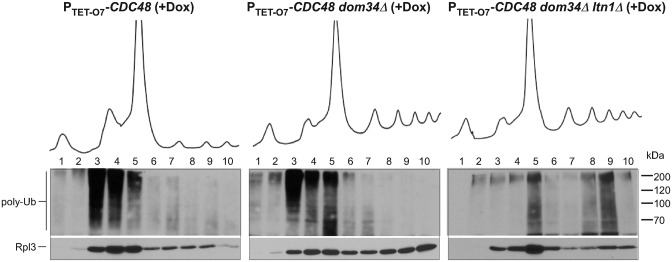
Having found that gradient analysis can differentiate Ltn1-modified NCs from those ubiquitinated through Ltn1-independent mechanisms, we sought to probe the upstream events that generate Ltn1 substrates. Splitting of ribosomal subunits during ribosome-associated QC in previous studies was attributed to the complex of the proteins Dom34 and Hbs1. Specifically, Dom34-Hbs1 was shown to promote separation of ribosomal subunits and drop-off of short peptidyl-tRNAs in an in vitro yeast translation system (6). Efficient splitting of ribosomes stalled on truncated mRNA sequences has been also demonstrated for the mammalian homologs of Dom34-Hbs1, Pelota-Hbs1 (7,40). However, the extent to which Dom34-Hbs1 contribute to the total pool of ubiquitinated NCs in cells, and specifically to Ltn1 substrates, is not known. To address this, we made a PTET-O7-CDC48 dom34Δ strain and analyzed the distribution of ubiquitinated NCs in ribosomal fractions after Cdc48 depletion. Surprisingly, deletion of DOM34 had only a modest effect on the relative level and distribution of ubiquitin conjugates, with significant accumulation still observed in the 60S fraction (Figure 2, middle; Supplementary Figure S2A). Introduction of an additional ltn1Δ deletion into the PTET-O7-CDC48 dom34Δ strain abolished accumulation of the ubiquitin signal in the 60S fraction, confirming that ubiquitination of the 60S·NC complexes in these cells was still dependent on Ltn1 (Figure 2, right; Supplementary Figure S2A). The unexpectedly minor impact of Dom34 on the accumulation of ubiquitinated 60S·NC complexes raised the possibility that additional factors may exist in cells to effect the splitting of ribosomes and generate substrates for the Ltn1-Cdc48 pathway.
Figure 2.
Deficiency in Dom34 does not significantly affect levels of ubiquitinated 60S·NCs. Lysates prepared from the indicated strains grown in the presence of Dox for 20 h to deplete Cdc48 were separated on 15–45% sucrose gradients and analyzed as described in Figure 1. Representative blots are shown. Experiment was repeated four times, see Supplemental Figure S2A for another experimental repeat with samples separated on one gel.
Canonical release factors are responsible for generating a major fraction of Ltn1-modified NCs
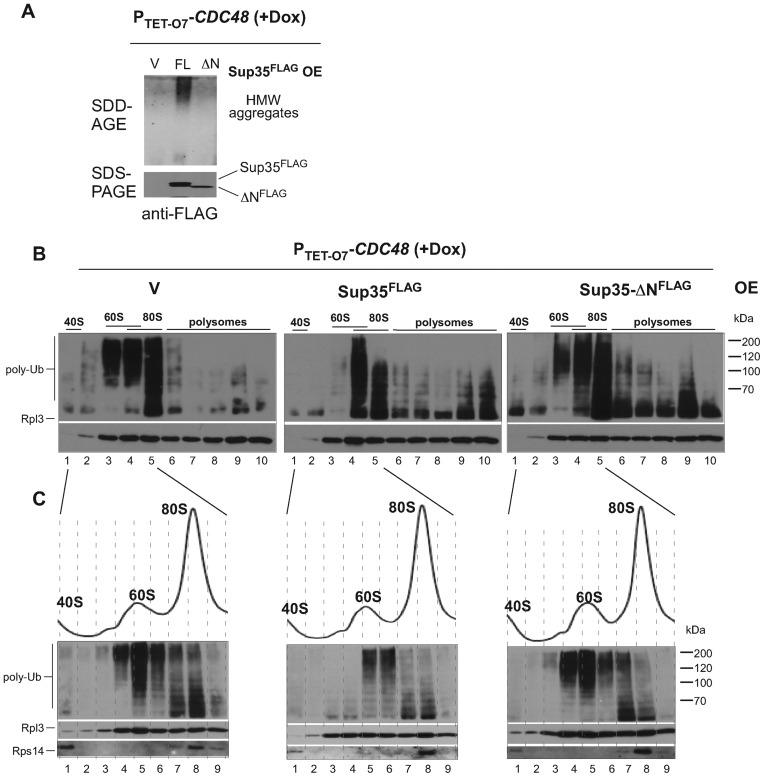
Dom34 and Hbs1 are structurally related to the canonical release factors Sup45 and Sup35 (termed eRF1 and eRF3 in other species). Recently, the Sup45–Sup35 complex has been shown to mediate premature translation termination on polylysine-containing luciferase reporters (28), prompting us to test the role of these factors in the generation of substrates for Ltn1 and Cdc48. We had to use partial depletion strategies since both SUP35 and SUP45 are essential. First, we used an established approach based on the partial inactivation of the endogenous Sup45–Sup35 complex via sequestration into prion-like aggregates of Sup35. These aggregates are formed with high efficiency when full-sized Sup35 is overproduced in cells bearing a prion form of another protein, Rnq1 (44–48). To generate Sup35 aggregates, we first overexpressed full-length