Abstract
Transcriptional interference (TI), where transcription from a promoter is inhibited by the activity of other promoters in its vicinity on the same DNA, enables transcription factors to regulate a target promoter indirectly, inducing or relieving TI by controlling the interfering promoter. For convergent promoters, stochastic simulations indicate that relief of TI can be inhibited if the repressor at the interfering promoter has slow binding kinetics, making it either sensitive to frequent dislodgement by elongating RNA polymerases (RNAPs) from the target promoter, or able to be a strong roadblock to these RNAPs. In vivo measurements of relief of TI by CI or Cro repressors in the bacteriophage λ PR–PRE system show strong relief of TI and a lack of dislodgement and roadblocking effects, indicative of rapid CI and Cro binding kinetics. However, repression of the same λ promoter by a catalytically dead CRISPR Cas9 protein gave either compromised or no relief of TI depending on the orientation at which it binds DNA, consistent with dCas9 being a slow kinetics repressor. This analysis shows how the intrinsic properties of a repressor can be evolutionarily tuned to set the magnitude of relief of TI.
INTRODUCTION
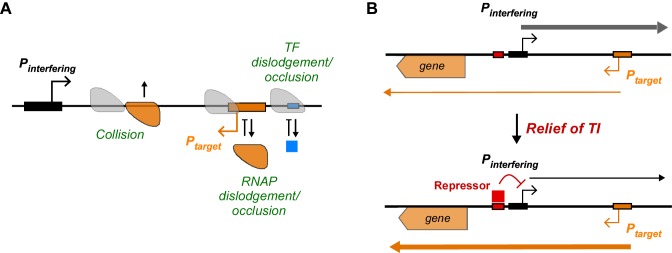
Transcriptional interference (TI), defined as the direct suppressive influence of one transcriptional process on another in cis, has been shown to play an important role in a variety of prokaryotic and eukaryotic gene regulatory systems (reviewed by (1–5)). An interfering promoter can exert TI on a target promoter by various mechanisms. Figure 1A shows TI mechanisms that can combine when the target and interfering promoter are non-overlapping and convergent, a frequent promoter juxtaposition in Escherichia coli and other bacteria (6–8), as well as in yeast (9) and multicellular organisms (10,11). Elongating RNA polymerases (RNAPs) initiated from the interfering promoter may inhibit the activity of the target promoter by (i) impeding the progress of elongating RNAPs, (ii) dislodging pre-initiating RNAP bound to the promoter, (iii) occluding binding of RNAP to the promoter or (iv) dislodging or inhibiting binding of a transcription factor needed for target promoter activity (Figure 1A).
Figure 1.
Transcriptional interference (TI) and its modulation. (A) The mechanisms of TI which are operative when a target promoter (orange) and an interfering promoter (black) are non-overlapping and convergent. Elongating RNAPs can remove TFs or promoter-bound RNAP from the DNA (dislodgement) or block their binding (occlusion). Head-to-head ‘collisions’ between elongating RNAPs can cause termination of one or both RNAPs. (B) Modulation of TI is a simple way to reverse the effect of a transcription factor. In this example, repression of the interfering promoter modulates TI so that the target promoter experiences an activating effect.
TI provides regulatory opportunities by allowing transcription factors to control a target promoter, indirectly, through regulation of the interfering promoter. Such regulation is also a simple way to reverse the effect of a transcription factor; a repressor can have an activating effect on a target promoter by repressing an interfering promoter to relieve TI (Figure 1B), while an activator can have a repressive effect on a target promoter by activating an interfering promoter to induce TI. Modulation of TI by regulation of the interfering promoter is known to be critical for gene regulation in a variety of systems, including bacteriophages, bacteria, yeast, Drosophila and mouse (12–20).
In many cases, this regulation involves competition between convergent promoters. Relief of TI by repression of the strong lytic promoter appears to be the primary mechanism for activation of the convergent lysogenic promoter by the immunity repressor proteins of P2-like bacteriophages (12,13). Repression of colicin genes by the E. coli LexA repressor relieves TI on expression of the convergent colicin immunity gene (14). The budding yeast FLO11 gene is regulated by a double TI system, where the upstream ICR1 promoter that inhibits FLO11 transcription is itself controlled by the convergent PWR1 promoter, with activation or repression of PWR1 inducing or relieving TI on ICR1, causing stimulation or inhibition of FLO11 transcription (19). The yeast a1/α2 repressor controls entry into meiosis by relief of TI, repressing an inhibitory convergent promoter downstream of the IME4 gene (17). In this example, and other cases involving convergent promoters, inhibition by TI can sometimes be supplemented by antisense RNA effects (5,7,21,22). The powerful combination of antisense RNA regulation and TI have encouraged the use of convergent promoters for synthetic biology applications (23,24).
Though it has been shown that promoters can be activated by relief of TI, the parameters that determine the efficacy of this mode of regulation are poorly understood. How does the activity of the target promoter respond to different levels of repression of the interfering promoter? Does the mechanism of TI matter? For convergent promoters, how do interactions between the elongating RNAPs from the target promoter and the repressor bound at the interfering promoter affect relief of TI? Elongating RNAPs from the target promoter may in theory dislodge the repressor and diminish its repression of the interfering promoter (25). If, on the other hand, the repressor is resistant to dislodgement, it may act as a roadblock to elongating RNAPs, preventing transcription from the target promoter reaching downstream genes (26,27).
To achieve a better quantitative understanding of relief of TI for convergent promoters, we first used stochastic simulations to systematically analyse how the mechanism of TI and the properties of the repressor might affect relief. Our modelling results indicated that relief of TI is strongly dependent on the binding kinetics of the repressor and its resistance to dislodgement by elongating RNAPs.
As a system for experimentally examining relief of TI, we used two convergent promoters in bacteriophage λ, a long-standing model for developmental switches (28). The λ PR and PRE promoters have opposing functions in λ's lysis-lysogeny decision. PR promotes lytic development by expressing genes needed for phage replication and virion production. PRE promotes lysogenic development by being the major early promoter for expression of the CI lysogenic repressor (29). PRE antagonizes PR indirectly, by expressing CI, which represses PR to shut off lytic development and establish lysogeny. We have shown that PR also fights back against PRE by using TI, causing a direct ∼5-fold inhibition of PRE activity (30).
Here we show that repression of PR by the λ CI protein can efficiently relieve TI on PRE. This effect can be quantitatively reproduced in a specific model of the λ system as long as CI has rapid DNA-binding kinetics, which prevents CI at PR being a significant roadblock to elongating RNAPs from PRE and also prevents diminished repression of PR resulting from its dislodgement by these RNAPs. In contrast, when the same λ PR promoter was repressed by a dCas9 complex that functions as a strong transcriptional roadblock with slow DNA-binding kinetics, relief of TI was abolished. Thus, repressors that give equal repression of the interfering promoter can have quite different effects on a convergent target promoter.
MATERIALS AND METHODS
Strains and reporters
All lacZ reporter constructs were integrated into the λ attB site of E. coli strain NK7049 (ΔlacIZYA)X74 galOP308 StrR Su– (31).
The construction of PR.(cro– PRE), PRE.(cro– PR) and PRE.(cro– PR–).lacZ reporters has been described previously (30), and the PRE.(cro+ PR) reporter was made in the same way. These constructs were present within a prophage of λRS45ΔYA, an imm21 lac reporter phage (31), modified by removal of the lacYA genes (32).
The roadblock reporters were derived from plasmid pIT-SL.P2 pC.lacZO2− (SpecR), and were integrated into the λ attP site of NK7049 using the CRIM system (33). pIT-SL.P2 pC.lacZO2− was derived from pIT-SL.lacZY (34) by deletion of 15 bp within the lacY gene, mutation of the lacO2 operator within the lacZ gene (35) and insertion of the bacteriophage P2 pC promoter (positions −62 to +40) and various restriction sites upstream of the RNaseIII cleavage site and the lacZO2– gene. The λ OR operator (OR1.OR2.OR3) was placed with the first bp of OR1 at the +102 position of P2 pC; the λ OL operator (OL1.OL2.OL3, with PL inactivated by mutations (32)) was placed with the first bp of OL1 at +102; the lacOid operator was placed with its first base at +102.
The CI repressor was expressed from Plac+ on single copy pZC320cI or high copy pZE15cI plasmids (AmpR; (32)), repressed by LacI expressed from a medium copy pUHA-1 plasmid (p15a origin, KanR; H. Bujard, Heidelberg University, Germany) and induced by IPTG. λ Cro was similarly supplied by pZE15cro for the roadblock assay.
