Abstract
The association of DSIF and NELF with initiated RNA Polymerase II (Pol II) is the general mechanism for inducing promoter-proximal pausing of Pol II. However, it remains largely unclear how the paused Pol II is released in response to stimulation. Here, we show that the release of the paused Pol II is cooperatively regulated by multiple P-TEFbs which are recruited by bromodomain-containing protein Brd4 and super elongation complex (SEC) via different recruitment mechanisms. Upon stimulation, Brd4 recruits P-TEFb to Spt5/DSIF via a recruitment pathway consisting of Med1, Med23 and Tat-SF1, whereas SEC recruits P-TEFb to NELF-A and NELF-E via Paf1c and Med26, respectively. P-TEFb-mediated phosphorylation of Spt5, NELF-A and NELF-E results in the dissociation of NELF from Pol II, thereby transiting transcription from pausing to elongation. Additionally, we demonstrate that P-TEFb-mediated Ser2 phosphorylation of Pol II is dispensable for pause release. Therefore, our studies reveal a co-regulatory mechanism of Brd4 and SEC in modulating the transcriptional pause release by recruiting multiple P-TEFbs via a Mediator- and Paf1c-coordinated recruitment network.
INTRODUCTION
The transcription by RNA polymerase II (Pol II) consists of several tightly coordinated steps. While the control of transcription initiation has been a long-held paradigm, recent evidence indicates that transcription elongation is another rate-limiting step for governing the rapid expression of inducible genes in metazoans (1–3). Genome-wide surveys in different systems reveal that for more than 30% of the transcriptionally active genes, the initiation has completed even at unstimulated state, while the Pol II is yet stalled at the promoter-proximal regions, 20–60 nucleotides downstream of transcription start site (4–10). Upon stimulation, the paused Pol II is rapidly released and progressed into productive elongation, leading to the synthesis of full-length mRNA. Moreover, most recent functional studies have revealed that genes regulated by this promoter-proximal pausing step are important in response to the developmental and environmental signals (8,11–13).
Although the precise mechanism for promoter–proximal Pol II pausing is still under debate, it is widely accepted that the association of two negative factors, DSIF (DRB sensitivity-inducing factor) and NELF (negative elongation factor), with the initiated Pol II at the promoter–proximal region is essential for Pol II pausing (1–3,14,15). While DSIF is a heterodimer of Spt4 and Spt5, NELF is a multisubunit complex consisting of NELF-A, -B, -C/D and -E (15). Biochemical studies reveal that the interaction of Spt5 and NELF-A with Pol II and the association of NELF-E with nascent mRNA may cooperatively induce Pol II pausing (15–18). This Pol II pausing not only acts as a quality checkpoint for 5′-capping of nascent mRNA, but also keeps the promoters in an open state (1,3,19). The release of paused Pol II depends on the promoter recruitment of positive transcription elongation factor P-TEFb, consisting of Cdk9 and Cyclin T. P-TEFb mediates phosphorylation of the C-terminal domain (CTD) of Pol II at Ser2, the Spt5 of DSIF and the NELF-E of NELF complex. These P-TEFb-mediated phosphorylations are prerequisite for the release of promoter–proximally paused Pol II (1–3,15,20–23).
The activity of P-TEFb is tightly regulated in cells, with majority of P-TEFb sequestrated in an inactive 7SK snRNP complex that contains 7SK snRNA, nuclear proteins HEXIM1/2, MePCE and LARP7 (3,24). In response to stress, P-TEFb is liberated from 7SK snRNP and is recruited to the promoters via P-TEFb recruitment factors (1,3,25–27). Ample evidence indicates that the bromodomain-containing protein Brd4 and the super elongation complex (SEC) are capable of recruiting P-TEFb to promoters (1,3). Brd4 belongs to BET family that contains two bromodomains and an extraterminal domain (28). Distinct from the other BET proteins, Brd4 contains a unique P-TEFb interaction domain (29). Upon stimulation, Brd4 binds to and recruits active form of P-TEFb to promoters to modulate Pol II processivity (30–32). SEC is a multisubunit complex consisting one of four AFF scaffold proteins (AFF1–AFF4), one of three ELL proteins (ELL1–ELL3), and an ENL (or its analogue AF9). Depending on cell type, the compositions of SEC can be varied to generate diverse subtypes of SEC (3,33). Same as Brd4, SEC is able to bind to and recruit P-TEFb to promoters via the interaction with Med26 subunit of Mediator (34), or Paf1 of polymerase-associated factor complex (Paf1c) (35). Besides Brd4 and SEC, several sequence-specific DNA-binding transcription factors have been reported to be able to recruit P-TEFb (36), but some of them were recently found to interact with Brd4/P-TEFb complex, rather than P-TEFb itself (37,38).
Although emerging evidence indicates that P-TEFb, Brd4 and SEC are essential for the transcription elongation, the detailed mechanism by which these three factors regulate transcription elongation remains unclear. Here, we show that Brd4 and SEC cooperatively regulate the transcriptional pause release by recruiting multiple P-TEFbs via a Mediator- and Paf1c-coordinated recruitment network. Moreover, we demonstrate that the phosphorylation of Ser2 of Pol II CTD is not essential for pausing-to-elongation transition.
MATERIALS AND METHODS
Materials
The detailed information of chemicals, antibodies and plasmids are described in Supplementary Information.
Preparation of LSF, HSF and LSEN with stepwise fractionation protocol
The low-salt fraction (LSF) that contains chromatin-free factors, low-salt extracted nuclei (LSEN) and high-salt fraction (HSF) that contain transcriptionally engaged factors were prepared with stepwise fractionation protocol (see Supplementary Information) (30,31).
Immunoprecipitation (IP) and Western blot (WB) analysis
Flag- or HA-tagged proteins and their associated factors were isolated by anti-Flag or anti-HA IPs from LSF, HSF or nuclear extract (NE) of transfected or infected HeLa cells as previously described (31,39). The levels of desired proteins in IP products, fractionated samples, NE or cell lysates were analyzed by WB with corresponding antibodies.
Mass spectrometry analysis
To identify the P-TEFb-associated transcriptional factors, the P-TEFb was isolated from HSF of F1C2 (Cdk9-f) cells by anti-Flag affinity purification and analyzed with liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS/MS) system (40,41). The normalized spectral abundance factors (NSAFs) were applied to calculate each detected protein to estimate relative protein levels (41). To identify the phospho-residues of NELF-A, HeLa cells expressing Flag-tagged NELF-A (NELF-A-f) were treated with solvent or 10 mM of HMBA for 2 h and subjected to high-salt buffer (0.5 M NaCl) extraction. The anti-Flag affinity-purified NELF-A-f from high-salt extracts was subjected to MS analysis for phosphorylated residues on NELF-A. Three phospho-residues, S374, T288 and T168, were most abundant in spectrum count of 73, 15 and 12 after HMBA treatment. The mass spectrometry data from this publication have been submitted to the PRIDE Archive database at http://www.ebi.ac.uk/pride/archive/ (see Supplementary Information for details).
In vitro kinase assay
For P-TEFb-mediated phosphorylation of T775 of Spt5 or NELF-A, P-TEFb was purified by anti-Flag antibody from HSF of F1C2 (Cdk9-f) cells. Unphosphorylated Spt5 or NELF-A was purified with anti-Flag affinity purification from the NE of HeLa cells expressing Spt5-f or NELF-A-f treated with 300 nM of Flavopiridol for 2 h. The in vitro kinase assay was performed as previously described (27) and the phosphorylation levels were detected by Western blot with a specific anti-phospho-Spt5 antibody. The phosphorylation of NELF-A-f by P-TEFb was detected by WB with antibodies against phospho-Ser (ph-Ser), phospho-Thr (ph-Thr) or phospho-NELF-A.
Luciferase assay
HeLa cells with an integrated HIV-LTR-luciferase reporter gene (HIV-LTR-Luc) were infected with lentiviruses expressing desired shRNA for 48–96 h, followed by incubation with 5 mM of HMBA for 4–6 h as indicated. Cell lysates were prepared and the luciferase activity was measured as previously described (25). Data from three replicates were averaged and presented as fold induction compared to untreated cells. All values were expressed as Mean±SD of three replicates. P-values were assessed using two-tailed Student's t-test.
qRT-PCR analysis of transcription elongation products
The quantitative real-time polymerase chain reaction (qRT-PCR) was performed as previously described (42). The qPCR primers corresponding to gene body (+600 bp downstream of transcription start site) were used for analyzing the level of transcription elongation products. The primer sequences are shown in the Supplementary Information. All values were expressed as Mean ± SD of three replicates. P-values were assessed using two-tailed Student's t-test.
Chromatin immunoprecipitation (ChIP)-qPCR
ChIP was performed in HeLa or HCT116 cells and immunoprecipitated DNA was analyzed by qRT-PCR as previously described (39,42). All values were expressed as Mean ± SD of three replicates. P-values were assessed using two-tailed Student's t-test. The primer sequences used for ChIP-qPCR analysis in this study are shown in the Supplementary Information.