Sup35 from the GAL1 promoter in PTET-O7-CDC48 cells. As a control, we used overexpression of Sup35-ΔN that lacks the N-terminal prion-domain responsible for aggregate formation but retains the ability to interact with Sup45 and act as a translation termination factor (49,50). As expected, galactose-induced expression of the full-length Sup35 led to the efficient formation of the high molecular weight aggregates of Sup35 (Figure 3A). Analysis of the ribosomal fractions in these cells (Figure 3B, middle; Supplementary Figure S2B) revealed a significant drop in the level of ubiquitinated products in the 60S fraction 3 and an altered accumulation pattern in the 60/80S fraction 4 and 80S fraction 5. In the control cells expressing Sup35-ΔN, aggregates failed to form (Figure 3A) and the distribution of the ubiquitinated species between the 60S and 80S fractions was similar to the empty-vector control (Figure 3B, right and left; Supplementary Figure S2B). To better understand the pattern of ubiquitination in the intermediate fraction 4, which contains both 60S and 80S ribosomes, we lowered concentration of sucrose in gradients and collected fractions of a smaller volume (Figure 3C). The improved separation of the 60S and 80S fractions confirmed a significantly lowered amount of 60S-associated ubiquitinated polypeptides in cells with aggregated Sup35 (compare fractions 4 and 5 between different panels in Figure 3C).
Figure 3.
Translational activity of conventional release factors promotes ubiquitination of 60S·NCs. (A) Induction of high molecular weight (HMW) aggregates of Sup35 in PTET-O7-CDC48 cells by overexpression (OE) of PGAL1-driven Sup35FLAG (full length, FL). Cells were shifted to galactose medium for 28 h to induce Sup35FLAG and Dox was added to cultures for an additional 20 h. Cellular lysates were resolved by electrophoresis on a 10% SDS-acrylamide gel to assess expression of Sup35FLAG (SDS-PAGE, bottom) and on a 1.8% SDS-agarose gel to detect its aggregates (SDD-AGE, top) by western blotting. Expression of Sup35-ΔN or empty vector (V) was used as controls. (Band C) Ribosomal species present in cellular lysates from the same cultures as analyzed in (A) were subjected to sedimentation through 15–45% (B) or 15–42% (C) sucrose gradients. Proteins isolated from individual fractions were separated on 10% SDS-acrylamide gels and probed with anti-ubiquitin, anti-Rpl3 and anti-Rps14 antibodies. Representative blots are shown; see Supplemental Figure S2B for another experimental repeat with samples separated on one gel.
To exclude the possibility that the release factors are involved in the generation of ubiquitinated NCs only in cells with deficient Cdc48 function, we overexpressed full-length Sup35 in a strain carrying wild-type CDC48 (Supplementary Figure S2C). Detection of ubiquitinated species in these cells required a much longer exposure of the blots compared to the PTET-O7-CDC48 cells used above, consistent with a lower overall abundance of ubiquitinated NCs in cells with normal Cdc48 function. Nonetheless, we observed a similar pattern of the specific reduction in the 60S-associated ubiquitin signal (Supplementary Figure S2D).