CII was expressed either from the low copy pZS41λcII plasmid (SpecR) or high copy pZE15λcII (AmpR; (30)). pZS41λcII was obtained by cloning a polymerase chain reaction (PCR) fragment containing the λ cII gene (λ positions 38357–38662) into the NheI and BamHI restriction sites downstream of the TetR-repressible pLTetO-1 promoter in pZS41 (36). TetR was supplied from a DNA fragment containing the pN25 promoter upstream of tetR, derived from DH5αZ1 (36) and chromosomally integrated as part of a CRIM-derived plasmid pIT-CH (ChlorR) into the HK022 attB site (33). CII expression from pZS41λcII was induced with 100 ng/ml anhydrotetracycline (ATc), the minimal concentration that gave maximal expression of the PRE.(cro– PR–).lacZ reporter. Expression of CII from pZE15λcII was repressed by LacI produced by pUHA1 and was induced by 130–300 μM IPTG. The activity of PR in the absence of CI was consistently 25% lower in the presence of CII than in the absence of CII, consistent with the non-specific inhibition of lacZ reporters by CII protein seen by (30). Sequences of all constructs are available on request.
The Streptococcus pyogenes dCas9 was expressed from pPC_Sp_dCas9 plasmid under the control of an intermediate strength promoter, PC. The pPC_Sp_dCas9 plasmid was derived from DS-ST1casN- (Addgene plasmid # 48659) by (i) replacing the Streptococcus thermophilus #1 dCas9 open reading frame with that of S. pyogenes dCas9 obtained from PCR amplification of pdCas9-bacteria (Addgene plasmid # 44249); (ii) removing the S. thermophilus #1 tracrRNA expression module; and (iii) replacing the spectinomycin resistance gene cassette with a kanamycin resistance gene cassette. The gRNA expression plasmid pgRNA-p15a was derived from pgRNA-bacteria (Addgene plasmid # 44251) by replacing the high-copy CloE origin of replication with the medium copy p15a origin. The spacer sequences were CGTGTTGACTATTTTACCTC (Top), GGTAAAATAGTCAACACGCA (Bottom) and AACTTTCAGTTTAGCGGTCT (control, which targets the RFP gene, not present in any of the strains used in this study), respectively.
LacZ assays
Microtiter plate-based LacZ assays were carried out as previously described (30). Briefly, fresh colonies on selective LB plates were resuspended in LB and used to inoculate 200 μl of LB containing the appropriate antibiotics and various concentrations of IPTG in a 96-well microtiter plate, and incubated overnight at 37°C with shaking. Overnight cultures were diluted to an OD600nm of ∼0.6 with LB before further diluting 3 μl into 97 μl of fresh LB plus appropriate antibiotic, 100 ng/ml ATc and various concentrations of IPTG. Cultures were incubated with shaking at 37°C until OD600nm reached mid-log phase (0.65–0.75), then added to a combined lysis-assay buffer, with each well of a microtiter plate containing the following: 20 μl culture, 30 μl LB, 150 μl of TZ8 (100 mM Tris–HCl, pH 8.0, 1 mM MgSO4, 10 mM KCl), 40 μl of ONPG (o-nitrophenyl-β-D-galactoside, 4 mg/ml in TZ8), 1.9 μl of 2-mercaptoethanol and 0.95 μl of polymyxin B (20 mg/ml; Sigma). Assays were performed on cultures started from independent colonies and repeated on at least three different days. Error bars represent 95% confidence intervals.
Stochastic simulations
Continuous, deterministic mathematical models can only be applied to simplified TI situations (37). We thus used stochastic simulations, with a discrete fixed time-step stochastic algorithm that is a combination of the TI simulation algorithm described in (30) and the roadblocking simulation algorithm described in (27). In the fixed time step approach, each step is set to the time taken for RNAP to move forward one base pair (1/60 s). All other events (see Figure 2A and B) are assigned a specific rate. If a particular event is possible, its occurrence during the next time step is decided by generating a random number between 0 and 1; if this number is less than 1–e–k, where k is the relevant rate, then that event occurs.
Figure 2.
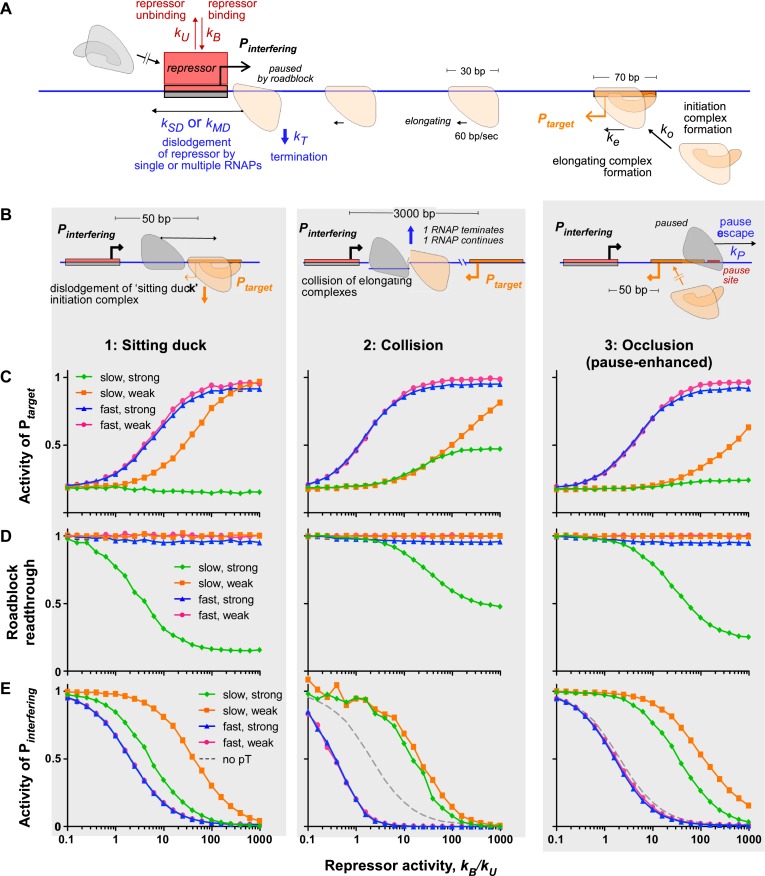
Stochastic modelling shows that TI is best relieved by transcription factors with fast kinetics. (A) Schematic and parameters of a model of TI between convergent promoters. (B) Three different TI scenarios are modelled in which each is primarily influenced by a different mechanism of interference. The parameters of each model are adjusted to produce the same overall level of interference (5.5-fold) in each scenario. (C) Fractional activity of the target promoter, Ptarget, for each scenario in (B). (D) The fraction of transcription from Ptarget that is able to ‘read-through’ the repressor roadblock at Pinterfering, for each scenario in (B). (E) Fractional activity of the interfering promoter, Pinterfering, for each scenario in (B). For (C, D and E), promoter activities and readthrough are shown as a function of increasing repressor activity, given by the ratio of binding to unbinding rate constants, kB/kU. Notably, the same equilibrium binding level kB/kU can be obtained with slow binding and unbinding kinetics (such as kU = 0.0005 s−1; green) or fast kinetics (such as kU = 0.5 s−1; red). Production rates of elongating polymerases ( = ko/(ko + ke), set with ko = ke) are 0.1 s–1 from Pinterfering and 0.01 s–1Ptarget (scenario 1) or 0.1 s–1 from both promoters (scenarios 2 and 3). The termination rate at a roadblock, kT, was fixed at 0.028 s–1 and the pause escape rate (scenario 3) kP = 0.032 s–1. For slow binding repressors (kU = 0.0005 s–1), the corresponding single RNAP dislodgement rates (kSD) and multiple RNAP dislodgement rates (kMD) are 0.0015 and 0.026 s–1 respectively, based on our study of roadblocking by Lac repressor (27). For fast binding repressors, instantaneous dislodgement was used. Repressors that dissociate slowly are more tightly bound and more difficult for RNAP to dislodge.
Rates are taken directly from (30) and (27) except where otherwise noted. For repressors that form a strong barrier to RNAP, the dislodgement rates by a single RNAP (kSD) or multiple stalled RNAPs (kMD) were set to 0.0015 and 0.026 s–1, equivalent to those estimated for LacI at a strong Oid site (27). For repressors that only form a weak barrier to RNAPs, instantaneous dislodgement was used. The termination rate for a roadblock-stalled RNAP (either stalled at a protein roadblock or stalled by a paused RNAP ahead of it) (27) was re-calibrated in this study to produce the same amount of roadblocking in the two-step promoter firing model (kT = 0.028 s−1) or the three-step CII-activated PRE model (kT = 0.066 s−1) as in the one-step promoter firing model, and thus is different from that reported by Hao et al. (27). RNAP bound at a promoter (the ‘sitting duck’) is assumed to not be a barrier to elongating RNAP. Although this is the case for RNAP bound to the pL promoter of phage 186 (38), and probably also for the λ PRE promoter, given the weak TI of PRE on PR (30), we cannot exclude the possibility that RNAP bound at λ PR is a barrier. However, we note that the high activity of λ PR means that bound but not elongating RNAPs at PR must be short lived.