ChIP-silver staining assay
To test the bulk occupancy of RNA Pol II on chromatin, the ChIP was carried out following the protocol of SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology). In brief, the formaldehyde cross-linked cells were extracted with low-salt buffers to remove the chromatin-free RNA and proteins. The nuclei were then incubated with Micrococcal Nuclease (MNase) (10 μg DNA per 1 unit MNase) to digest genomic DNA to the optimal length (1–2 nucleosomes), followed by sonication to break down the nuclear membrane in the presence of 0.1% SDS. Before ChIP, an aliquot of lysate was removed for WB of the levels of nucleosomal histone H3. ChIP was performed with anti-Rpb1 (Pol II) antibody (40 μg DNA per 1 μg antibody). An aliquot of chromatin immunoprecipitated (ChIPed) DNA was resolved by polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining with the protocol of PlusOne DNA Silver Staining Kit (Cat#: 17-6000-30, Amersham Biosciences).
ChIP-Seq analysis
For ChIP-Seq analysis, the above ChIPed DNAs were subjected to high-throughput sequencing with Illumina HiSeq2500 in RiboBio Co. Ltd. (Guangzhou, China). After quality assessment, the raw ChIP-seq reads were trimmed for adaptor sequences and retained 46 bp from the 3′-end. Reads were then mapped to human genome (GRCh37/hg19) using Bowtie 1.1.1 (43) with 3-mismatches. Enriched Pol II binding peaks were identified and then annotated by HOMER v4.7 with default settings (44,45) (http://biowhat.ucsd. edu/homer). To calculate Pol II pause ratio (TR), the number of ChIP-seq reads over input background at promoter region (-50 to +300 bp from TSS) and gene body (+300 bp to the end of genes) of each gene was counted by the analyzeRNA.pl command of HOMER with ‘-tss’ option, and then normalized by the concentration of input DNA for ChIP. Only gene length > 500 bp were analyzed. In order to assess the statistical significance of Pol II pausing, the Benjamini–Hochberg method corrected Fisher's exact tests were performed to control the false discovery (P-value < 0.005). The criterion of Pol II pausing was defined as TR (the relative ratio of promoter read density/gene body read density) >4.0 (6). The ChIP-seq data from this publication have been submitted to the GEO database at http://www.ncbi.nlm.nih.gov/geo/ (see Supplementary Information for details).
RESULTS
Brd4 and SEC are responsible for global recruitment of P-TEFb
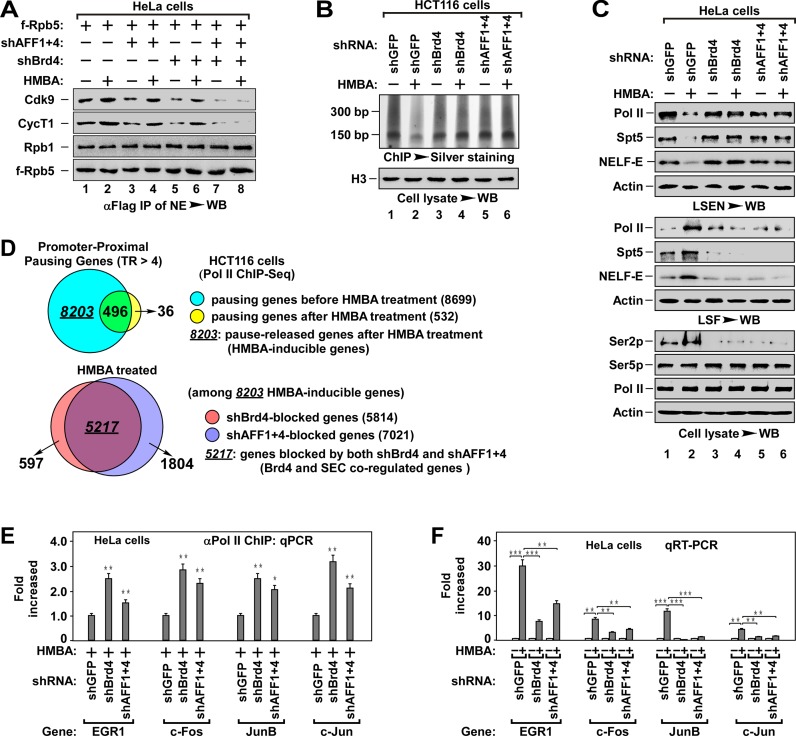
Both Brd4 and SEC have been shown to recruit P-TEFb (1). To compare the difference of these two factors in P-TEFb recruitment, we analyzed Pol II-associated P-TEFb in Brd4 and/or SEC knockdown cells. We used combinatorial knockdown of AFF1 and AFF4 to deplete SEC functions in HeLa and HCT116 cells since only AFF1 and AFF4 of SEC expressed in these cells (Supplementary Figure S1A). Depletion of Brd4 or SEC alone only partially reduced the Pol II-associated P-TEFb, whereas depletion of Brd4 and SEC together abolished almost all the P-TEFb recruitment even in the presence of hexamethylene bisacetamide (HMBA) (Figure 1A), a chemical capable of activating P-TEFb (25). These data indicate that Brd4 and SEC are involved in the global P-TEFb recruitment.
Figure 1.
Brd4 and SEC cooperatively regulate promoter–proximal pausing release. (A) The anti-Flag IPs derived from NEs of HeLa cells with indicated f-Rpb5 cDNA and shRNA(s) co-transfection and HMBA treatment were analyzed by WB for the levels of Pol II-associated P-TEFb. The f-Rpt5-bound Rpb1 was also tested to indicate the levels of intact Pol II. shAFF1+4: shAFF1+shAFF4. (B) ChIP-Silver staining assay for the effect of Brd4 or AFF1+4 knockdown on the accumulation of Pol II on genomic DNA. The Pol II-bound genomic DNAs from HCT116 cells with indicated shRNA(s) infection and HMBA treatment were chromatin-immunoprecipitated (ChIPed), followed by PAGE resolving and silver staining assay for the accumulation of Pol II on chromatin (top). The histone H3 in cell lysates was examined by WB as a loading control (bottom). (C) WB analysis for the effect of Brd4 or AFF1+4 knockdown on HMBA-induced pause release of Pol II. The low-salt extracted nuclei (LSEN), top, low-salt fraction (LSF), middle and cell lysates (bottom) were prepared from HeLa cells with indicated shRNA(s) infection and HMBA treatment. (D) The ChIPed DNAs in (B) were subjected to high-throughput sequencing analysis. Venn diagrams display the proportion of promoter–proximal pausing genes before and after HMBA treatment (top) and the proportion of Brd4 or AFF1+4 knockdown-induced pausing genes among the 8203 HMBA-inducible genes under HMBA treatment. Pausing gene: TR > 4.0, P-value < 0.005. (E) The Pol II-bound genomic DNAs were ChIPed from HeLa cells with indicated shRNA(s) infection and HMBA treatment were analyzed by qPCR for the enrichment of Pol II on promoter region of representative genes. The level in shGFP infected cells was set to 1.0. All values were expressed as Mean ± SD of three replicates. *P < 0.05, **P < 0.01; P-values were assessed using two-tailed Student's t-test. (F) Total RNAs isolated from HeLa cells with indicated shRNA(s) infection and HMBA treatment were analyzed by qRT-PCR for mRNA levels of representative genes with primers matching elongation region. The level in untreated cells was set to 1.0. All values were expressed as Mean ± SD of three replicates after normalized to actin. **P < 0.01, ***P < 0.001; P-values were assessed using two-tailed Student's t-test.
Both of Brd4 and SEC are required for the release of promoter–proximally paused Pol II
Since depletion of Brd4 or SEC only partially blocked P-TEFb recruitment (Figure 1A), we suspected that such depletion could also partially block the signal-induced release of paused Pol II. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) and DNA gel resolving and silver staining assay (ChIP-silver staining assay) to examine the release of Pol II (Figure 1B and Supplementary Figure S1B). In untreated cells, bulk of Pol II accumulated on genomic DNA and this accumulation was remarkably reduced by HMBA treatment (Figure 1B, lane 1 and 2). Unexpectedly, this HMBA-induced release was almost completely blocked by the depletion of either Brd4 or SEC (lane 3 to 6), indicating that Brd4 and SEC are required for the release of the paused Pol II. To further confirm this, we fractionated cells into LSF, which contains chromatin-free proteins, and LSEN, which contains the transcriptionally engaged factors, with a stepwise fractionation protocol (See Supplementary Figure S1C) (30,31). Immunoblotting analysis of LSEN showed that Pol II, DSIF (Spt5) and NELF (-E) were enriched in the LSEN of untreated cells (Figure 1C), and this enrichment was markedly decreased in LSEN after HMBA treatment (lane 2). Interestingly, the decrease was associated with an increase in LSF (lane 2). Of note, the phosphorylated Ser2 (Ser2p) of Pol II CTD was increased during HMBA treatment (Figure 1C, lane 2, bottom), indicating that the decrease of Pol II in LSEN is likely due to the efficient transcription induced by HMBA. Consistent with the results of ChIP-silver staining assay, depletion of Brd4 or SEC almost completely blocked HMBA-induced pause release (Figure 1C, lane 3–6). These data suggest that both Brd4 and SEC are required for the efficient release of the paused Pol II.