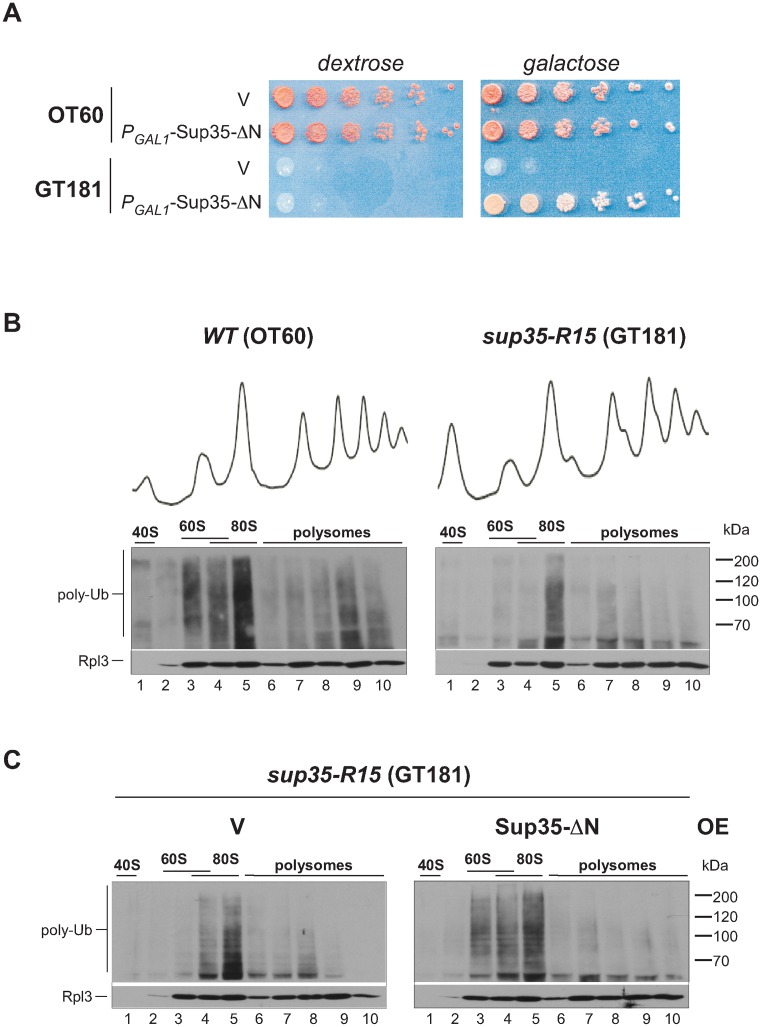
To test the role of the release factors in the generation of Ltn1-Cdc48 substrates through a different approach, we next created a strain carrying a hypomorphic sup35 allele (see Materials and Methods for details). Reduced activity of Sup35 in the newly generated sup35-R15 strain resulted in a temperature sensitive growth phenotype (Supplementary Figure S3) that was rescued by expression of the translationally active Sup35-ΔN (Figure 4A). We examined the distribution of ubiquitinated NCs across ribosomal fractions in the sup35-R15 strain and its parental wild-type strain after shifting these cells from 25°C to a semi-permissive temperature (30°C) for 4 h (Figure 4B). We found that the ubiquitin signal in the 60S fraction was greatly reduced compared to wild-type cells, although these cells continued to display the robust accumulation of ubiquitinated 80S complexes, resembling the effect of the ltn1Δ deletion (compare Figure 4B, right with Figure 1C). The accumulation of ubiquitinated products in the 60S fraction was restored when the translationally competent Sup35-ΔN was supplied by expression from a plasmid (Figure 4C). Taken together, these results suggest that the translational activity of the canonical release factors gives rise to a substantial portion of Ltn1-modified NCs.
Figure 4.
Reduced ubiquitination level of 60S·NCs in the GT181 strain harboring the hypomorphic sup35-R15 allele. (A) GT181 (sup35-R15) and isogenic wild-type control strain OT60 were transformed with an empty vector (V) or an expression construct for the translationally active N-terminally truncated Sup35-ΔN under the control of a galactose-inducible promoter (PGAL1-Sup35-ΔN). Serial 5-fold dilutions were spotted on dextrose- and galactose-containing synthetic media plates and incubated at 30°C for 3 days. (B) Wild-type and sup35-R15 strains were grown in YPDA at 25°C to mid-log phase and shifted to 30°C for 4 h. Cell lysates were centrifuged through sucrose gradients and analyzed as in Figure 1. (C) sup35-R15 cells were transformed with an empty vector (V) or PGAL1-Sup35-ΔN expression construct. Sup35-ΔN was induced by growth in galactose-containing medium for 16 h followed by shift to 30° in YPDA for 4 h. Polyubiquitinated NCs present in ribosomal fractions were analyzed as in Figure 1. Each strain was analyzed a minimum of three times; representative blots are shown.