In the PR–PRE specific model (Figures 3 and 6), promoter firing is modelled as a three-step process in order to accommodate activation of PRE by CII, as described in (30). The slightly lower TI seen in the experiments described here, a 5-fold difference (compared to 5.5-fold in (30)), was fitted by assuming a somewhat higher CII concentration produced by the ATc induction system, which increases PRE activity 1.5-fold. Rates of CI binding, kB were 0.0034, 0.0089, 0.0163, 0.0355, 0.0986, 0.2437, 0.5454, 0.9458, 2.8461 s−1, for pZC320cI with 0, 2.5, 5, 10, 20, 40, 80, 160 μM IPTG or pZE15cI with 200 μM IPTG, respectively, calculated using a fixed unbinding rate (kU = 0.5 s−1) and the IPTG to CI conversion measurements of (32). The rate of RNAP termination at tR1 was taken as 0.168 s−1 (30). Note that there are three discrete termination sites at tR1, which are at positions +352, +372 and +402, respectively.
Figure 3.
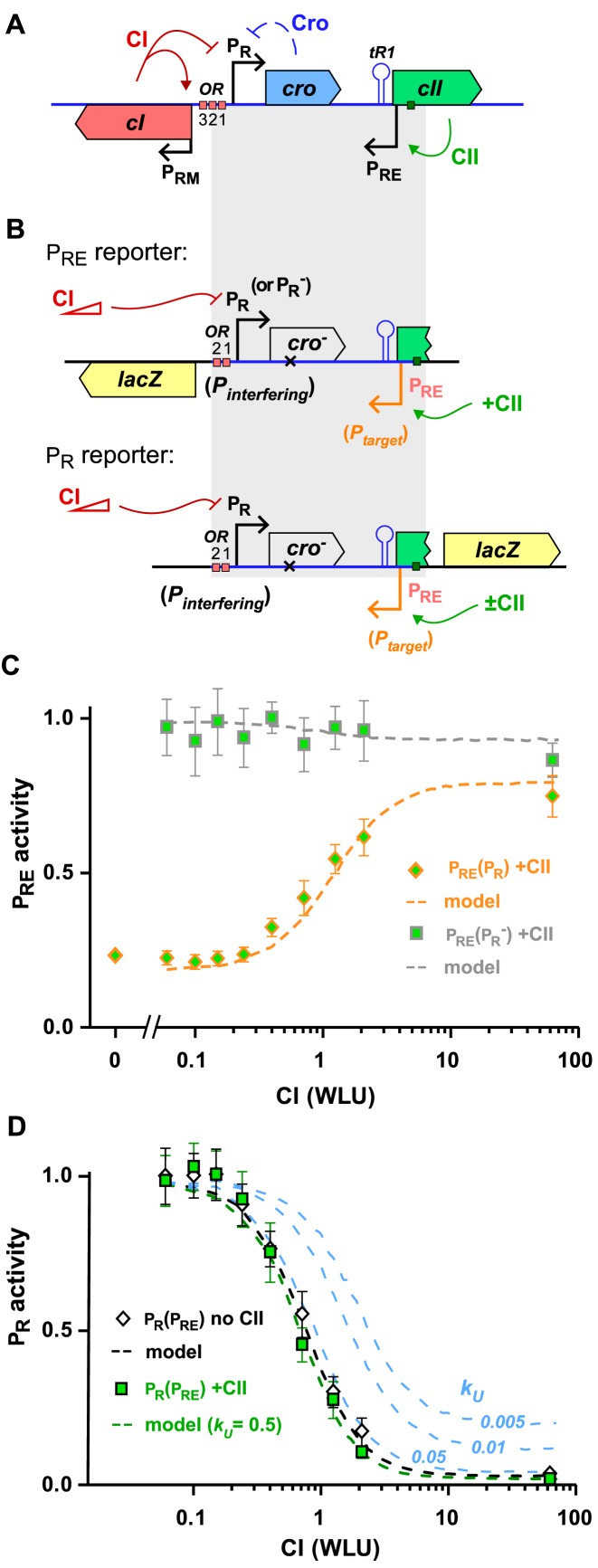
CI repression of λ PR activates PRE by relief of TI. (A) Map of the λ CI-CII region. (B) Structure of the reporter constructs used to measure PRE and PR activity. (C) Activity of the CII-activated PRE in the PRE.(cro− PR).lacZ and PRE.(cro− PR−).lacZ reporters with increasing CI repression of PR. CI levels are expressed in wild-type lysogenic units (WLU), where 1 WLU is the amount of CI produced by a wild-type lambda lysogen. Activity is normalized to the highest LacZ units obtained in the PRE.(cro− PR−).lacZ reporter (n = 9). (D) CI repression of PR in the PR.(cro− PRE). lacZ reporter in the presence or absence of PRE activity (±CII). LacZ activities of each promoter were normalized by their value in the absence of CI (n = 9). Model fits in (C and D) were obtained by incorporation of λ-specific details (30) into the relief of pause-enhanced occlusion model of Figure 2 (see text).
Figure 6.
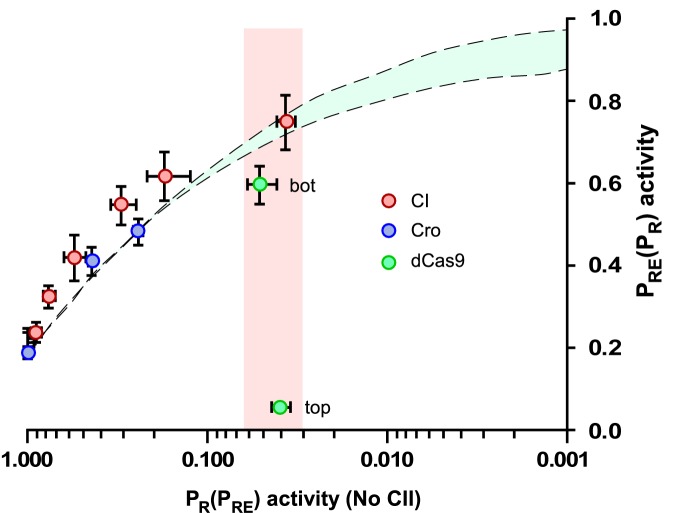
Relief of TI is modulated by the intrinsic properties of the repressors. Relief of TI is plotted as PRE versus PR activity (in the absence of CII). Data on relief of TI by CI is from Figure 3C and D, and dCas9 from Figure 5C and D. Relief of TI at PRE by Cro repression of PR was measured with Cro expressed either from the chromosomal LacZ reporter itself (1xcro) or from the reporter and additionally from a second chromosomal PR.cro expression construct (2xcro) (see main text for details). Vertical and horizontal error bars are 95% confidence intervals in PRE and PR activity, respectively (n = 9 for CI and dCas9, n = 6 for Cro). Cyan shaded region shows simulated PRE activity at different magnitude of PR repressions for fast kinetics repressors of PR, kU = 0.5 s–1 with either strong or weak barrier parameters (Figure 2).
The order of attempted events in the simulation is as follows:
Repressor binding or unbinding. Binding only occurs if the operator is not already occupied by a repressor, and none of the operator positions is overlapped by RNAP. Bound repressor occupies all 20 bp of the operator site. A bound repressor could either dissociate spontaneously from the operator with a rate kU or be removed from the operator by RNAP(s) with rates kSD or kMD.
RNAP movement. Starting with the RNAP furthest from the promoter, each RNAP (paused or not) attempts to move forward by one time step. If the RNAP is blocked either by repressor or by a paused RNAP, it only moves forward if itself or the leading RNAP dislodges the repressor, through application of the kSD or kMD rate (depending on whether there is one or more RNAP stalled at the roadblock). Thus a successful dislodgement will enable all RNAP queued at the roadblock to advance. Any RNAP that is unable to move forward is considered paused. If the back of an RNAP passes the end of the DNA (that is positions 1 and 420 respectively for left- and right-moving RNAPs), a new transcript is counted and that RNAP is eliminated from the DNA.
Termination. If two convergent RNAPs collide, one is removed at random and the other remains. A paused RNAP can be removed from the system with rate kT.
Promoter firing. A new open complex is loaded at PR with rate ko = 0.19 s−1 if there is no RNAP overlapping positions 1– 65 of the DNA (the first base pair of DNA in the model is at −51 of PR) or at PRE with rate ko = 0.0006 s−1 if there is no RNAP overlapping positions 356–420 of the DNA. Loading of a new RNAP at PRE is accelerated to 0.20 s−1 in the presence 100 ng/ml ATc (equivalent to ∼2000 CII tetramers). An open complex is converted to an elongating complex with rate ke (100 s−1 and 0.26 s−1 respectively for PR and PRE). Once an open complex is converted to an elongating complex, the size of RNAP is reduced from 65 to 30 bp, occupying −16 to +14 of the promoter.