Brd4 and SEC cooperatively regulate the release of promoter–proximally paused Pol II
To test above notion, we performed Pol II ChIP-Seq to analyze the effect of depletion of Brd4 or SEC on HMBA-induced pause release (Figure 1D and Supplementary Table S1). Of 8699 genes with promoter–proximally paused Pol II (pausing genes, TR > 4, P-value < 0.005), HMBA treatment induced the release of paused Pol II in 8203 genes (94.3%), indicating that HMBA is able to induce global pause release (see Supplementary Figure S1D). Among 8203 HMBA-induced genes, knockdown of Brd4 (shBrd4) inhibited pause release in 5814 genes (70.9%) and depletion of SEC (shAFF1+4) inhibited 7021 genes (85.6%). Importantly, among shBrd4-inhibited genes (5814), 5217 genes (89.7%) were overlapped with SEC-inhibited genes (7021, 74.3%) (Figure 1D). Therefore, these data indicate that Brd4 and SEC cooperatively regulate the release of promoter–proximally paused Pol II in most pausing genes. By ChIP-qPCR and qRT-PCR analysis of the representative pausing genes (46,47), we found that depletion of Brd4 or SEC inhibited HMBA-induced release of paused Pol II and the transcription elongation (Figure 1E and F). These data further confirm the notion that Brd4 and SEC cooperatively regulate the pause release. Of note, the relative lower efficiency of pause release inhibition in Brd4 and SEC knockdown cells might be due to the inefficient shRNA(s) knockdown (see Supplementary Figure S1E).
HMBA induces the association of P-TEFb with transcription factors
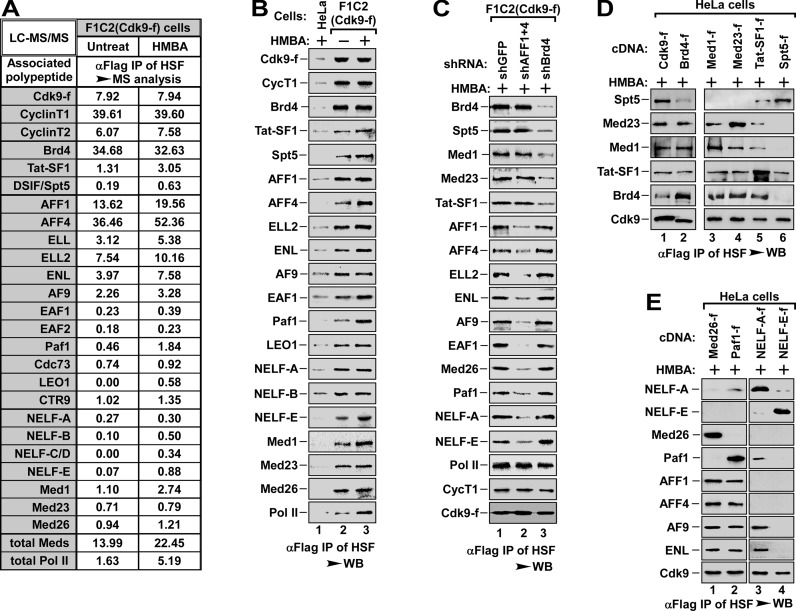
To understand the co-regulatory mechanism by Brd4 and SEC in HMBA-induced pause release, we began to explore how Brd4 and SEC mediated P-TEFb recruitment, an essential step for the pause release. We first carried out affinity purification of the transcriptionally engaged Cdk9-f/P-TEFb complexes from HSF (0.3 M salt extract of LSEN, see Supplementary Figure S1C) of F1C2 (Cdk9-f) cells, a HeLa-based cell line stably expressing Flag-tagged Cdk9 (25), followed by mass spectrometry and WB analysis (Figure 2A and B). In line with the stimulatory effect of HMBA on pause release (Figure 1D), HMBA induced the association of P-TEFb with a set of critical transcription factors (Figure 2A and B). Since knockdown of MePCE, a key component of inactive 7SK snRNP (24), also induced the association of P-TEFb with transcription factors (Supplementary Figure S2A), indicating that HMBA-induced association (Figure 2A and B) is due to the HMBA-induced activation of P-TEFb from 7SK snRNP (25,31).
Figure 2.
Brd4 and SEC recruit P-TEFbs to DISF and NELF, respectively. (A) Cdk9-f immunoprecipitates with anti-Flag affinity resin from HSF of F1C2 (Cdk9-f) cells without or with HMBA treatment were analyzed with LC-MS/MS for Cdk9-associated proteins. The IPs from HeLa cells were served as negative control. The relative abundance of P-TEFb-bound proteins was calculated with NSAF method and the proteins with significant increase after HMBA treatment were shown. (B) The IPs were prepared as in (A) and analyzed by WB to confirm the interaction between P-TEFb and its associated factors as indicated in (A). The IPs from HeLa cells were served as negative control. (C) P-TEFb and its associated factors were purified with anti-Flag affinity resin from HSF of F1C2 (Cdk9-f) cells with shGFP (as control), shBrd4 or shAFF1+4 infection and HMBA treatment as indicated and analyzed by WB for the indicated proteins. (D and E) Anti-Flag immunoprecipitates from HSF of HeLa cells with indicated cDNA transfection and HMBA treatment were analyzed by WB for the indicated proteins.
The P-TEFb recruited by Brd4 and SEC targets DSIF and NELF, respectively
Since DSIF and NELF are important factors for the Pol II pausing, we next investigated whether they are involved in Brd4 and SEC co-regulated release of paused Pol II. We knocked down Brd4 or AFF1+4 in F1C2 (Cdk9-f) cells and immunoprecipitated Cdk9-f/P-TEFb complexes from HSF in 0.3 M salt concentration as in Figure 2B. Immunoblotting analysis of the immunoprecipitated complexes revealed that depletion of Brd4 specifically abolished the association of P-TEFb with DSIF (Spt5), Med1 and Med23 subunits of Mediator, and Tat-SF1, whereas depletion of SEC specifically impaired the association of P-TEFb to NELF (-A and -E), Med26 of Mediator, and Paf1 subunit of Paf1c complex (Figure 2C), indicating that the transcription factors associated with Brd4/P-TEFb complex are different from those associated with SEC/P-TEFb complex. Intriguingly, two critical subunits of middle module of Mediator, Med1 and Med26, bound to Brd4/P-TEFb and SEC/P-TEFb, respectively. To further confirm these interactions, we performed immunoprecipitation with different salt concentration to determine whether these interactions could be affected by salt concentration. Immunoblotting analysis of the immunoprecipitated complexes showed that 0.3 M salt concentration disrupted the association between Med1 and Med26, but not the binding of Brd4/P-TEFb to Med1 or SEC/P-TEFb to Med26, whereas 0.15 M had no effect on these interactions (Supplementary Figure S2B). These data suggest that Med1 and Med26 bind to different P-TEFb complexes, even though they coexist in the same middle module of Mediator. Consistently, knockdown of Med26 did not affect the binding of Brd4/P-TEFb to Med1, and depletion of Med1 had no effect on the interaction of SEC/P-TEFb with Med26 (Supplementary Figure S2C). Together, these data demonstrate that P-TEFb recruited by Brd4 or SEC interacts with Med1 or Med26, respectively.
To further analyze these interactions, we compared the anti-Flag immunoprecipitates derived from HSF of HeLa cells expressing Med1-f, Med23-f, Tat-SF1-f or Spt5-f, with the immunoprecipitates from HSF of Cdk9-f and Brd4-f cells as positive control (Figure 2D). Interestingly, while DSIF (Spt5) only bound to P-TEFb (Figure 2D, lane 6), Med1, Med23 and Tat-SF1 bound to both Brd4 and P-TEFb (lane 2 to 5), implying that Med1, Med23 and Tat-SF1 might be involved in the recruitment of Brd4/P-TEFb complex, with Spt5/DSIF being the end point of P-TEFb. Immunoblotting analysis of the immunoprecipitates derived from HSF of HeLa cells expressing Med26-f, Paf1-f, NELF-A-f or NELF-E-f showed that all four proteins bound to P-TEFb, but only Med26 and Paf1 bound to scaffold protein AFF1 and AFF4 (Figure 2E). These data implicate that Med26 and Paf1 might be engaged in the recruitment of SEC/P-TEFb, with NELF complex being the end point of P-TEFb. Surprisingly, only NELF-A, but not NELF-E, bound to ENL/AF9 (Figure 2E, lane 3 and 4), suggesting that NELF-A and NELF-E might interact with different SEC subtypes. Similar to Med1 and Med26 (Supplementary Figure S2B), 0.3 M salt concentration disrupted the interaction between NELF-A and NELF-E, but not the binding of NELF-A to both P-TEFb and ENL or the association of NELF-E to P-TEFb (Supplementary Figure S2D). More specifically, knockdown of NELF-E failed to disrupt the binding of NELF-A to both P-TEFb and ENL, and depletion of NELF-A also failed to change the binding of NELF-E to P-TEFb (Supplementary Figure S2E), indicating that the SEC/P-TEFb subtypes in associating with NELF-A are different from those in association with NELF-E (see below for details).