Accumulation of ubiquitinated 60S-NCs in a tRNA-free form
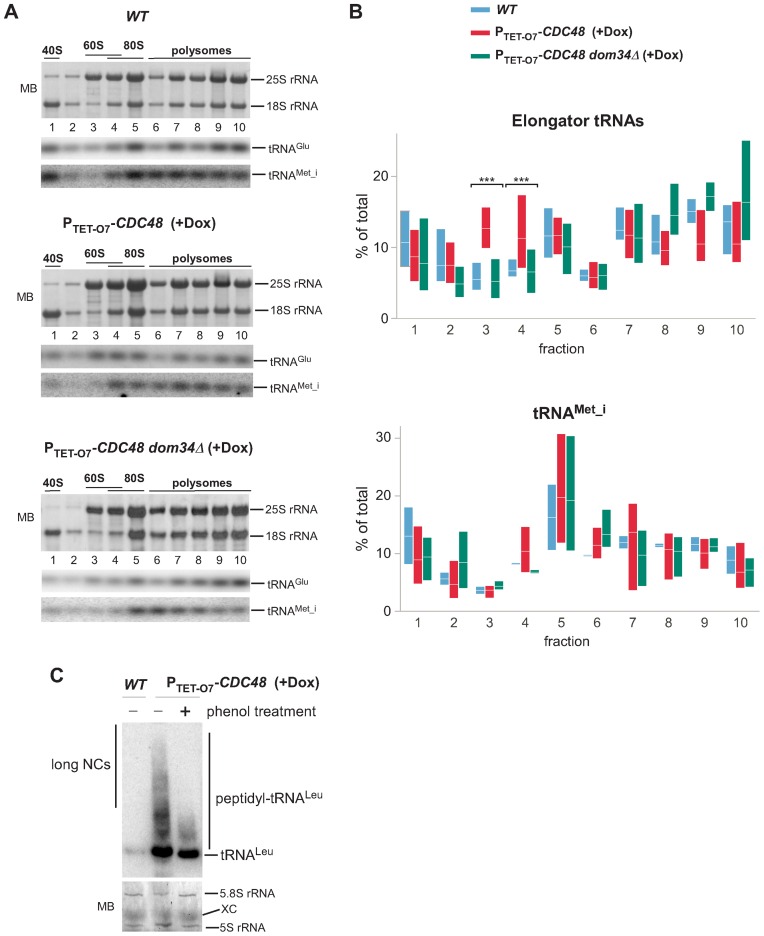
During canonical translation termination, the release factor complex Sup45–Sup35 promotes the cleavage of tRNAs from NCs (51,52). In contrast, the splitting of ribosomal subunits by the release-like factors Dom34–Hbs1 occurs without accompanying peptidyl-tRNA hydrolysis (6) since Dom34 lacks the GGQ motif required for catalysis (53,54). Previous biochemical and cryo-electron microscopy studies obtained with model substrates revealed the presence of peptidyl-tRNA in 60S subunits in cells harboring either a temperature-sensitive cdc48-3 allele (4) or catalytically inactive Ltn1 (11,13). Recent studies have shown that the exposed tRNA moiety in tRNA-linked 60S·NC species can interact with Rqc2 (11,13), which, along with Ltn1, is part of the RQC complex (2,10). Considering the possibility that Sup45–Sup35 can also generate Ltn1 substrates (Figures 3 and 4), we wondered to what extent peptidyl-tRNAs contribute to the total pool of the ubiquitinated species associating with 60S ribosomal complexes. To address this, we examined sedimentation profiles of tRNAs after treating individual ribosome-containing gradient fractions with proteinase K, which hydrolyzes the ester bond between tRNAs and the conjugated peptides (55). We determined the relative amounts of four different elongator tRNAs (tRNAGlu, tRNALeu, tRNAVal, tRNAThr) in each gradient fraction by quantitative northern hybridizations. By comparing tRNA distributions in different strains (Figure 5A–B, Supplementary Figure S4), we were able to observe a decrease in elongator tRNA amounts associated with polysomes (fractions 6–10) after Cdc48 depletion, expected from partially repressed translation in these cells, and increase in polysome-associated tRNAs in dom34Δ cells, as was previously described (56). Remarkably, the tRNA distribution data indicated a significant increase in the amount of elongator tRNAs in the 60S fraction 3 in Cdc48-depleted cells relative to wild-type cells (Figure 5B, Wilcoxon rank sum test, P = 2 × 10−4) and in fraction 4 containing 60S and 80S (Wilcoxon rank sum test, P = 4 × 10−4). In contrast to the elongator tRNA species, we did not observe a similar enrichment in the parallel analysis of the initiator tRNAMet_i, which is not present in peptidyl-tRNAs (Figure 5A and B, Supplementary Figure S4A). We also observed enrichment in elongator tRNAs in the 60S fraction 3 in ltn1Δ cells (Supplementary Figure S4B; Wilcoxon rank sum test, P = 4 × 10−3), although this effect was less pronounced than in Cdc48-depleted cells.
Figure 5.
Enrichment of elongator tRNAs in 60S fractions in Cdc48-depleted cells depends on Dom34. We treated individual ribosome-containing sucrose gradient fractions with proteinase K to separate tRNAs from the conjugated peptides. Next, the extracted RNAs were sequentially analyzed by northern hybridization of the same membrane with probes that detect tRNAMet_i, tRNAGlu, tRNALeu, tRNAVal, tRNAThr. The 18S and 25S rRNAs were analyzed on all membranes to verify alignment of the gradient fractions. (A) A representative example of the distribution of rRNA and tRNAs in gradient fractions is shown; see Supplementary Figure S4 for full hybridization sets. (B) The percentage of tRNAs in each fraction relative to the total was determined using phosphorimager quantification of the hybridization signals. The floating bars represent the full range of values obtained in each fraction (min to max) and the crossing lines indicate the mean. Four elongator tRNAs (top) or tRNAMet_i (bottom) were quantified in gradients prepared from 2 biological replicates of wild-type cells, 3 replicates of PTET-O7-CDC48 and three replicates of dom34Δ PTET-O7-CDC48 cells. (C) RNA in ribosomes pelleted from the 60S gradient fractions of the indicated strains was resolved on an acid-urea polyacrilamide gel and analyzed by northern hybridization using a radioactively labeled probe against tRNALeu. Treatment with phenol (‘+’) was used to selectively remove long peptidyl-tRNAs. Prior to hybridization, the membrane was stained with methylene blue (MB) to control loading by visualizing 5S and 5.8S rRNAs in the 60S subunits. XC, the xylene cyanol band.