In a typical run, the simulation was allowed to continue for 106 s for each of the following three conditions with CI ranging from 0 to 70 WLU: (i) PR–PRE, +CII; (ii) PR–PRE, No CII; (iii) PRE only, +CII. From the simulations, roadblock readthrough for RNAP from PRE at CI-bound PR was calculated as the ratio of transcripts produced for (PRE only, +CII, +CI) to (PRE only, +CII, No CI). The relief of TI of PR on PRE by CI was calculated as the ratio of transcripts produced for (PRE-PR, +CII, +CI) to (PRE only, +CII, No CI). Programs were written in FORTRAN (available on request) and were executed on a standard PC.
RESULTS
Modelling relief of TI by repression of a convergent promoter
To better understand how relief of TI might depend on the properties of the repressor and the mechanism of TI, we performed stochastic simulations with a model incorporating binding and unbinding of a repressor to the DNA and its interactions with elongating RNAPs, as well as the RNAP–RNAP interactions between convergent prokaryotic promoters that generate TI (Figure 2A).
We examined four combinations of properties of the repressor: slow versus fast DNA binding kinetics, and formation of a strong or weak barrier to elongating RNAPs. The repressor was treated as a single species that binds over the Pinterfering promoter, with an effective on-rate kB (s–1) (the product of the repressor concentration (M) and its on-rate constant kon (M–1s–1)), and an off-rate kU (s–1). The ratio of kB and kU determines the equilibrium occupancy of the repressor binding site (occupancy = kB/(kB + kU)), thus the same occupancy can be achieved if kB and kU are both high (fast kinetics) or both low (slow kinetics). In the simulations, slow repressor kinetics were obtained with kU = 0.0005 s−1, which is roughly equivalent to that estimated for Lac repressor unbinding from its Oid site, one of the strongest known repressor–DNA interactions, while fast kinetics were obtained with kU = 0.5 s−1, which is similar to Lac repressor unbinding from its weak O3 binding site (27). Interaction of the repressor with elongating RNAPs was treated as in our previous study of Lac repressor roadblocking in vivo (27). A single elongating RNAP that encounters the repressor stalls and can then either be terminated with rate kT or can resume elongation if (i) it can actively dislodge the repressor, with a rate kSD, or (ii) if the repressor spontaneously dissociates (Figure 2A). A queue of two or more RNAPs stalled at the roadblock can cooperate, dislodging the repressor with a higher rate kMD (each stalled RNAP remains at risk of termination). In the simulations, low dislodgement rates, equivalent to those estimated for LacI at Oid (kSD = 0.0015 and kMD = 0.026 s–1, (20)), were used for the repressor forming a strong barrier to RNAP, while instantaneous dislodgement was used to give a weak barrier.
Relief of TI by these four repressor classes was simulated for three different TI scenarios (Figure 2B), each designed to maximize the effect of one of the three major TI mechanisms found in studies involving moderate strength promoters in E. coli (30,37,38): (i) ‘sitting duck’ TI, where a non-elongating RNAP bound at the promoter is removed or inactivated by an elongating RNAP from the opposing promoter; (ii) collision TI, where encounters between elongating RNAPs moving in opposite directions on the DNA lead to dissociation of one or both RNAPs; and (iii) occlusion TI, where an elongating RNAP passing over the opposing promoter blocks access to that promoter by free RNAP (Figure 2B). Firing of the target promoter Ptarget and the interfering promoter Pinterfering was modelled simply as a two-step process (37), with the rate of formation of the bound initiation complex given by ko and its conversion to the elongating complex by ke. In the simulations, ko was set equal to ke for each promoter. The overall rates of production of elongating RNAPs from Ptarget and Pinterfering, kt and ki, are thus equal to ko/2 (and ke/2) (37).
We adjusted parameters for each TI scenario to obtain ∼5.5-fold TI by Pinterfering on Ptarget. In the sitting duck scenario, we used a 10-fold difference in promoter strength (ki = 0.1 s−1, kt = 0.01 s−1) to maximize sitting duck interference, and a short distance between the promoters (50 bp) to minimize collisions (37). Occlusion is not significant when Pinterfering is of moderate strength (37). In the collision scenario, collision TI was strengthened by increasing the interpromoter distance to 3 kb, and sitting duck TI was weakened by having Pinterfering and Ptarget of equal strength (ki = kt = 0.1 s−1). In the occlusion scenario, a short interpromoter distance was used and occlusion was enhanced by introducing a pause site for RNAP just upstream of Ptarget (30). RNAP from Pinterfering that pauses at this site overlaps the binding site for RNAP at Ptarget to inhibit RNAP binding. A trailing RNAP originating from Pinterfering can cooperate with paused RNAP to re-initiate elongation. To give 5.5-fold TI, the rate of spontaneous exit from the paused state kP was set at 0.032 s−1 (pause lifetime = 31 s), a value consistent with in vivo measurements (30). Paused RNAPs were not subject to termination.
For each of these repressor-TI combinations, we simulated how much relief of TI was obtained as the activity of the repressor was increased. Repressor activity was increased in the model by increasing kB, which is equivalent to increasing repressor concentration, since kB = kon × [repressor] and kon is a constant. We plot repressor activity as kB/kU to enable comparison of repressors with different kinetics but the same equilibrium binding activity (Figure 2C–E).
Strong relief of TI was seen in all TI scenarios, with the target promoter able to be restored to full activity at high repressor activity. However, relief of TI was strongly dependent on the properties of the repressor. Repressors with fast kinetics gave the most effective and sensitive relief of TI, whether or not they are a barrier to RNAP (Figure 2C). Repressors with slow kinetics and weak barrier activity could give complete relief of TI but only at higher repressor concentrations. The combination of slow kinetics and strong barrier activity strongly inhibited relief of TI.
As anticipated, the inhibition of relief of TI is due to two factors: roadblocking by the repressor and sensitivity of repression to dislodgement. Only the slow-kinetics strong-barrier repressor produces effective roadblocking, reducing the fraction of RNAP from Ptarget that pass (read through) its binding site (Figure 2D) and effectively limits the scope for relief of TI. The fast-kinetics strong-barrier repressor does not cause a decrease in readthrough, even though it can block RNAP elongation, because it naturally dissociates quickly, allowing most stalled RNAPs to pass before they are terminated.
Dislodgement sensitivity inhibits relief of TI because it weakens repression of the interfering promoter, and is seen for both slow-kinetics repressors (Figure 2E). Dislodgement sensitivity is the only mechanism reducing relief of TI by a slow-kinetics weak-barrier repressor, as it does not cause roadblocking (Figure 2D). The slow-kinetics strong-barrier repressor is less dislodgement sensitive simply because it is often not dislodged by the RNAPs. Dislodgement of slow-kinetics repressors by RNAPs from Ptarget has a large fold effect on their DNA occupancy because they spontaneously unbind at a low rate, and consequently dislodgement can make a large difference to their overall rate of leaving the DNA (25). In contrast, fast-kinetics repressors spontaneously unbind frequently, so that dislodgement events make little difference to their overall rate of leaving the DNA. Thus, dislodgement effects become significant when the rate of production of RNAP from the target promoter exceeds the kU of the repressor. At very high repressor concentrations, the absolute magnitude of the repressor dislodgement effect becomes less, due to fast re-binding of the repressor (Figure 2C), causing the relief of TI to be limited solely by the roadblocking effect of the repressor.
Some differences in the relief of TI responses are apparent for the different TI mechanisms. These result primarily from the effect of Ptarget strength on dislodgement and the effect of both Ptarget strength and the interpromoter distance on roadblocking. Repression of Pinterfering is more affected by dislodgement when Ptarget is stronger (collision and occlusion scenarios). Roadblocking decreases when Ptarget is stronger (collision and occlusion scenarios) and when the interpromoter distance is larger (collision scenario), as these factors allow more cooperation by multiple RNAPs to overcome the barrier. The collision scenario gave enhanced relief of TI compared to the other scenarios, even for the fast-kinetics repressors, for which roadblocking and dislodgement sensitivity are absent (Figure 2C). In the collision scenario there is strong reciprocal TI between Pinterfering and Ptarget, and reductions in the activity of one promoter are magnified because it becomes less able to defend itself against TI from the other promoter (37). This causes repression of Pinterfering to be hyper-sensitive, occurring at roughly 10-fold lower repressor concentrations than in the other scenarios (Figure 2E). Repression of Pinterfering reduces the TI experienced by Ptarget but also allows Ptarget to exert stronger interference on Pinterfering, reducing Pinterfering activity further and allowing increased relief of TI.
CI repression can relieve TI by λ PR on PRE
The modelling indicates that efficient relief of TI should be achievable by strong repression of the interfering promoter, provided that the repressor is not a substantial roadblock to RNAPs and its repression is not strongly disrupted by dislodgement by RNAPs. To quantitatively examine relief of TI in a natural context, we used the PR and PRE convergent promoters of bacteriophage λ, a system in which PR exerts strong TI on PRE, primarily through the pause-enhanced occlusion mechanism (30).