Collectively, these data demonstrate that Brd4 or SEC recruits different pool of P-TEFb to target DSIF or NELF, respectively, for the co-regulated pause release of Pol II.
Brd4 recruits P-TEFb to DSIF via a recruitment pathway consisting of Med1, Med23 and Tat-SF1
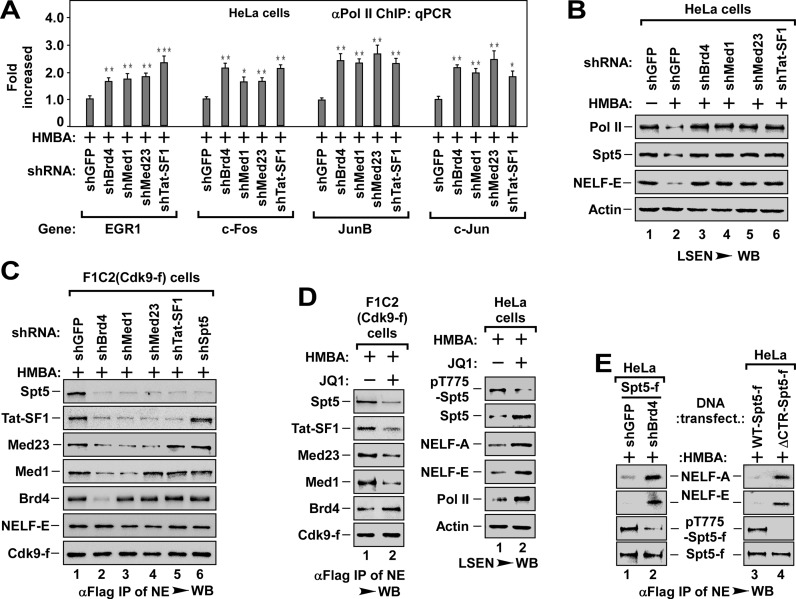
To investigate the role of Med1, Med23 and Tat-SF1 in Brd4/P-TEFb recruitment, we first evaluated the effect of knockdown of these factors on pause release. Similar to Brd4 knockdown (Figure 1C, E and F), depletion of any one of these factors blocked HMBA-induced pause release (Figure 3A and B) and transcription elongation (Supplementary Figure S3A), suggesting that Med1, Med23 and Tat-SF1 have a non-redundant role in pause release, very likely, through recruiting Brd4/P-TEFb to DSIF.
Figure 3.
The P-TEFb targeting DSIF is recruited by Brd4 via a recruitment pathway consisting of Med1, Med23 and Tat-SF1. (A) The Pol II-bound genomic DNAs ChIPed from HeLa cells with indicated shRNA infection and HMBA treatment were analyzed by qPCR for the enrichment of Pol II on the promoter region of representative genes as in Figure 1E. The level in shGFP infected cells was set to 1.0. All values were expressed as Mean ± SD of three replicates. *P < 0.05, **P < 0.01, ***P < 0.001; P-values were assessed using two-tailed Student's t-test. (B) LSEN from HeLa cells with indicated shRNA infection and HMBA treatment were analyzed by WB as in Figure 1C. (C) Effect of depletion of Brd4, Med1, Med23 or Tat-SF1 on the recruitment of P-TEFb to DSIF (Spt5) and NELF (-E). Anti-Flag immunoprecipitates from NEs of F1C2 (Cdk9-f) cells with indicated shRNA infection and HMBA treatment were analyzed by WB for the indicated Cdk9-f/P-TEFb-associated proteins. (D) Effect of JQ1 on P-TEFb recruitment to Spt5/DSIF (left panel) and P-TEFb-mediated phosphorylation of Spt5 (pT775-Spt5, right panel). The anti-Flag immunoprecipitates from NEs of F1C2 (Cdk9-f) (left panel) and the cell lysates of HeLa cells (right panel) with indicated JQ1 and HMBA treatment were analyzed by WB for the indicated proteins. (E) Effect of Spt5 phosphorylation on the dissociation of NELF. The anti-Flag immunoprecipitates from NEs of HeLa cells with indicated Spt5-f cDNA and shRNA co-transfection (left) or with WT- or ΔCTR-Spt5-f transfection (right) were analyzed by WB for the levels of NELF-A and NELF-E and the levels of phosphorylated Spt5 (pT775-Spt5).
To prove this hypothesis, we knocked down these factors one by one in F1C2 (Cdk9-f) cells and examined the interaction of P-TEFb with relevant factors (Figure 3C). As shown in Figure 3C, depletion of Brd4 blocked the binding of P-TEFb to Med1, Med23, Tat-SF1 and DSIF (Figure 3C, lane 2). Interestingly, depletion of Med1 blocked the binding of P-TEFb to Med23, Tat-SF1 and DSIF, but not to Brd4 (lane 3), whereas depletion of Med23 blocked the binding of P-TEFb to Tat-SF1 and DSIF, but not to Brd4 and Med1 (lane 4). Finally, depletion of Tat-SF1 only abolished the binding of P-TEFb to DSIF (lane 5). Similarly, pre-treatment of the cells with Brd4 specific inhibitor JQ1 (28,48) also blocked the recruitment of P-TEFb to DSIF by impairing the association of Brd4/P-TEFb with Med1, Med23 and Tat-SF1 (Figure 3D, left panel). Importantly, the recruitment of P-TEFb to NELF was not affected in those knockdown cells (Figure 3C). Together with the data in Figure 2C and D, these data indicate that Brd4 recruits P-TEFb to DSIF via a specific recruitment pathway consisting of Med1, Med23 and Tat-SF1.
P-TEFb-mediated phosphorylation of Spt5 is essential for the dissociation of NELF from Spt5/DSIF
P-TEFb is capable of phosphorylating C-terminal region (CTR) of Spt5 (22). Therefore, we generated an antibody that specifically recognized P-TEFb-mediated phosphorylation of T775 of Spt5 CTR (pT775-Spt5) (Supplementary Figure S3B). Consistent with the inhibitory effect on pause release (Figure 3B), depletion of Brd4, Med1, Med23 or Tat-SF1 all blocked Spt5 phosphorylation (Supplementary Figure S3C). Moreover, pre-treatment with JQ1 not only blocked Spt5 phosphorylation, but also impaired the dissociation of NELF and the pause release (Figure 3D, right panel), suggesting that the phosphorylation of Spt5 might be essential for the dissociation of NELF. Further supporting this, we found that depletion of Brd4 or expression of CTR-deleted Spt5-f (ΔCTR-Spt5-f) blocked the dissociation of NELF from Spt5/DSIF by inhibiting Spt5 phosphorylation (Figure 3E).
Taken together, the data in Figure 3 reveal that Brd4 recruits P-TEFb to DSIF via a recruitment pathway consisting of Med1, Med23 and Tat-SF1. These data also indicate that P-TEFb-mediated Spt5 phosphorylation is required for the dissociation of NELF from DSIF and Pol II (see Supplementary Figure S3D for illustrated model).
SEC recruits P-TEFb to NELF-A and NELF-E via Paf1c and Med26, respectively
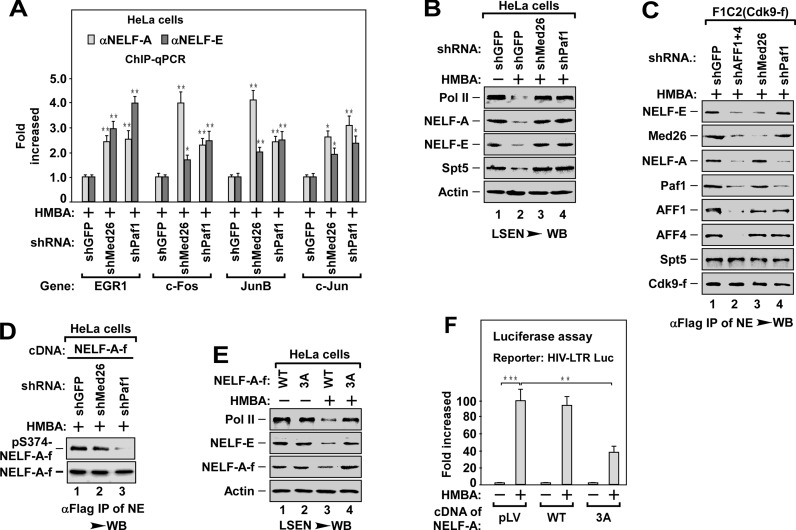
Since depletion of SEC blocked the binding of P-TEFb to Med26 and Paf1 (Figure 2C), we next evaluated whether these two factors might contribute to a SEC/P-TEFb recruitment pathway. Depletion of Med26 or Paf1 blocked HMBA-induced pause release by impairing the dissociation of NELF (-A and -E) from promoter regions (Figure 4A and B). These data indicate that both Med26 and Paf1 are essential for pause release, most likely, by mediating the recruitment of SEC/P-TEFb to NELF.