To confirm that the observed increase in elongator tRNAs in the 60S fraction of Cdc48-depleted cells reflects accumulation of peptidyl-tRNA species (11–13), we precipitated 60S-containing complexes from PTET-O7-CDC48 cells and wild-type control cells by ultracentrifugation and resolved them by electrophoresis on acid-urea polyacrylamide gels. Hybridization with a radioactively labeled tRNA probe revealed the increased quantities of peptidyl-tRNAs in the 60S complexes isolated from Cdc48-depleted, but not wild-type cells (Figure 5C). Treatment of the peptidyl-tRNAs with a phenol-containing reagent prior to electrophoresis selectively removed the top part of the smear of peptidyl-tRNAs (Figure 5C), consistent with the property of tRNAs conjugated to peptide chains longer then 80 residues to partition to the organic phase (36). Together with the gradient tRNA distribution analysis (Figure 5A and B, Supplementary Figure S4), these results support the notion that tRNA-linked NCs accumulate in the 60S subunits in Cdc48-depleted cells, in agreement with the current models of RQC (18).
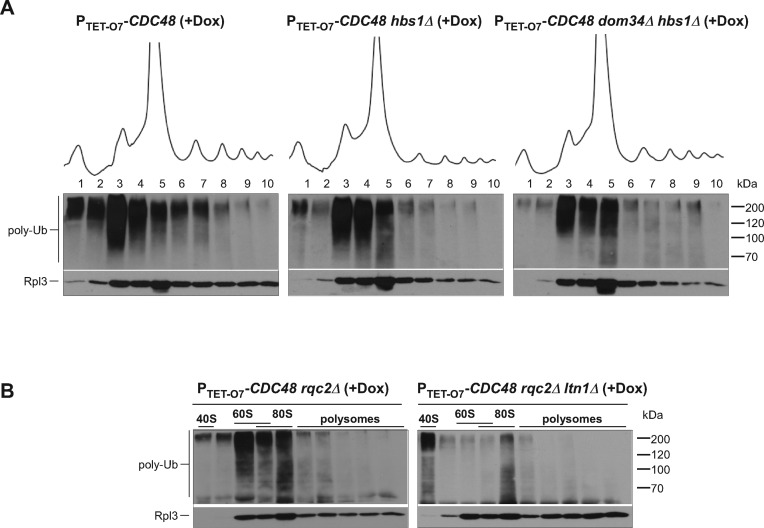
If Dom34 is responsible for the generation of tRNA-linked NCs, as suggested by previous studies (6,7), its lack in the cell should abolish accumulation of elongator tRNAs in the 60S fraction. As predicted, introducing a dom34Δ deletion into PTET-O7-CDC48 cells prevented the increase in elongator tRNAs in fractions 3 and 4 after Cdc48 depletion (Figure 5A–B, Supplementary Figure S4A; Wilcoxon rank sum test, P < 1 × 10−4). However, as shown above, the western blot analysis of the gradient fractions indicates that large amounts of ubiquitin conjugates continue to accumulate in the 60S fraction in dom34Δ cells (Figure 2). We confirmed that accumulation of 60S-associated ubiquitinated species also occurred in hbs1Δ cells after Cdc48 depletion (Figure 6A), even though like in dom34Δ cells, elongator tRNAs did not accumulate in the 60S fraction in hbs1Δ cells (Supplementary Figure S4C). These data are consistent with the idea that many 60S·NC complexes targeted for Ltn1-Cdc48-mediated degradation do not contain tRNA. To further substantiate this result, we evaluated how much the total ubiquitination of the 60S·NC complexes depends on Rqc2, which binds to the tRNA in 60S·NC-tRNA complexes (12,13). We reasoned that if most Ltn1 substrates in the cell are derived from Sup45–Sup35 activities, Rqc2 would be expected to impact a relatively minor pool of 60S·NC-tRNA species that are products of Dom34-Hbs1. Indeed, the distribution of ubiquitinated NCs in the rqc2Δ PTET-O7-CDC48 strain was virtually unaffected relative to the parental PTET-O7-CDC48 strain (Figure 6B, left), whereas the ltn1Δ rqc2Δ double deletion abolished the formation of ubiquitinated 60S species (Figure 6B, right).
Figure 6.
Accumulation of 60S-associated ubiquitinated products does not require Hbs1, Dom34 or Rqc2. The indicated strains were grown in the presence of Dox for 20 h, lysed and ribosomes were separated by centrifugation through a sucrose gradient. Proteins were extracted from each fraction and analyzed as in Figure 1. Gradients were repeated three times with hbs1Δ PTET-O7-CDC48 and dom34Δ hbs1Δ PTET-O7-CDC48 cells, and three times with rqc2Δ PTET-O7-CDC48 and ltn1Δ rqc2Δ PTET-O7-CDC48 cells. Representative blots are shown.