PR and PRE are separated by 320 bp and are involved in the decision between lytic or lysogenic development. PR, one of the early lytic promoters of λ, is essential for lytic development. PRE is the major early promoter for the gene encoding the lysogenic repressor CI. PRE activity requires activation by the CII protein, and is needed for establishment of lysogeny after infection ((39,40); Figure 3A). During establishment of lysogeny, CI expressed from PRE should repress PR, potentially relieving TI on PRE and increasing transcription of the cI gene. Thus, here PRE is the target promoter and PR is the interfering promoter subject to repression.
We assayed PR or PRE transcription using chromosomally integrated lacZ reporters (30). The use of a single reporter gene necessitated different constructs for assay of PR or PRE but was preferred due to the high sensitivity of LacZ detection. The inserted λ DNA fragment contains no active genes (the cro gene carried mutations in the helix-turn-helix DNA binding motif), and has the PRM promoter truncated, but contains the OR1 and OR2 CI binding sites for PR repression as well as the CII binding site at PRE (Figure 3B). The tR1 terminator, which gives 66% termination of PR transcription in these reporters in the absence of the λ N anti-terminator protein (30), was also present. We also used a fragment carrying PR inactivated by mutation (PR–; (30)) for measuring the activity of un-interfered PRE. Previously, we found 5.5-fold TI by PR on PRE, primarily due to RNAP from PR pausing at the tR1 terminator and occluding PRE. TI is asymmetrical, with transcription from CII-activated PRE not substantially interfering with PR. This is explained by the promoters being of similar strengths (low sitting duck TI), the short interpromoter distance (low collision TI) and a lack of roadblocking by CII bound at PRE (30). Thus, the PR–PRE situation is similar to our occlusion scenario (Figure 2B).
We activated PRE in the reporter strains by expressing a fixed level of CII. Continued high level CII expression is toxic to cells (41) and we thus induced CII expression from the pLTetO1 promoter on a low copy plasmid under the control of chromosomally expressed TetR repressor, with an empty expression plasmid used as the no-CII control.
A range of CI levels were provided from IPTG induction of the single copy pZC320cI plasmid or the multicopy pZE15cI plasmid (‘Materials and Methods'section). These plasmids provide a wide range of CI concentrations up to ∼80 WLU (wild-type lysogenic units, where 1 WLU is the level of CI in a wild-type λ lysogen; (32)).
An almost 5-fold TI by unrepressed PR on PRE was seen by comparison of the activity of PRE in the presence or absence of PR (no-CI, PRE.(cro– PR).lacZ versus PRE.(cro– PR–).lacZ, Figure 3C), similar to the 5.5-fold seen previously (30). Increasing repression of PR by CI produced increased PRE activity up to ∼80% of its uninhibited level. In contrast, high CI levels gave no increase of PRE transcription in the absence of PR activity (PRE.(cro– PR–).lacZ, Figure 3C), demonstrating that CI activation of PRE is by relief of TI, rather than direct stimulation of PRE.
This relief of TI is possible because CI is neither a substantial roadblock to RNAPs from PRE, nor is its repression of PR sensitive to dislodgement by these RNAPs. The slight decrease in PRE transcription in the absence of PR activity (PRE.(cro– PR–).lacZ, Figure 3C) indicates that CI binding at PR causes at most 13 ± 6% roadblocking of PRE transcription. A lack of dislodgement sensitivity is shown by repression of PR by CI being unaffected by the activity of PRE (+CII versus no CII, Figure 3D).
λ CI is not a strong roadblock to RNAP
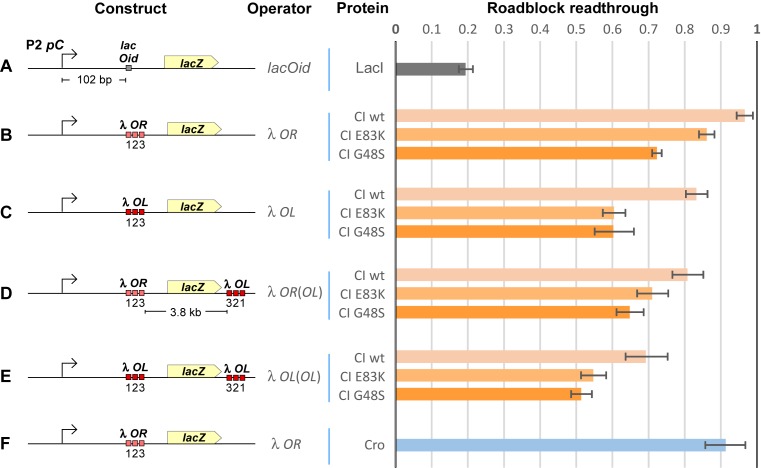
The slight decrease in activity of the PRE.(cro– PR–).lacZ reporter (Figure 3C) suggests that CI binding to PR is at most a weak roadblock to RNAPs from PRE. However, the PR– mutation in this reporter alters three basepairs of OR1, which is expected to reduce its CI affinity to a level similar to OR2 (42) and is likely to reduce roadblocking. To more precisely determine the roadblocking potential of CI, we examined a number of different situations in our roadblock assay system (27), placing CI binding sites between the weak PC promoter of phage P2 and a lacZ reporter gene (Figure 4). Use of a weak promoter maximizes roadblocking, since it reduces the probability of multiple RNAPs cooperating to overcome the roadblock (26,27). A high level of CI, sufficient to occupy all CI binding sites (43), was supplied with pZE15cI and 200 μM IPTG. In this assay system, Lac repressor binding to its single, strong Oid site reduced readthrough of the roadblock to 20% (Figure 4A). In contrast, CI binding to the wild-type OR site (containing operators OR1.2.3, without PR) allowed over 95% of RNAPs to pass without being terminated (Figure 4B).
Figure 4.
Assay of roadblocking by λ CI and Cro. Roadblock readthrough is defined as the LacZ activity obtained in the presence of the DNA-binding protein, which binds a site between the promoter and the LacZ reporter, divided by the LacZ activity obtained in the absence of the DNA-binding protein. Means and 95% confidence limits of the ratios are shown. (A) The interaction between LacI and its operator Oid presents a strong roadblock to transcription. LacI was supplied from pUHA-1, with pUHA-1ΔlacI used for the no protein control (n = 4). (B–E) Wild-type λ CI or DNA-binding enhanced CI mutants, CI(E83K) or CI(G48S) (45), were supplied by IPTG induction (100 μM) of pZE15cI/cI(E83K)/cI(G48S) (n = 6) (these reporter strains also carried pUHA1 for LacI-mediated control of CI expression). (F) λ Cro binding to OR is also a weak roadblock to transcription. λ Cro was similarly supplied by pZE15cro, with pZE15 serving as the no protein control (n = 9).
In our relief of TI reporters, OR is the only CI binding site. However, in the natural λ context, CI binding to OR is assisted by a DNA looping interaction with CI bound at OL 2.3 kb away (44). We found that the presence of OL downstream of lacZ, 3.8 kb away from OR, improved CI roadblocking, reducing readthrough to 81% (Figure 4D). CI roadblocking was also increased by increasing the strength of its DNA interaction in other ways. First, roadblocking was increased by replacing OR with the stronger OL set of binding sites, both in the absence or presence of looping to the distal OL site (83 or 69% readthrough, respectively, Figure 4C and E). Second, two mutants of CI that bind DNA more than 10-fold more strongly than wild-type, CI(E83K) and CI(G48S) (45), decreased readthrough for all constructs (Figure 4B–E). However, the maximal roadblocking, obtained with the CI(G48S) mutant at the looped OL sites, reducing readthrough to 50%, was still much weaker than the roadblocking by Lac repressor at Oid.
Modelling relief of TI by λ CI
The low level of roadblocking by CI and its insensitivity to dislodgement are consistent with its strong relief of TI. However, we wished to test whether our data was quantitatively comparable to our model of relief of TI. We thus altered the model to incorporate various specific details of the λ PR–PRE system used in our previous simulations of PR–PRE interference (30). The new model is similar to the pausing-enhanced occlusion scenario of Figure 2, but with some added complexities. It uses a three-step promoter firing model for PR and PRE, including RNAP in a closed complex at the promoter in equilibrium with free RNAP. It includes CII tetramerization and DNA binding, with CII activating PRE by increasing formation of the closed complex and also increasing isomerization to the open complex (46). Because the relative response of PRE activity to CII concentration was unaffected by PR activity, CII binding kinetics were assumed to be fast such that CII is neither sensitive to dislodgement, nor acts as a roadblock (30). There are three RNAP pause sites at tR1, with common rates of pause exit and termination at each site (47), with a trailing RNAP causing a paused leading RNAP to exit the pause (30).
CI repression of PR was implemented by assuming that OR is occupied by a CI complex with a fixed unbinding rate kU, and with specific empirically-derived kB values for each CI concentration. The repression of PR in the absence of PRE activity (PR.(cro– PRE) reporter in the absence of CII; Figure 3D) is thus determined by the ratio of kU and kB values at each concentration, but this does not constrain the absolute value of kU. However, we expected the lack of dislodgement sensitivity in CI repression of PR would require fast kinetics. Indeed, running the model with different kU values showed that only unbinding rates ≥ ∼0.05 s–1 reproduce CI repression of PR in the presence of PRE activity (Figure 3D).