Figure 4.
SEC recruits P-TEFbs to NELF-A and NELF-E via Paf1c and Med26, respectively. (A) Effect of depletion of Med26 and Paf1 on HMBA-stimulated dissociation of NELF-A and NELF-E. The DNAs ChIPed from HeLa cells with indicated shRNA(s) infection and HMBA treatment were analyzed by qPCR for the enrichment of NELF-A and NELF-E on the promoter region of representative genes as in Figure 1E. The level in shGFP infected cells was set to 1.0. All values were expressed as Mean ± SD of three replicates. *P < 0.05, **P < 0.01; P-values were assessed using two-tailed Student's t-test. (B) LSEN prepared from HeLa cells with indicated shRNA(s) infection and HMBA treatment were analyzed by WB as in Figure 1C. (C) Effect of depletion of Med26 and Paf1 on P-TEFb recruitment to NELF. The anti-Flag immunoprecipitates from NEs of F1C2 (Cdk9-f) cells with indicated shRNA infection and HMBA treatment were analyzed by WB for the levels of the indicated Cdk9-f/P-TEFb-associated proteins. (D) Effect of depletion of Med26 and Paf1 on P-TEFb-mediated phosphorylation of NELF-A. The anti-Flag immunoprecipitates from HeLa cells with indicated NELF-A-f cDNA and shRNA co-transfection and HMBA treatment were analyzed by WB for the levels of phosphorylated NELF-A (pS374-NELF-A). (E) Effect of overexpression of 3A-NELF-A on HMBA-induced pause release. The LSEN prepared from HeLa cells with WT- and 3A-NELF-A-f infection and HMBA treatment as indicated were analyzed by WB as in Figure 1C. (F) HeLa cells stably integrated a HIV-LTR-luciferase reporter gene (HIV-LTR-Luc) were overexpressed with WT- or 3A-NELF-A, followed by HMBA treatment as indicated. The cell lysates prepared from the cells were subjected to luciferase assay. The level in untreated cells was set to 1.0. All values were expressed as Mean ± SD of three replicates. *P < 0.01, **P < 0.001; P-values were assessed using two-tailed Student's t-test.
We next examined the effect of Med26 and Paf1 knockdown on the binding of P-TEFb to NELF in F1C2 (Cdk9-f) cells. Unexpectedly, depletion of Med26 only blocked the association of P-TEFb with NELF-E, but not with NELF-A and Paf1, whereas depletion of Paf1 abolished the binding of P-TEFb to NELF-A, but not to NELF-E and Med26 (Figure 4C). Moreover, in the absence of NELF-A or NELF-E, which did not change the binding of P-TEFb to NELF-E or to NELF-A, respectively (Supplementary Figure S4A, lane 1 and 4), depletion of Med26 specifically abolished the recruitment of P-TEFb to NELF-E (lane 2), whereas depletion of Paf1 only impaired the recruitment of P-TEFb to NELF-A (lane 6). These data indicate that Med26 specifically mediates the recruitment of P-TEFb to NELF-E, whereas Paf1c mediates the recruitment of P-TEFb to NELF-A.
P-TEFb-mediated phosphorylation of NELF-A is essential for pause release
P-TEFb has been shown to phosphorylate NELF-E (21), but whether NELF-A could be phosphorylated by P-TEFb is not clear. To answer this question, we first determined whether NELF-A could be phosphorylated in cells by mass spectrometry. Analyzing mass spectrometry data of immunoprecipitated NELF-A from HeLa cells stably expressing NELF-A, we identified three phospho-residues, including threonines (T) 168, 288 and serine (S) 374 (Supplementary Figure S4B). We next substituted all three amino acids to alanines to generate a phosphorylation-defective mutant of NELF-A (T168A/T288A/S374A, designated as 3A-NELF-A). In vitro kinase assay followed by immunoblotting analysis with antibody against phospho-Ser (ph-Ser) or phospho-Thr (ph-Thr) revealed that P-TEFb directly phosphorylated WT NELF-A but not 3A-NELF-A (Supplementary Figure S4C). Phosphorylated NELF-A could also be detected in cultured cells by an antibody that specifically recognizes phosphorylated S374 of NELF-A (pS374-NELF-A) (Supplementary Figure S4D).
Consistent with the inhibitory effect of knocking down Paf1 on the binding of P-TEFb to NELF-A (Figure 4C), depletion of Paf1, but not Med26, inhibited the phosphorylation of NELF-A at S374 (Figure 4D). Moreover, overexpression of 3A-NELF-A blocked HMBA-induced pause release (Figure 4E) and the transcription of HIV-LTR-driven luciferase reporter gene (HIV-LTR-Luc, Figure 4F). These data suggest that Paf1c-delivered P-TEFb could phosphorylate NELF-A, leading to the pause release of Pol II.
The P-TEFb targeting NELF-A or NELF-E is recruited by different SEC subtypes
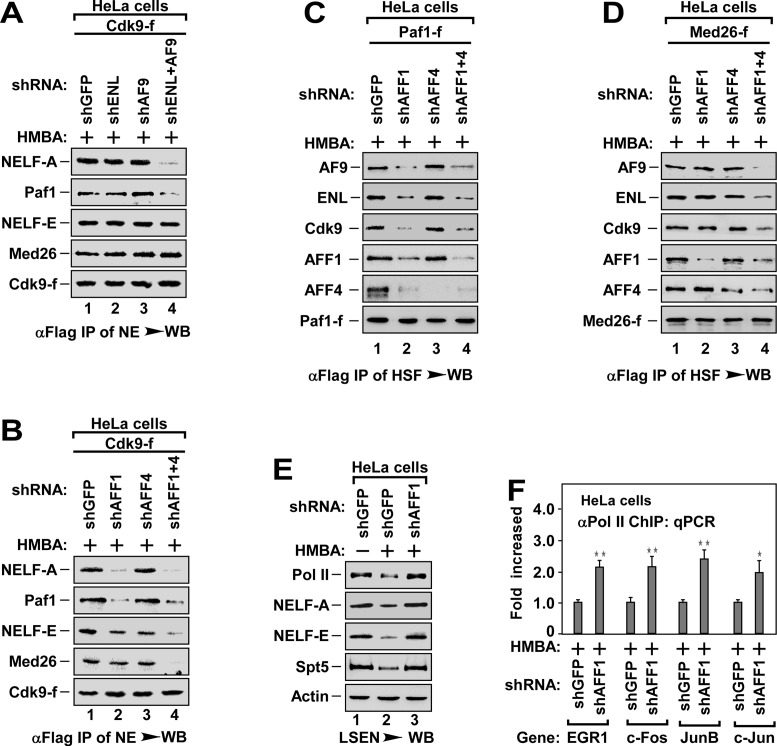
Given that SEC is essential for recruiting P-TEFb to NELF (Figure 4) and there are diverse SEC subtypes in cells (33), we next determined the functional difference of SEC subtypes in P-TEFb recruitment. Since ENL and AF9 bound to NELF-A, but not to NELF-E (Figure 2E and Supplementary Figure S2D and E), we first assessed the role of ENL and AF9 in P-TEFb recruitment. Depletion of ENL or AF9 alone had little effect on the binding of P-TEFb to NELF-A and Paf1 (Figure 5A). However, depletion of both ENL and AF9 at the same time significantly inhibited the binding of P-TEFb to NELF-A and Paf1 (Figure 5A). The binding of P-TEFb to NELF-E and Med26 was not affected by the depletion of ENL and/or AF9 (Figure 5A). These data indicate that the SEC subtype containing ENL or AF9 has redundant role in recruiting P-TEFb to NELF-A, but has no function in recruiting P-TEFb to NELF-E.
Figure 5.
The P-TEFbs targeting NELF-A and NELF-E are recruited by distinct SEC subtypes. (A and B) The anti-Flag immunoprecipitates from NEs of HeLa cells with Cdk9-f cDNA and shRNA(s) co-transfection and HMBA treatment as indicated were analyzed by WB for the levels of indicated Cdk9-f/P-TEFb-associated proteins. (C and D) The anti-Flag immunoprecipitates derived from NEs of cells with indicated cDNA and shRNA(s) co-transfection and HMBA treatment were analyzed by WB for the levels of indicated proteins in association with Paf1c-f (C) or Med26-f (D). (E) LSEN of HeLa cells with indicated shRNA infection and HMBA treatment were analyzed by WB for the levels of indicated proteins. (F) ChIP-qPCR analysis for the enrichment of Pol II at the promoter region of representative genes in HeLa cells with indicated shRNA infection and HMBA treatment as in Figure 1E. The level in shGFP infected cells was set to 1.0. All values were expressed as Mean ± SD of three replicates. *P < 0.05, **P < 0.01; P-values were assessed using two-tailed Student's t-test.