Based on these data, we conclude that only a small fraction of ubiquitinated 60S·NC complexes accumulating in Cdc48-depleted cells contain peptidyl-tRNA and are contigent on Dom34/Hbs1-mediated release. These results support a model in which faulty translation products can be targeted for Ltn1-mediated ubiquitination and Cdc48-assisted degradation through canonical release mechanisms that involve tRNA hydrolysis.
DISCUSSION
Much information on ribosome-associated QC has been obtained through analysis of translation products derived from model mRNA substrates that either lack a stop codon or encode sequences (e.g. polylysine) that induce ribosome stalling. In this work, we took a different approach by examining the total population of ubiquitinated nascent polypeptide chains associated with different ribosomal complexes. We made use of the observation that deficiency of Cdc48 results in the accumulation of the ubiquitin-labeled proteome in cells (57), including polypeptides that are modified cotranslationally (4), to assess upstream steps in ribosome-associated degradation. In agreement with prior work utilizing translation reporters, our data indicate that the E3 ligase Ltn1 carries out most, if not all 60S-associated substrate ubiquitination in yeast cells. Importantly, our results establish that Dom34-Hbs1 are not the only factors responsible for generating Ltn1-Cdc48 substrates. Rather, our data point to the canonical release factors Sup45–Sup35 as the important mediators in ribosome-associated degradation that supply the bulk of 60S·NC substrates for Ltn1 (Figure 7).
Figure 7.

Two alternative pathways generate substrates for Ltn1-Cdc48. Our data are consistent with a model for Ltn1-Cdc48-mediated translational surveillance in which one upstream pathway utilizes release factors Sup45–Sup35 and captures ribosomes stalled at any position on mRNA, giving rise to 60S·NC complexes polyubiquitinated by Ltn1 (left). Another upstream pathway requires release-like factors Dom34-Hbs1 and operates on ribosomes stalled at the 3′ end of non-stop and truncated messages (right). The Dom34-Hbs1-generated complex, 60S·NC-tRNA, is recognized by the tRNA-binding protein Rqc2 and Ltn1. Prior to Cdc48-mediated release of the ubiquitinated NC, tRNA undergoes hydrolysis to allow NC passage through the exit tunnel of 60S.
The existence of more than one pathway leading to Cdc48-mediated clearance of NCs could reflect the need to target different types of arrested ribosomes. Recent studies of model stalling substrates have suggested that Sup35-Sup45 can terminate translation at internal polylysine segments (28). By contrast, Dom34 targets ribosomes arrested on truncated mRNAs and near the ends of 3′ UTRs, but does not appear to facilitate the splitting of ribosomes on several types of coding sequences known to cause stalling (26,58). Consistently, recent in vitro studies (7,8), supported by structural data (59), revealed that subunit dissociation by Dom34-Hbs1 requires a short length of the mRNA's 3′ extension relative to the codon in the P-site, 23 and 9 nucleotides for yeast and human Dom34-Hbs1, respectively. Thus, one possibility is that Dom34-Hbs1 evolved to recognize a specific type of substrates that cannot be efficiently resolved through canonical release factors, such as ribosomes reaching the 3′ end of nonstop messages or inactive 80S ribosomes (60,25). The existence of alternative mechanisms to resolve translational stalls may explain why DOM34 and HBS1 are nonessential in yeast, unlike the key components of the translation rescue systems in bacteria (61).
Another specialized function of Dom34-Hbs1 may be to generate tRNA-linked 60S·NC complexes ((6), Figure 5A, B). It was recently shown that tRNA in these complexes recruits Rqc2, which promotes synthesis of non-templated C-terminal extensions of NCs (CAT tails), required for the transmission of the translation-stress signal to Hsf1 (10,13). Because cotranslational clearance of misfolded peptides and rescue of stalled ribosomes is a process that must take place in cells at all times, some way to avoid continuous translation-stress signaling in clearly needed. A plausible scenario consistent with our data is that most of the ‘routine’ clearance of aberrant NCs is performed by the release factors and accompanied by peptidyl-tRNA hydrolysis, thereby carrying no intrinsic proteotoxic signaling function. This could also explain why mutations in tRNA-interacting residues of Rqc2 (13) or rqc2Δ deletion only weakly affect the clearance of model reporters (10) and recruitment of Cdc48 to 60S·NC complexes (2).