Simulations using a fast kU = 0.5 s–1, together with weak barrier parameters (kSD = 0.07 s−1 and kMD = 0.08 s−1) extrapolated from the LacI roadblocking model (27), reproduced the weak roadblocking of PRE transcription in the PRE.(cro– PR) reporter (Figure 3D; we note that stronger barrier parameters would be needed if CI has a kU = 0.5 s–1).
Using these parameters, simulations of PRE activity in the presence of PR reproduced the observed relief of TI in the PRE.(cro– PR).lacZ reporter due to increasing repression of PR by CI (Figure 3C). Thus, the CI data can be explained by fast DNA-binding kinetics for CI at OR.
Relief of TI by λ Cro
PR is also repressed by the λ Cro protein, which binds to the same operators used by CI (34,48). We thus examined whether Cro was able to give relief of TI in this system.
We tested the effect of Cro by replacing the mutant cro gene on the reporters with the cro+ gene (1xcro). We also expressed a higher level of Cro by combining the cro+ reporter constructs with an additional PR.cro+ expression construct (no lacZ gene) inserted elsewhere on the E. coli chromosome (2xcro). In the absence of CII, the presence of an intact cro gene on the PR.(cro+ PRE).lacZ reporter reduced PR activity roughly 2-fold compared with the cro– reporter (1xcro/cro– relative PR activity 0.44 ± 0.02; errors are 95% confidence limits), consistent with previous measurements of Cro autoregulation in cis (34,48). The additional Cro supplied in trans to the cro+ reporter resulted in ∼4-fold reduction in PR activity (2xcro/cro– relative PR activity 0.24 ± 0.01).
Cro repression of PR was not affected by the activity of PRE, indicating a lack of sensitivity to dislodgement. In the presence of CII, the ratio of the activities of the PR.(cro+ PRE).lacZ reporter and its cro– counterpart (1xcro/cro–) was 0.45 ± 0.04, very similar to the 0.45 ± 0.02 seen in the absence of CII. Similarly, the 2xcro/cro– ratio for the PR reporters in the presence of CII was 0.23 ± 0.02, close to the 0.24 ± 0.01 seen in the absence of CII.
We tested Cro's roadblocking ability using the pC.OR.lacZ roadblocking reporter (Figure 4). Cro was expressed from pZE15cro, giving Cro levels capable of repressing PR over 2.5-fold (to 0.39 ± 0.04 of unrepressed activity in the PR (cro– PRE) reporter. The 92% readthrough seen (Figure 4F) indicates that Cro at OR is unlikely to be a substantial roadblock to transcription from PRE.
With these CI-like properties, Cro should also give relief of TI. In the absence of Cro, the PRE (cro–.PR) reporter gave 0.18 ± 0.01 maximal activity (relative to the PRE (cro– PR–) reporter), showing 5.6-fold TI of PR on PRE. With Cro supplied in cis (1xcro), the activity of PRE increased over 2-fold to 0.41 ± 0.03 of maximal (PRE (cro+ PR) reporter). When Cro was supplied both in cis and in trans (2xcro), PRE activity increased further to 0.48 ± 0.03 of maximal. Thus, as expected, repression of PR by Cro also activates PRE by relieving TI.
Regulating relief of TI by dCas9
Although the λ CI and Cro repressors are both capable of giving efficient relief of TI, our modelling predicts that relief of TI is not inevitable. In particular, repressors with slow binding/unbinding kinetics should give poor relief of TI.
To test this experimentally, we used a CRISPR dCas9 protein to repress PR. Cas9 proteins mutated to remove their DNA cleavage capability (dCas9) have been used as repressors of bacterial promoters by co-expressing the dCas9 protein and a guide RNA (gRNA) that targets the complex to the promoter DNA (49). We were thus able to program dCas9 to target the λ PR promoter in our reporters without the need to alter the existing sequences.
Furthermore, the dCas9-gRNA complex appears to have slow DNA-binding kinetics. The unbinding rate for S. pyogenes dCas9 (SpdCas9), measured in vitro by bio-layer interferometry was very low, ∼5 × 10−5 s−1 (50). The ability of SpdCas9 to cause strong transcriptional roadblocking in vivo (49,51), is also indicative of slow kinetics (Figure 2). Interestingly, SpdCas9 roadblocking appears to depend on the orientation with which it binds DNA relative to the direction of transcription, with strong roadblocking only observed when the RNA guide is complementary to the non-template DNA strand (49,51).
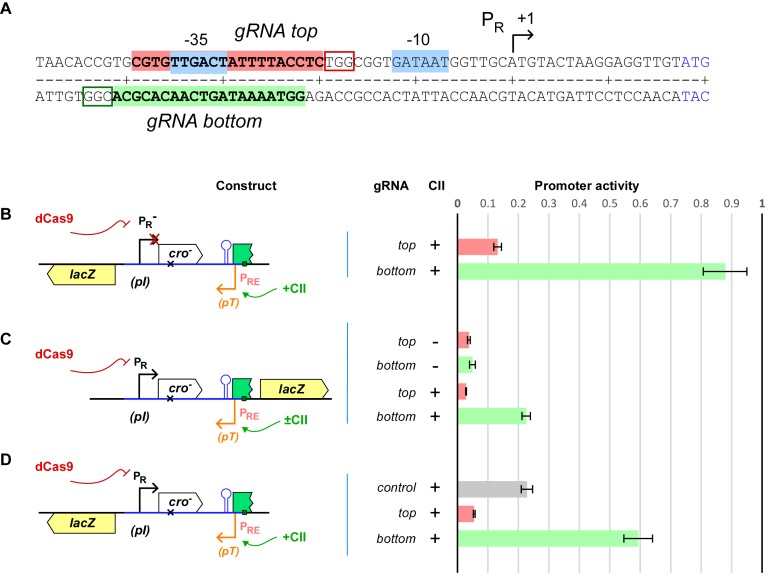
We transformed our PR and PRE reporter strains with a plasmid that constitutively expresses SpdCas9. We also independently expressed either of two gRNAs (designated top and bottom) that target the same region of the PR promoter but on opposite DNA strands (Figure 5A). The RNA in the dCas9–gRNA-top complex (dCas-top) binds to the bottom strand of PR (the non-template strand for PRE), while the RNA in the dCas9–gRNA-bottom complex (dCas-bot) binds to the top strand of PR (the template strand for PRE). Repression of PR in the absence of CII (PRE inactive) was strong for both dCas9-top and dCas-bot (Figure 5C, ∼96% repression) indicating strong binding and competition with RNAP binding in both cases.
Figure 5.
Modulating relief of TI by dCas9. (A) λ PR sequence and spacer sequence for top and bottom strand gRNA. The PAM sequences (NGG) are boxed. (B) Activity of the CII-activated PRE in the presence of dCas9 without the convergent PR promoter. Activity is normalized to the LacZ units obtained in the same reporter in the presence of control gRNA (n = 9). (C) dCas9 repression of PR in the PR.(cro− PRE).lacZ reporter in the presence or absence of PRE activity (±CII). LacZ activities of each promoter were normalized by their value obtained with scrambled gRNA (n = 9). (D) Activity of the CII-activated PRE in the PRE.(cro− PR).lacZ reporters in the presence of dCas9 and either scrambled or top or bottom strand gRNAs. Activity is normalized to the highest LacZ units obtained in the PRE.(cro− PR−).lacZ reporter with control gRNA (n = 9).
In contrast to repression, roadblocking by dCas9 showed a strong orientation bias, consistent with previous reports (49,51). Binding of the gRNA to the non-template strand of the PRE promoter (dCas9-top) reduced transcriptional readthrough to ∼13%, while binding to the template strand of DNA only weakly inhibited PRE transcription to ∼88% (dCas9-bot).
Repression of PR by the two dCas9 orientations was affected very differently by the activity of PRE, indicating different dislodgement sensitivities. In the presence of CII-activated PRE, PR repression by dCas9-top was unaffected, while repression by dCas9-bot was significantly weakened, with PR activity increasing ∼4.4-fold (Figure 5C).
These different roadblocking and dislodgement behaviours are consistent with dCas9-top being a slow-kinetics strong-barrier repressor and dCas9-bot being a slow-kinetics weak-barrier repressor (Figure 2). dCas9-top prevents passage of RNAPs from PRE because it rarely dissociates spontaneously and strongly resists active dislodgement by those RNAPs, thus its repression of PR remains strong and it causes strong roadblocking. In contrast, dCas9-bot is a weak barrier to RNAPs from PRE, so forms no roadblock, and its slow binding kinetics mean that its frequent dislodgement by these RNAPs can significantly reduce its occupation of its binding site, leading to partial derepression of PR when PRE is active.