Since both AFF1 and AFF4 bound to Med26 and Paf1 (Figure 2E), we next evaluated the effect of depletion of AFF1 and/or AFF4 on P-TEFb recruitment. Interestingly, depletion of AFF1, but not AFF4, impaired the binding of P-TEFb to Paf1 and NELF-A (Figure 5B). The interactions of P-TEFb with Med26 and NELF-E were abolished only when both AFF1 and AFF4 were depleted (Figure 5B and Supplementary Figure S5A). Consistently, depletion of AFF1, but not AFF4, impaired the P-TEFb-mediated NELF-A phosphorylation (Supplementary Figure S5B). Examining the factors in association with Paf1-f or Med26-f indicated that depletion of AFF1 alone blocked the binding of Paf1 to P-TEFb, ENL and AF9, but not the binding of Med26 to NELF-E (Figure 5C and D, lane 2). Depletion of AFF1 and AFF4 together abolished the binding of both Paf1 and Med26 to P-TEFb (lane 4). These data suggest an essential role of AFF1 but not AFF4 in pause release. Consistent with this notion, depletion of AFF1 alone inhibited HMBA-induced pause release (Figure 5E and F) and transcription elongation (Supplementary Figure S5C).
Collectively, the data in Figures 4 and 5 reveal that both AFF1-SEC and AFF4-SEC subtypes are able to recruit P-TEFb to NELF-E via Med26, whereas both AFF1-ENL-SEC and AFF1-AF9-SEC subtypes are responsible for the recruitment of another P-TEFb to NELF-A via Paf1c. In addition to the phosphorylation of NELF-E (21), P-TEFb-mediated phosphorylation of NELF-A is also required for the dissociation of NELF from paused Pol II (see Supplementary Figure S5D for illustrated model).
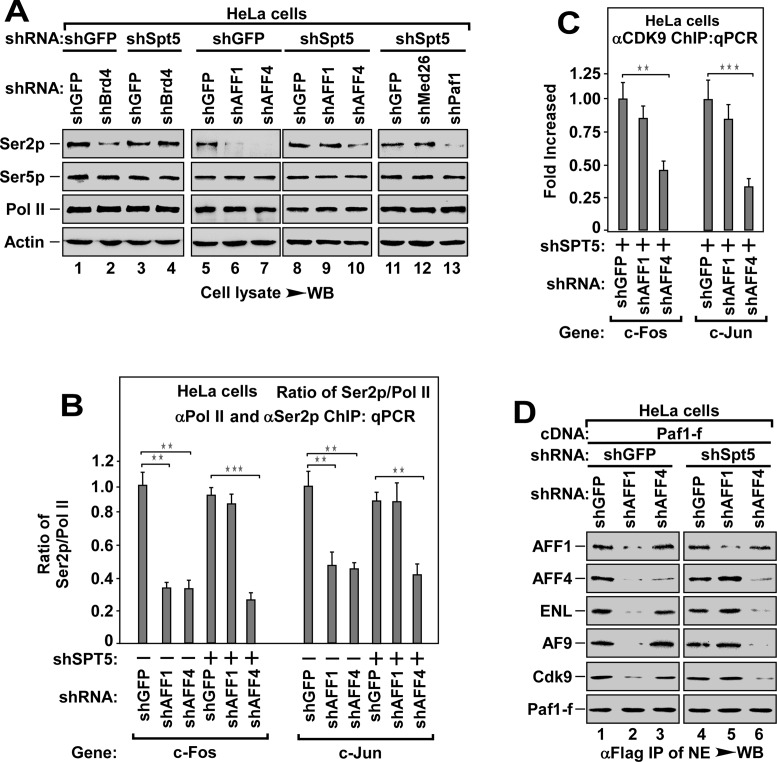
The P-TEFb responsible for phosphorylation of Pol II Ser2 is recruited by AFF4-ENL (AF9)-SEC with Paf1c being a docking site
The phosphorylation of Ser2 of Pol II CTD by P-TEFb has long been shown to be a prerequisite for pause release (14). Given that multiple P-TEFbs are recruited to Spt5/DSIF and NELF (-A and -E), we investigated whether depletion of Spt5/DSIF and NELF (A+E) could affect Ser2 phosphorylation. Interestingly, depletion of Spt5 or NELF (A+E) did not affect Ser2 phosphorylation, but led to the transcription elongation even without HMBA stimulation (Supplementary Figure S6A and B). Thus, we determined the effect of depletion of Brd4, AFF1 or AFF4 on Ser2 phosphorylation in the presence or absence of Spt5. In the presence of Spt5, depletion of any one of these three factors inhibited Ser2 phosphorylation (Figure 6A). However, in the absence of Spt5 with the enhanced transcription elongation (Supplementary Figure S6B), only AFF4 depletion inhibited Ser2 phosphorylation (Figure 6A, lane 10). This effect could also be observed in the absence of NELF (A+E) (Supplementary Figure S6C). ChIP-qPCR assay showed that depletion of AFF4, but not AFF1, decreased the levels of Pol II Ser2 phosphorylation at the termination regions (Figure 6B), likely due to the impairment of P-TEFb recruitment (Figure 6C). Of note, although both Med26 and Paf1 bound to and recruited SEC/P-TEFb (Figure 5), only depletion of Paf1, but not Med26, abolished Ser2 phosphorylation in the absence of Spt5 (Figure 6A, lane 13). Moreover, in the absence of Spt5, depletion of AFF4, but not AFF1, blocked the binding of Paf1-f to P-TEFb, ENL and AF9 (Figure 6D). Collectively, these data demonstrate that AFF4-ENL-SEC and AFF4-AF9-SEC subtypes are responsible for recruiting P-TEFb for Ser2 phosphorylation using Paf1c as a docking site.
Figure 6.
The P-TEFb responsible for the phosphorylation of Pol II Ser2 is recruited by AFF4-ENL (AF9)-SEC with Paf1c being the docking site. (A) WB analysis of the levels of phosphorylated Ser2 (Ser2p) and Ser5 (Ser5p) in HeLa cell lysates with indicated shRNAs co-infection. (B) Effect of depleting AFF1 or AFF4 in the absence or presence of Spt5 on the enrichment of Ser2p Pol II at the termination region of tested genes, The DNAs ChIPed with anti-Ser2p or anti-Pol II antibody from HeLa cells with indicated shRNAs co-infection were analyzed by qPCR. The ratio of Ser2p Pol II to bulk Pol II was shown. All values were expressed as Mean ± SD of three replicates. **P < 0.01, ***P < 0.001; P-values were assessed using two-tailed Student's t-test. (C) Effect of depleting AFF1 or AFF4 in the absence of Spt5 on the recruitment of P-TEFb at the termination region of tested genes. The DNAs ChIPed from HeLa cells with indicated shRNAs co-infection were analyzed by qPCR as in (B). All values were expressed as Mean ± SD of three replicates. **P < 0.01, ***P < 0.001; P-values were assessed using two-tailed Student's t-test. (D) The anti-Flag immunoprecipitates from NEs of HeLa cells with Paf1-f cDNA and shRNA co-transfection and HMBA treatment as indicated were analyzed by WB for the levels of indicated Paf1-f-associated proteins.
The P-TEFb-mediated Ser2 phosphorylation is not essential for pause release
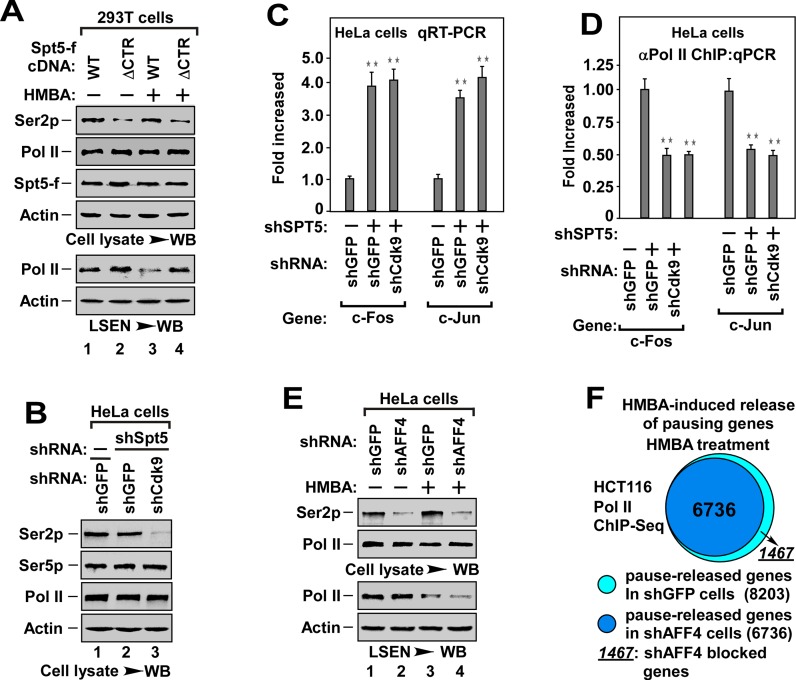
Since depletion of Brd4 and AFF1 abolished Ser2 phosphorylation only in the presence of Spt5/DSIF (Figure 6A), and Brd4 and AFF1 are required for pause release (Figures 3–5), it raised the possibility that Ser2 phosphorylation might occur after pause release. Indeed, overexpression of ΔCTR-Spt5, which blocked pause release (Figure 7A, bottom, lane 4), inhibited Ser2 phosphorylation (top). In addition, depletion of Cdk9 markedly abolished Ser2 phosphorylation (Figure 7B), but did not impair the transcription elongation and pause release in the absence of Spt5/DSIF (Figure 7C and D). Furthermore, depletion of AFF4 markedly abolished Ser2 phosphorylation (Figure 7E, top), but only slightly blocked HMBA-induced pause release even in the presence of DSIF (Figure 7E, bottom). Consistently, among 8203 HMBA-inducible genes, AFF4 depletion only blocked the release of promoter-proximally paused Pol II in 1467 genes (17.9%) (pausing genes, TR > 4, P-value < 0.005), as indicated by Pol II ChIP-Seq analysis (Figure 7F and Supplementary Table S2). Taken together, these data demonstrate that P-TEFb-mediated Ser2 phosphorylation is dispensable for promoter-proximal pause release.