Considering that the canonical release factors are capable of efficiently discriminating against sense codons under normal circumstances (6,62), an important question for the future studies is whether these factors function in QC by mediating detection of aberrant translation events such as ribosome slippage, which could lead to termination on premature stop codons, or their function involves a previously uncharacterized type of interactions with ribosomes, distinct from conventional termination. Intriguingly, there is prior evidence that Sup45–Sup35 may be capable of binding to stalled ribosomal complexes that contain a sense codon in the A site (63,64,28). Another interesting question concerns the activity of Ltn1 on 60S·NC substrates in the absence of tRNA or Rqc2. Given the highly flexible structure of Ltn1 (65), this ubiquitin ligase could have additional modes of interaction with ribosomal complexes in addition to the previously observed conformation in cryo-EM structures in which its N-terminus contacts Rqc2 at the subunit interface (11–13). Indeed, previous in vitro studies of these proteins' mammalian homologs suggest that Ltn1-mediated ubiquitination of aberrant translation products may occur, albeit with reduced efficiency, without Rqc2 (40).
An unconventional activity of Sup45–Sup35 on stalled ribosomes may also have ramifications for gene expression. The switches between soluble and aggregated [PSI+] prion states of Sup35 impart complex phenotypes, which have been attributed mainly to altered read-through of stop codons, but it is clear now that the full spectrum of the observed effects cannot be fully accounted for by this mechanism (66–68). We suggest that deficiency in functional Sup45–Sup35 conferred by [PSI+] elements or conditions that affect Sup35 aggregation such as aging and heat stress (69,70) may lead to changes in the proteome via increased synthesis of proteins with built-in elongation stalling sites. This could serve as a regulatory mechanism contributing to survival under unfavorable conditions. Additionally, environmental insults that induce ribosome stalling might elicit differential responses depending on whether the release factors are available to trigger premature termination on selected mRNAs, with the subsequent destruction of the partially synthesized polypeptides via the Ltn1–Cdc48 pathway.
Supplementary Material
Acknowledgments
We are thankful to S.G. Inge-Vechtomov (St. Petersburg State University, St. Petersburg, Russia) for the sup35/45 tester strains, to D. Bedwell (University of Alabama, Birmingham, USA) for anti-Sup35C antibodies and to John Woolford (Carnegie Mellon University, Pittsburgh, USA) for anti-Rps14 antibodies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UMDNJ Foundation (to N.S.); National Institutes of Health [GM093294 to T.A.C. and GM074091 to D.G.P.]; National Science Foundation [MCB-1516872 to Y.O.C.]. Funding for open access charge: Rowan University intramural support funds (to N.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang F., Canadeo L.A., Huibregtse J.M. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie. 2015;114:127–133. doi: 10.1016/j.biochi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Defenouillère Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A., et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duttler S., Pechmann S., Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma R., Oania R.S., Kolawa N.J., Deshaies R.J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F., Durfee L.A., Huibregtse J.M. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoemaker C.J., Eyler D.E., Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisareva V.P., Skabkin M.A., Hellen C.U.T., Pestova T.V., Pisarev A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoemaker C.J., Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.-P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I., et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.-W., Zhou S., King D., Shen P.S., Weibezahn J., et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyumkis D., Oliveira dos Passos D., Tahara E.B., Webb K., Bennett E.J., Vinterbo S., Potter C.S., Carragher B., Joazeiro C.A.P. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15981–15986. doi: 10.1073/pnas.1413882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao S., Brown A., Santhanam B., Hegde R.S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen P.S., Park J., Qin Y., Li X., Parsawar K., Larson M.H., Cox J., Cheng Y., Lambowitz A.M., Weissman J.S., et al. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bengtson M.H., Joazeiro C.A.P. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao S., von der Malsburg K., Hegde R.S. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C., Glynne R.J., Wu H., Masliah E., Joazeiro C.A.P., et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.-L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandman O., Hegde R.S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrova L.N., Kuroha K., Tatematsu T., Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito-Harashima S., Kuroha K., Tatematsu T., Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B., Han Y., Qian S.-B. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowder J.J., Geigges M., Gibson R.T., Fults E.S., Buchanan B.W., Sachs N., Schink A., Kreft S.G., Rubenstein E.M. Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J. Biol. Chem. 2015;290:18454–18466. doi: 10.1074/jbc.M115.663559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von der Malsburg K., Shao S., Hegde R.S. The ribosome quality control pathway can access nascent polypeptides stalled at the Sec 61 translocon. Mol. Biol. Cell. 2015;26:2168–2180. doi: 10.1091/mbc.E15-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Elzen A.M.G., Schuller A., Green R., Séraphin B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 2014;33:265–276. doi: 10.1002/embj.201386123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda R., Ikeuchi K., Nomura S., Inada T. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells. 2014;19:1–12. doi: 10.1111/gtc.12106. [DOI] [PubMed] [Google Scholar]