As expected from these different properties, the two dCas9 complexes gave quite different relief of TI. In the absence of any PR-targeted repressor, the PRE.(cro– PR).lacZ reporter showed the usual ∼5-fold TI of PR on PRE (dCas9 + control gRNA Figure 5D). Repression of PR with dCas9-top resulted in not only no relief of TI, but also a further repression of PRE (Figure 5D). This is because, although dCas9-top is capable of repressing PR activity, any relief of TI on PRE is outweighed by strong transcriptional roadblocking of the PRE RNAPs. We suspect that the lower PRE transcription seen in the PR+ case (∼6%, Figure 5D) compared with the roadblock-only PR– case (∼13%, Figure 5B) is due to a combination of roadblocking and TI.
In contrast to dCas9-top, repression of PR with dCas9-bot, which causes only a small amount of roadblocking, resulted in substantial relief of TI, with PRE transcription reaching 60% of maximal (Figure 5D). However, this relief of TI was slightly lower than that seen with CI. Figure 6 shows the fractional relief of TI obtained for all four repressors—CI, Cro, dCas9-top and dCas9-bot—plotted against their repressive activity against PR (in the absence of PRE activity). While CI and Cro show strong relief of TI, at least as high as the maximum expected for repressors with fast kinetics, dCas9-bot gives sub-optimal relief of TI, and dCas9-top gives no relief of TI (Figure 6).
DISCUSSION
Modulation of gene expression by relief of TI
The magnitude of TI between two promoters can be tuned by evolutionary adjustment of promoter kinetics, relative promoter positioning and RNAP pausing (30,37,38). For any given TI scenario, the degree of relief of TI is also readily tunable by adjusting the concentration of the transcription factor and the affinity of its binding sites, since the main determinant of relief of TI is the simple magnitude of the regulatory effect exerted by the transcription factor on the interfering promoter (Figure 2).
However, for convergent promoters, where RNAPs from the target promoter pass over the transcription factor binding site at the interfering promoter, the kinetics of the transcription factor and its ability to resist dislodgement by these RNAPs become critical. As we have shown, repressors with fast binding kinetics, such as CI and Cro, are optimal for relief of TI, as they do not cause significant roadblocking of RNAPs from the target promoter and their repression of the target promoter is not sensitive to dislodgement by these RNAPs. When the repressors have slow binding kinetics, the degree of relief of TI depends on how easily they are dislodged by elongating RNAPs. An easily dislodged repressor, such as dCas9-bot, gives suboptimal relief of TI due to weakening of its repression of the interfering promoter, however, modelling indicates that this defect can be overcome at higher repressor concentrations. In contrast, a slow kinetics repressor that is a strong barrier, such as dCas9-top, can cause strong roadblocking and prevent relief of TI.
Our modelling indicated that the mechanism of TI had little effect on relief of TI except when there is strong reciprocal collisional TI. For promoters that fire every 10 s and are 3 kb apart, this effect can increase repression of the interfering promoter ∼5-fold (Figure 2E) and significantly enhance relief of TI at low repressor concentrations. This mechanism may be particularly important in relief of TI in eukaryotic systems, since convergent interfering promoters are often considerable distances apart (17,19).
Repressor kinetics, dislodgement sensitivity and barrier activity
Genomic DNA must be efficiently transcribed by RNAP but is also bound by many proteins with important functions. It can thus be expected that genome function requires that bound proteins do not often significantly impede RNAP, and also that the binding of these proteins is not often significantly diminished by the passage of RNAP. Our work shows that this combination of properties, a lack of roadblocking and a lack of dislodgement sensitivity, can be achieved by fast DNA-binding kinetics. This provides a simple mechanism to allow DNA-binding proteins and elongating RNAP to operate independently of each other.
Our assay of dislodgement sensitivity of repression provides a novel method to measure repressor binding kinetics in vivo. Examination of fluorescently-tagged DNA-binding proteins in living cells with techniques such as fluorescence recovery after photobleaching (FRAP) and single-molecule tracking reveal a broad range of binding kinetics for different proteins (52). Lac repressor binding and unbinding to a strong operator occurs on the scale of multiple minutes (53), while many eukaryotic transcription factors have residence times on the scale of tens of seconds (54). Our estimate of kU > 0.05 s–1 for CI unbinding from OR1.OR2 means that CI spontaneously unbinds from OR1.OR2 at least once every 20 s in vivo. We note that this value is primarily dependent on our estimate that fully activated PRE is producing elongating RNAPs at a rate of ∼0.1 s–1, a value obtained by calibration with well-characterized promoters (30). A kU = 0.05 s–1 for CI is substantially faster than unbinding rates obtained for CI at OR1 alone in vitro (45,55). However, fast CI kinetics in vivo are supported by rates of the order of seconds for transitions between different CI-bound states at OR123/OL123 derived from measurements of single-cell distributions of CI and PRM transcripts (56). Thus, CI kinetics may be faster in vivo than in vitro. λ CII activation of PRE was also previously found to be insensitive to dislodgement by RNAPs from PR, implying that CII also has fast binding kinetics (30). The dislodgement sensitivity of dCas9-bot repression indicates substantially slower kinetics than CI, while Cro's lack of sensitivity suggests CI-like fast kinetics. A more systematic analysis of dislodgement sensitivity of these proteins should allow quantitation of their kinetic properties. Eukaryotic transcription factors can also be sensitive to dislodgement by elongating RNAPs (15). The large, cooperative protein complexes typically involved in regulation of eukaryotic transcription seem likely to spontaneously unbind from DNA slowly, potentially making dislodgement sensitivity a more general feature in these systems.
In our study of LacI roadblocking (27), we found that higher affinity LacI binding sites gave stronger roadblocking in a manner that could not be explained by increased occupation alone. Instead, the results could be explained by invoking a direct relationship between the spontaneous unbinding rate of the repressor (kU) and its rates of dislodgement by RNAPs (kSD and kMD). That is, the more likely it is that the repressor spontaneously unbinds from the DNA, the easier it is for elongating RNAPs to actively dislodge it. While this seems plausible, our results with dCas9 roadblocking cannot be explained in this way. The two orientations of dCas9 repressed PR to a similar degree, implying similar kU values, yet their ability to be dislodged by RNAP was quite different. Our results and the consistent orientation effects seen by others (49,51), indicate that RNAPs approaching from the PAM-binding side dislodge dCas9 poorly compared to those approaching from the 5′ RNA side. This suggests that dislodgement can be a multi-step process, with the order with which DNA contacts are broken by RNAP being important. The dCas9–gRNA–DNA complex is an atypical repressive complex, being asymmetrical and with both DNA–protein and DNA–RNA interactions spread over a large contact region (∼20 bp). It is not known whether roadblocking asymmetry occurs for more simple repressors.
A potential role for relief of TI in the λ lysis-lysogeny decision
In the natural λ context, expression of CI from PRE means that relief of TI by CI should provide CI positive feedback: CI production from PRE gives increased CI repression of PR, relieving PR's TI on PRE and thus further increasing CI expression. Positive feedback provides ultrasensitivity, providing a more switch-like response (57,58) that might aid the λ gene circuitry to make a clear decision between the lytic and lysogenic pathways.
There are two previously established mechanisms of positive feedback by CI, direct stimulation of PRM by CI binding at OR2 (59,60)) and a double-negative loop in which CI repression of PR represses Cro, which in turn represses PRM (34,59,61). However, neither of these positive feedback loops seems likely to operate early after infection. λ phages with a mutant CI that is defective in PRM activation (and with compensating mutations that increase PRM activity) are not obviously defective in the lysis-lysogeny decision (62). Also, mutations of OR that reduce Cro repression of PRM have little effect on the establishment of lysogeny (34). This lack of impact of CI's direct and indirect regulation of PRM is explained by the observation that the vast majority of CI produced after infection comes from PRE rather than PRM (29). Thus, the ability of CI to stimulate PRE by relief of TI is likely to be the predominant form of CI positive feedback after infection and may provide a decisive commitment to lysogeny. Although the 4- to 5-fold regulatory effects of PR–PRE TI and its relief by CI are small compared to what can be achieved with direct promoter repression or activation, a 4- to 5-fold change in CI expression can be expected to have a large impact on the λ lysis-lysogeny decision (43).
Relief of TI in developmental switches
A number of examples of relief of TI, ranging from bacteriophages to fungi to mammals, involve developmental decisions (12,13,17–20). Our studies with the canonical λ system suggest clues as to why relief of TI may be particularly useful in cell fate choices.
First, developmental decisions often require the switching on of one set of genes while at the same time switching off another set of genes. Relief of TI provides a simple way for a single regulator to achieve this mutually exclusive expression, since it can allow the regulator to act directly as a repressor and indirectly as an activator. For example, the yeast a1/α2 repressor actively represses many haploid-specific genes in diploid cells and is also able to activate diploid-specific genes indirectly via relief of TI (17,18). The alternative one-regulator mechanism is to use a dual-function regulator that can directly repress or directly activate transcription, like λ CI. Remarkably, λ thus appears to employ both of these ‘minimalist’ strategies for switching between lytic and lysogenic transcription: CI is a dual function repressor-activator and uses relief of TI for indirect activation.