Figure 7.
The P-TEFb-mediated Ser2 phosphorylation is dispensable for pause release. (A) WB analysis of the effect of ΔCTR-Spt5 on HMBA-induced Ser2p and pause release. The cell lysates (top) and LSEN (bottom) from 293T cells with indicated WT- or ΔCTR-Spt5-f transfection and HMBA treatment were analyzed for the levels of indicated proteins. (B) WB analysis of Ser2p, Ser5p and indicated proteins in cell lysates of HeLa cells with co-infection of shSpt5 and shCdk9 as indicated. (C) qRT-PCR analysis for the mRNA levels of representative genes in cells with indicated shRNA(s) co-infection as in Figure 1F. All values were expressed as Mean ± SD of three replicates. **P < 0.01; P-values were assessed using two-tailed Student's t-test. (D) Effect of depletion of Cdk9 and Spt5 on the enrichment of Pol II on the promoter region of tested genes. The DNAs ChIPed from HeLa cells co-infected with indicated shRNA(s) were analyzed by qPCR as in Figure 1E. All values were expressed as Mean ± SD of three replicates. **P < 0.01; P-values were assessed using two-tailed Student's t-test. (E) WB analysis of the effect of knockdown AFF4 on Ser2p and pause release. LSEN and cell lysates from HeLa cells with shRNA(s) infection and HMBA treatment were analyzed for the levels of indicated proteins. (F) ChIP-Seq analysis of the effect of AFF4 knockdown on HMBA-induced pause release as in Figure 1D. Diagrams display the proportion of pause-released genes in HCT116 cells with or without AFF4 knockdown under HMBA treatment. The criterion of Pol II pausing was defined as TR (the relative ratio of promoter read density/gene body read density) > 4.0, P-value < 0.005.
DISCUSSIONS
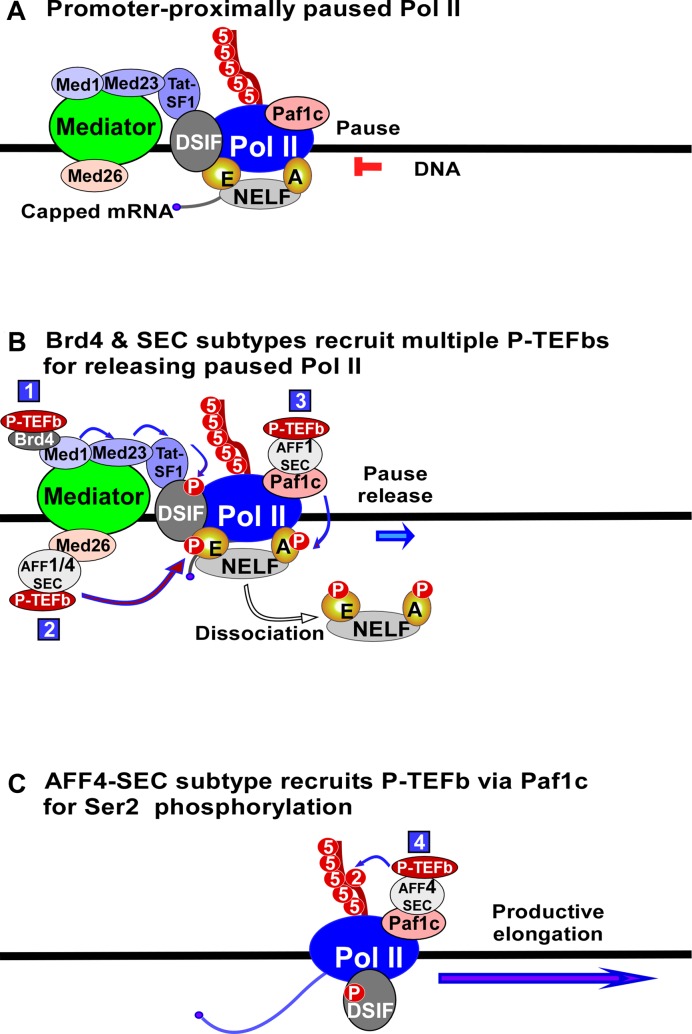
P-TEFb, Brd4 and SEC play essential roles in regulating transcription elongation. However, the detailed mechanism by which Brd4 and SEC recruit P-TEFb for pause release remains elusive. In the present study, we demonstrate that Brd4 and SEC cooperatively regulate transcriptional pause release by recruiting multiple P-TEFbs via different recruitment mechanisms (Figure 8). To release a promoter-proximally paused Pol II, Brd4 recruits the first P-TEFb to DSIF via a specific recruitment pathway consisting of Med1, Med23 and Tat-SF1, leading to the phosphorylation of Spt5 (Figure 8). Meanwhile, AFF1-SEC or AFF4-SEC recruits the second P-TEFb to NELF-E via Med26, and AFF1-ENL (AF9)-SEC recruits the third P-TEFb to NELF-A via Paf1c, leading to the phosphorylation of NELF-E and NELF-A and the dissociation of NELF from paused Pol II (Figure 8). Finally, AFF4-ENL (AF9)-SEC recruits the P-TEFb to Paf1c to phosphorylate Pol II CTD at Ser2 (Figure 8). Thus, the transcriptional pause release is regulated by the cooperation of multiple P-TEFbs which are recruited by Brd4 and SEC subtypes via a Mediator- and Paf1c-coordinated recruitment network.
Figure 8.
Model of Brd4 and SEC cooperatively regulating pause release of Pol II. A model depicting the co-regulation processes of transcriptional pause release by Brd4 and SEC through recruiting multiple P-TEFbs via Mediator- and Paf1c-coordinated recruitment network. The number in blue square denotes the P-TEFb recruited by distinct mechanism as indicated.
The mechanisms of multiple P-TEFbs-mediated pause release
It has been widely accepted that P-TEFb-mediated phosphorylation of Ser2 of Pol II, the Spt5 of DSIF and the NELF-E of NELF is a prerequisite for the release of promoter–proximally paused Pol II (1–3,15,23). In this study, we found that these three different phosphorylations are mediated by three ‘appointed’ P-TEFbs (Figure 8). Moreover, in addition to NELF-E, NELF-A is also phosphorylated by an ‘appointed’ P-TEFb (Figure 5). Therefore, it requires at least four ‘appointed’ P-TEFbs to convert paused transcription to productive elongation (Figure 8).
The dissociation of NELF from paused Pol II is a key step for pause release (15,23). Since NELF-A associates with Pol II, and NELF-E binds to nascent mRNA (15,16,18), it is reasonable to assume that both NELF-A and NELF-E have to be phosphorylated in order to dissociate NELF from promoter–proximally paused Pol II. Importantly, besides the phosphorylation of NELF-A and NELF-E, our data demonstrated that the phosphorylation of Spt5 CTR was also essential for the dissociation of NELF from paused Pol II (Figure 3). Given that the association of NELF with Pol II depends on its interaction with DSIF (16), it is conceivable that the phosphorylation of Spt5 might facilitate the disruption of the interaction between DSIF and NELF and the dissociation of NELF from DSIF (Figure 3). This feature might provide a plausible answer to how DSIF is transited from a negative factor to a positive factor in transcription elongation after the phosphorylation of Spt5 CTR (22). Moreover, the necessity of P-TEFb-mediated phosphorylation of both Spt5 and NELF for the dissociation of NELF from paused Pol II could be the underlying mechanism for Brd4- and SEC-mediated co-regulation of pause release.