- 28.Chiabudini M., Tais A., Zhang Y., Hayashi S., Wölfle T., Fitzke E., Rospert S. Release factor eRF3 mediates premature translation termination on polylysine-stalled ribosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2014;34:4062–4076. doi: 10.1128/MCB.00799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Chernoff Y.O., Lindquist S.L., Ono B., Inge-Vechtomov S.G., Liebman S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 31.Newnam G.P., Wegrzyn R.D., Lindquist S.L., Chernoff Y.O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirmer E.C., Homann O.R., Kowal A.S., Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol. Biol. Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagliocco F.A., Moore P.A., Brown A.J. Polysome analysis. Methods Mol. Biol. 1996;53:297–311. doi: 10.1385/0-89603-319-8:297. [DOI] [PubMed] [Google Scholar]
- 34.Mansour F.H., Pestov D.G. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal. Biochem. 2013;441:18–20. doi: 10.1016/j.ab.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestov D.G., Lapik Y.R., Lau L.F. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol. 2008 doi: 10.1002/0471143030.cb2211s39. Chapter 22, Unit 22.11. [DOI] [PubMed] [Google Scholar]
- 36.Janssen B.D., Diner E.J., Hayes C.S. Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Methods Mol. Biol. 2012;905:291–309. doi: 10.1007/978-1-61779-949-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kryndushkin D.S., Alexandrov I.M., Ter-Avanesyan M.D., Kushnirov V.V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 38.Allen K.D., Wegrzyn R.D., Chernova T.A., Müller S., Newnam G.P., Winslett P.A., Wittich K.B., Wilkinson K.D., Chernoff Y.O. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.-T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Shao S., Hegde R.S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodersen D.E., Clemons W.M., Carter A.P., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 42.Cabañas M.J., Vázquez D., Modolell J. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur. J. Biochem. FEBS. 1978;87:21–27. doi: 10.1111/j.1432-1033.1978.tb12347.x. [DOI] [PubMed] [Google Scholar]
- 43.Ganoza M.C., Kiel M.C. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 2001;45:2813–2819. doi: 10.1128/AAC.45.10.2813-2819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chernoff Y.O., Inge-Vechtomov S.G., Derkach I.L., Ptyushkina M.V., Tarunina O.V., Dagkesamanskaya A.R., Ter-Avanesyan M.D. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 1992;8:489–499. doi: 10.1002/yea.320080702. [DOI] [PubMed] [Google Scholar]
- 45.Derkatch I.L., Bradley M.E., Hong J.Y., Liebman S.W. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 46.Osherovich L.Z., Weissman J.S. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 47.Vishveshwara N., Bradley M.E., Liebman S.W. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liebman S.W., Chernoff Y.O. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ter-Avanesyan M.D., Kushnirov V.V., Dagkesamanskaya A.R., Didichenko S.A., Chernoff Y.O., Inge-Vechtomov S.G., Smirnov V.N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 50.Paushkin S.V., Kushnirov V.V., Smirnov V.N., Ter-Avanesyan M.D. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee H.H., Kim Y.-S., Kim K.H., Heo I., Kim S.K., Kim O., Kim H.K., Yoon J.Y., Kim H.S., Kim D.J., et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 54.Graille M., Chaillet M., van Tilbeurgh H. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. J. Biol. Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 55.Vidales F.J., Bernabeu C., Ballesta J.P. Peptidyl transfer ribonucleic acid hydrolase activity of proteinase k. Biochemistry. 1979;18:4155–4158. doi: 10.1021/bi00586a016. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharya A., McIntosh K.B., Willis I.M., Warner J.R. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol. Cell. Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai R.M., Li C.C. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 58.Guydosh N.R., Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker T., Armache J.-P., Jarasch A., Anger A.M., Villa E., Sieber H., Motaal B.A., Mielke T., Berninghausen O., Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 60.Strunk B.S., Novak M.N., Young C.L., Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Himeno H., Nameki N., Kurita D., Muto A., Abo T. Ribosome rescue systems in bacteria. Biochimie. 2015;114:102–112. doi: 10.1016/j.biochi.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 62.Chavatte L., Frolova L., Laugâa P., Kisselev L., Favre A. Stop codons and UGG promote efficient binding of the polypeptide release factor eRF1 to the ribosomal A site. J. Mol. Biol. 2003;331:745–758. doi: 10.1016/s0022-2836(03)00813-1. [DOI] [PubMed] [Google Scholar]
- 63.Janzen D.M., Frolova L., Geballe A.P. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell. Biol. 2002;22:8562–8570. doi: 10.1128/MCB.22.24.8562-8570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doronina V.A., Wu C., de Felipe P., Sachs M.S., Ryan M.D., Brown J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lyumkis D., Doamekpor S.K., Bengtson M.H., Lee J.-W., Toro T.B., Petroski M.D., Lima C.D., Potter C.S., Carragher B., Joazeiro C.A.P. Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1702–1707. doi: 10.1073/pnas.1210041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.True H.L., Lindquist S.L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 67.True H.L., Berlin I., Lindquist S.L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 68.Baudin-Baillieu A., Legendre R., Kuchly C., Hatin I., Demais S., Mestdagh C., Gautheret D., Namy O. Genome-wide translational changes induced by the prion [PSI+] Cell Rep. 2014;8:439–448. doi: 10.1016/j.celrep.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 69.Saarikangas J., Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife. 2015;4:e06197. doi: 10.7554/eLife.06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace E.W.J., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.