Second, in those cases where convergent promoters direct transcription of the alternative developmental programs, as for PR and PRE in λ, relief of TI may be a way of ‘hard-wiring’ mutual exclusivity, such that the rise of one developmental program is closely synchronized with the fall of the other.
Acknowledgments
We thank other Shearwin lab members and Kim Sneppen (University of Copenhagen) for discussions. Michael Crooks helped with reporter constructions.
FUNDING
Australian Research Council Discovery Early Career Researcher Award [DE150100091 to N.H.]; Australian Research Council Discovery Grants [DP110101420 to I.B.D., K.E.S., DP150103009 to K.E.S., I.B.D.]; NHMRC [APP1100653]; James S. McDonnell Foundation Postdoctoral Fellowship (to A.C.P.). Funding for open access charge: Australian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shearwin K.E., Callen B.P., Egan J.B. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer A.C., Egan J.B., Shearwin K.E. Transcriptional interference by RNA polymerase pausing and dislodgement of transcription factors. Transcription. 2011;2:9–14. doi: 10.4161/trns.2.1.13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazo A., Hodgson J.W., Petruk S., Sedkov Y., Brock H.W. Transcriptional interference: an unexpected layer of complexity in gene regulation. J. Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 4.Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 5.Bordoy A.E., Chatterjee A. Cis-antisense transcription gives rise to tunable genetic switch behavior: a mathematical modeling approach. PLoS One. 2015;10:e0133873. doi: 10.1371/journal.pone.0133873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sesto N., Wurtzel O., Archambaud C., Sorek R., Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat. Rev. Microbiol. 2013;11:75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 8.Wade J.T., Grainger D.C. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014;12:647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- 9.Xu Z., Wei W., Gagneur J., Clauder-Munster S., Smolik M., Huber W., Steinmetz L.M. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Liu X.S., Liu Q.R., Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha S., Haggard-Ljungquist E., Nordstrom K. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 1987;6:3191–3199. doi: 10.1002/j.1460-2075.1987.tb02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd I.B., Shearwin K.E., Sneppen K. Modelling transcriptional interference and DNA looping in gene regulation. J. Mol. Biol. 2007;369:1200–1213. doi: 10.1016/j.jmb.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 14.Lloubes R., Baty D., Lazdunski C. The promoters of the genes for colicin production, release and immunity in the ColA plasmid: effects of convergent transcription and Lex A protein. Nucleic Acids Res. 1986;14:2621–2636. doi: 10.1093/nar/14.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens J.A., Wu P.Y., Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbin V., Maniatis T. Role of transcriptional interference in the Drosophila melanogaster Adh promoter switch. Nature. 1989;337:279–282. doi: 10.1038/337279a0. [DOI] [PubMed] [Google Scholar]
- 17.Hongay C.F., Grisafi P.L., Galitski T., Fink G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 18.van Werven F.J., Neuert G., Hendrick N., Lardenois A., Buratowski S., van Oudenaarden A., Primig M., Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bumgarner S.L., Dowell R.D., Grisafi P., Gifford D.K., Fink G.R. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latos P.A., Pauler F.M., Koerner M.V., Senergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E., et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee A., Johnson C.M., Shu C.C., Kaznessis Y.N., Ramkrishna D., Dunny G.M., Hu W.S. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee A., Drews L., Mehra S., Takano E., Kaznessis Y.N., Hu W.S. Convergent transcription in the butyrolactone regulon in Streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS One. 2011;6:e21974. doi: 10.1371/journal.pone.0021974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann S.A., Kruse S.M., Arndt K.M. Long-range transcriptional interference in E. coli used to construct a dual positive selection system for genetic switches. Nucleic Acids Res. 2016;44:e9. doi: 10.1093/nar/gkw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brophy J.A., Voigt C.A. Antisense transcription as a tool to tune gene expression. Mol. Syst. Biol. 2016;12:854. doi: 10.15252/msb.20156540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakanishi H., Mitarai N., Sneppen K. Dynamical analysis on gene activity in the presence of repressors and an interfering promoter. Biophys. J. 2008;95:4228–4240. doi: 10.1529/biophysj.108.132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epshtein V., Toulme F., Rahmouni A.R., Borukhov S., Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao N., Krishna S., Ahlgren-Berg A., Cutts E.E., Shearwin K.E., Dodd I.B. Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res. 2014;42:8861–8872. doi: 10.1093/nar/gku627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ptashne M. A Genetic Switch: Phage λ revisited. 3rd edn. NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 29.Reichardt L., Kaiser A.D. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. U.S.A. 1971;68:2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer A.C., Ahlgren-Berg A., Egan J.B., Dodd I.B., Shearwin K.E. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons R.W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 32.Dodd I.B., Perkins A.J., Tsemitsidis D., Egan J.B. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haldimann A., Wanner B.L. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert R.A., Dodd I.B., Egan J.B., Shearwin K.E. Cro's role in the CI Cro bistable switch is critical for λ's transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–2472. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller J., Oehler S., Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 36.Lutz R., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sneppen K., Dodd I.B., Shearwin K.E., Palmer A.C., Schubert R.A., Callen B.P., Egan J.B. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- 38.Callen B.P., Shearwin K.E., Egan J.B. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Jain D., Kim Y., Maxwell K.L., Beasley S., Zhang R., Gussin G.N., Edwards A.M., Darst S.A. Crystal structure of bacteriophage lambda cII and its DNA complex. Mol. Cell. 2005;19:259–269. doi: 10.1016/j.molcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Schmeissner U., Court D., Shimatake H., Rosenberg M. Promoter for the establishment of repressor synthesis in bacteriophage lambda. Proc. Natl. Acad. Sci. U.S.A. 1980;77:3191–3195. doi: 10.1073/pnas.77.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer K., Jensen K.F., Poulsen P., Oppenheim A.B., Gottesman M. Isolation of Escherichia coli rpoB mutants resistant to killing by lambda cII protein and altered in pyrE gene attenuation. J. Bacteriol. 1987;169:5289–5297. doi: 10.1128/jb.169.11.5289-5297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarai A., Takeda Y. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodd I.B., Shearwin K.E., Perkins A.J., Burr T., Hochschild A., Egan J.B. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui L., Murchland I., Shearwin K.E., Dodd I.B. Enhancer-like long-range transcriptional activation by lambda CI-mediated DNA looping. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2922–2927. doi: 10.1073/pnas.1221322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson H.C., Sauer R.T. Lambda repressor mutations that increase the affinity and specificity of operator binding. Cell. 1985;42:549–558. doi: 10.1016/0092-8674(85)90112-6. [DOI] [PubMed] [Google Scholar]
- 46.Shih M.C., Gussin G.N. Role of cII protein in stimulating transcription initiation at the lambda PRE promoter. Enhanced formation and stabilization of open complexes. J. Mol. Biol. 1984;172:489–506. doi: 10.1016/s0022-2836(84)80019-4. [DOI] [PubMed] [Google Scholar]
- 47.Roberts E.A., Eisenbraun T.L., Andrews C.L., Bear D.G. 3′-end formation at the phage lambda tR1 rho-dependent transcription termination site. Biochemistry. 1991;30:5429–5437. doi: 10.1021/bi00236a015. [DOI] [PubMed] [Google Scholar]
- 48.Svenningsen S.L., Costantino N., Court D.L., Adhya S. On the role of Cro in lambda prophage induction. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4465–4469. doi: 10.1073/pnas.0409839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 51.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller F., Stasevich T.J., Mazza D., McNally J.G. Quantifying transcription factor kinetics: at work or at play. Crit. Rev. Biochem. Mol. Biol. 2013;48:492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
- 53.Elf J., Li G.W., Xie X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phair R.D., Scaffidi P., Elbi C., Vecerova J., Dey A., Ozato K., Brown D.T., Hager G., Bustin M., Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Guo L., Golding I., Cox E.C., Ong N.P. Quantitative transcription factor binding kinetics at the single-molecule level. Biophys. J. 2009;96:609–620. doi: 10.1016/j.bpj.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sepulveda L.A., Xu H., Zhang J., Wang M., Golding I. Measurement of gene regulation in individual cells reveals rapid switching between promoter states. Science. 2016;351:1218–1222. doi: 10.1126/science.aad0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becskei A., Seraphin B., Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrell J.E., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 59.Meyer B.J., Maurer R., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J. Mol. Biol. 1980;139:163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- 60.Jain D., Nickels B.E., Sun L., Hochschild A., Darst S.A. Structure of a ternary transcription activation complex. Mol. Cell. 2004;13:45–53. doi: 10.1016/s1097-2765(03)00483-0. [DOI] [PubMed] [Google Scholar]
- 61.Johnson A.D., Meyer B.J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michalowski C.B., Little J.W. Positive autoregulation of cI is a dispensable feature of the phage lambda gene regulatory circuitry. J. Bacteriol. 2005;187:6430–6442. doi: 10.1128/JB.187.18.6430-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]