It is commonly believed that the phosphorylation of Pol II CTD at Ser2 is essential for the release of promoter–proximally paused Pol II (15). Different from this notion, our data indicate that Ser2 phosphorylation is dispensable for pause release (Figure 7 and 8). First, in the absence of Spt5/DSIF, depletion of Cdk9 abolished Ser2 phosphorylation, but it did not affect the pause release and transcription elongation (Figure 7B to D). Second, in the presence of DSIF, depletion of AFF4 severely impaired Ser2 phosphorylation, but only slightly affected the release of promoter–proximally paused Pol II (Figure 7E and F). Finally, in the presence of DSIF, depletion of AFF1, which is required for the recruitment of P-TEFb to NELF-A for pause release (Figure 5), blocked the binding of Paf1 to AFF4 (Figure 6D), which is essential for the recruitment of P-TEFb to Paf1c for Ser2 phosphorylation (Figure 6). However, in the absence of DSIF, AFF1 depletion did not affect the binding of Paf1 to AFF4 (Figure 6D). These data indicate a sequential binding of AFF1 and AFF4 to Paf1c, i.e. the interaction of AFF1 with Paf1c is prior to the binding of AFF4 to Paf1c. Although the mechanism for such a sequential binding is currently unclear, this binding manner could be the underlying mechanism dictating the occurring sequence of pause release and Ser2 phosphorylation. Collectively, our data demonstrate that Ser2 phosphorylation occurs after pause release.
The mechanisms of P-TEFb recruitment by Brd4 and SEC
How Brd4 mediates the recruitment of P-TEFb to promoter region is a question of interest to many researchers. While previous studies reported the interaction of P-TEFb with Med1, Med23, Tat-SF1 or DSIF (22,49–51), our current data indicate that these factors are functionally connected to compose a Brd4/P-TEFb recruitment pathway which targets DSIF to regulate the pause release (Figure 8). However, how Brd4/P-TEFb complex transfers within this recruitment pathway and finally to DSIF is currently unclear. Since JQ1, an inhibitor interfering with the interaction of Brd4's bromodomains with its acetylated substrates (48), blocked the binding of Brd4/P-TEFb to Med1 (Figure 3D), it is likely that the binding of Brd4 to Med1 is through the association of Brd4's bromodomains with acetylated Med1. This might provide an insight into the inhibitory mechanism of JQ1 on Brd4's biological functions (28). It has to be noted that while this Brd4/P-TEFb recruitment process may represent a general mechanism, it is possible that other recruitment mechanisms might also exist (37,38,52), since depletion of Brd4 did not completely abolish HMBA-induced release of paused Pol II (Figure 1D).
The compositions of SEC vary with diverse subtypes depending on the cell types (1,33). However, how each SEC subtype differentially regulates transcription elongation is still unknown. Our data reveal the functional differences of AFF1, AFF4 and ENL (AF9) in regulating pause release (Figure 8), but the role of ELLs in this process remains undetermined. Consistent with the observation that ENL (AF9) binds to Paf1c (35), we found that SEC subtypes containing ENL (AF9) recruited P-TEFbs via association with Paf1c, resulting in different functional consequences. For example, AFF1-ENL (AF9)-SEC recruited P-TEFb to phosphorylate NELF-A, whereas AFF4-ENL (AF9)-SEC recruited P-TEFb to phosphorylate Ser2 of Poll II (Figures 5 and 6). Besides Paf1c, Med26 has also been reported to serve as a docking site for SEC/P-TEFb (34). We found that Med26 recruited AFF1- and AFF4-SEC/P-TEFb to NELF-E (Figure 5). Thus, together with Brd4/P-TEFb's recruitment pathway, Mediator- and Paf1c-coordinated P-TEFb recruitment creates a network that allows Brd4 and SEC to cooperatively regulate the release of transcriptionally paused Pol II.
In this study, we identified a mechanism by which Mediator and Paf1c coordinate with Brd4 and SEC subtypes to form a P-TEFb-recruitment network that dictates the transcriptional pause release. Whether and how this general mechanism applies to signal-specific or gene-specific release of pausing genes during stimulation will be the subject of future investigation. More importantly, the observation that AFF4 depletion abolishes Ser2 phosphorylation, but not the promoter–proximal pause release, provides an intriguing approach to define the specific functions of P-TEFb in other transcription-coupled events, such as splicing, 3′-end processing and termination.
Supplementary Material
Acknowledgments
The authors thank Analysis and Testing Center of School of Life Sciences of Xiamen University for MS analysis and Dr Zhong, Chuangqi for help of the calculation of MS data.
Author contributions: R.C, and R.L. designed the project. X.L. and X.Z. performed most of the experiments. Y.L., and K.Z. designed and performed MS analysis. M.L., X.L., B.Y., H.Y. and Y.W. designed and performed ChIP-Seq, ChIP-silver staining and ChIP-qPCR analysis. Y.W. and M.R. performed qRT-PCR analysis. Y.W., K.Z., Y.H., Y.W., Y.C. and M.C. performed cDNA cloning and shRNA preparation. Y.L. and X.L. carried out kinase assay. R.C. and L.C. write the manuscript. All authors commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81361120386, 31570751, 31270809 and 30930046 to R.C., 31501096 to M.L., 81171192 to R.L.]; 973 program [2013CB917802 to R.C.]; NIH-CA179511 [to L.F.C.]; Fundamental Research Funds for the Central Universities [20720150053 to M.L.]; NSFC for Fostering Talents in Basic Research [J1310027 to Y.C]. Funding for open access charge: National Natural Science Foundation of China [81361120386, 31570751, 31270809 and 30930046 to R.C., 31501096 to M.L., 81171192 to R.L.]; 973 program [2013CB917802 to R.C.]; NIH-CA179511 [to L.F.C.]; Fundamental Research Funds for the Central Universities [20720150053 to M.L.]; NSFC for Fostering Talents in Basic Research [J1310027 to Y.C].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwak H., Lis J.T. Control of transcriptional elongation. Annu. Rev. Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Q., Li T., Price D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriques T., Gilchrist D.A., Nechaev S., Bern M., Muse G.W., Burkholder A., Fargo D.C., Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams L.H., Fromm G., Gokey N.G., Henriques T., Muse G.W., Burkholder A., Fargo D.C., Hu G., Adelman K. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol. Cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Kraus W.L., Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem. Sci. 2015;40:516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxwell C.S., Kruesi W.S., Core L.J., Kurhanewicz N., Waters C.T., Lewarch C.L., Antoshechkin I., Lis J.T., Meyer B.J., Baugh L.R. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Rep. 2014;6:455–466. doi: 10.1016/j.celrep.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi Y., Shibata H., Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Missra A., Gilmour D.S. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D.K., Hasegawa J., Omori M., Inukai N., et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi Y., Inukai N., Narita T., Wada T., Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn S.H., Kim M., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 21.Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Peterlin B.M. Transcription elongation takes central stage: the P-TEFb connection. Cell Cycle. 2010;9:2933–2934. doi: 10.4161/cc.9.15.12698. [DOI] [PubMed] [Google Scholar]
- 24.Xue Y., Yang Z., Chen R., Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R., Liu M., Li H., Xue Y., Ramey W.N., He N., Ai N., Luo H., Zhu Y., Zhou N., et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R., Yang Z., Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 27.Yik J.H., Chen R., Nishimura R., Jennings J.L., Link A.J., Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 28.Shi J., Vakoc C.R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisgrove D.A., Mahmoudi T., Henklein P., Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., et al. Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 2011;39:9592–9604. doi: 10.1093/nar/gkr698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X., Lu X., Liu R., Ai N., Cao Z., Li Y., Liu J., Yu B., Liu K., Wang H., et al. Histone cross-talk connects protein phosphatase 1alpha (PP1alpha) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. J. Biol. Chem. 2014;289:23154–23167. doi: 10.1074/jbc.M114.570812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Luo Z., Lin C., Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi H., Parmely T.J., Sato S., Tomomori-Sato C., Banks C.A., Kong S.E., Szutorisz H., Swanson S.K., Martin-Brown S., Washburn M.P., et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He N., Chan C.K., Sobhian B., Chou S., Xue Y., Liu M., Alber T., Benkirane M., Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E636–645. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang B., Yang X.D., Zhou M.M., Ozato K., Chen L.F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart H.J., Horne G.A., Bastow S., Chevassut T.J. BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1. Cancer Med. 2013;2:826–835. doi: 10.1002/cam4.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R., Liu M., Zhang K., Zhou Q. Isolation and functional characterization of P-TEFb-associated factors that control general and HIV-1 transcriptional elongation. Methods. 2011;53:85–90. doi: 10.1016/j.ymeth.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X., Tian L., Li J., Zhang Y., Han V., Li Y., Xu X., Li H., Chen X., Chen J., et al. Investigation of receptor interacting protein (RIP3)-dependent protein phosphorylation by quantitative phosphoproteomics. Mol. Cell. Proteomics. 2012;11:1640–1651. doi: 10.1074/mcp.M112.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paoletti A.C., Parmely T.J., Tomomori-Sato C., Sato S., Zhu D., Conaway R.C., Conaway J.W., Florens L., Washburn M.P. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N.J., Alber T., Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A., et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donner A.J., Ebmeier C.C., Taatjes D.J., Espinosa J.M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong Y.W., Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 50.Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 51.Wang W., Yao X., Huang Y., Hu X., Liu R., Hou D., Chen R., Wang G. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription. 2013;4:39–51. doi: 10.4161/trns.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.