Abstract
Nonsense mutations introduce premature termination codons and underlie 11% of genetic disease cases. High concentrations of aminoglycosides can restore gene function by eliciting premature termination codon readthrough but with low efficiency. Using a high-throughput screen, we identified compounds that potentiate readthrough by aminoglycosides at multiple nonsense alleles in yeast. Chemical optimization generated phthalimide derivative CDX5-1 with activity in human cells. Alone, CDX5-1 did not induce readthrough or increase TP53 mRNA levels in HDQ-P1 cancer cells with a homozygous TP53 nonsense mutation. However, in combination with aminoglycoside G418, it enhanced readthrough up to 180-fold over G418 alone. The combination also increased readthrough at all three nonsense codons in cancer cells with other TP53 nonsense mutations, as well as in cells from rare genetic disease patients with nonsense mutations in the CLN2, SMARCAL1 and DMD genes. These findings open up the possibility of treating patients across a spectrum of genetic diseases caused by nonsense mutations.
INTRODUCTION
Nonsense mutations change an amino acid codon to a premature termination codon (PTC). mRNAs bearing PTCs typically show decreased stability or form defective truncated proteins, resulting almost invariably in profound loss of function and severe forms of disease (1). Nonsense mutations have been identified in most of the >5000 human genetic diseases identified to date and account overall for about 11% of the causative mutations (1). In sporadic cancer, nonsense mutations similarly account for ∼11% of mutations in tumor suppressor genes such as TP53.
It has been known for decades that genetic and epigenetic alterations can partially restore the function of genes carrying nonsense mutations, a phenomenon termed nonsense suppression (2–4). Nonsense suppression or PTC readthrough can also be produced by exogenous chemicals. High concentrations of certain aminoglycoside antibiotics were found in 1979 to suppress several nonsense alleles in yeast (5,6) and subsequently in a reporter gene introduced into mammalian cells (7).
Aminoglycosides exert their PTC readthrough activity by binding at the decoding center of the eukaryotic ribosome. Binding alters the ability of translation termination factors to accurately recognize a PTC. Consequently, aminoglycosides increase the frequency of pairing of near-cognate aminoacyl-tRNAs to the PTC and enables formation of full-length protein (8,9). In 1997, the aminoglycoside gentamicin was found to induce CFTR protein expression from the CFTR gene harboring nonsense mutations in bronchial epithelial cells from patients with cystic fibrosis (10). This raised hopes of using PTC readthrough drugs to treat diseases caused by nonsense mutations.
The therapeutic potential of gentamicin has been investigated in preclinical models of genetic disease and in patients. Studies in mice bearing the human CFTR G542X transgene demonstrated increased CFTR function upon gentamicin administration (11). Gentamicin treatment also elevated CFTR chloride conductance in patients with CFTR nonsense mutations (12,13). However, improvements were small and patient response was variable (14). PTC readthrough by gentamicin was also demonstrated in mdx mice (15) harboring nonsense mutations in their Dmd gene to model human Duchenne muscular dystrophy (DMD). The first small trial in DMD patients showed no effect and two others showed dystrophin expression in some patients (16) but the level of expression was insufficient for patient improvement. The lack of potency of gentamicin and its recognized nephrotoxicity and ototoxicity at high dose discouraged its further development.
Major efforts have been put into developing aminoglycoside derivatives with reduced toxicity (17,18) and discovering non-aminoglycoside readthrough compounds such as negamycin, tylosin, RTC13, RTC14, GJ71, GJ72 and ataluren (19–23). These compounds increased protein production in several cell culture and animal disease models, but often at the detection limit of western blotting for endogenous protein expression and with variable responses between genes, cell lines, and PTC mutations.
Ataluren is the sole new compound to have advanced through clinical trials. It has an excellent safety profile compared with gentamicin. Ataluren's PTC readthrough activity has been challenged based on artifactual activity in luciferase reporter assays of the type used for its discovery, and lack of demonstrable readthrough activity in many assays (24–26). Nevertheless, it has shown activity in several animal models, including increased dystrophin expression and muscle function in the mdx mouse (27) and CFTR protein expression and improved chloride conductance in the intestine of the G542X-hCFTR mouse (20). Ataluren has undergone Phase 3 clinical trials (28,29). It has been given conditional approval for DMD treatment by the European Medicines Agency (30) but received a Refuse to File Letter from the US Food and Drug Administration.
Overall, currently available PTC readthrough compounds elicit readthrough in only a subset of genetic disease systems tested and the levels of readthrough are low, typically achieving no more than 5% of wild-type (WT) protein levels. This study was undertaken to identify new compounds capable of broader and more efficacious PTC readthrough activity. We report the identification and characterization of compounds that do not show readthrough activity in human cells when used as single agents but that strongly potentiate the readthrough activity of aminoglycoside antibiotics.
MATERIALS AND METHODS
Yeast cells
Saccharomyces cerevisiae B0133-3B ([PSI]+ MATa can1-100 cyc1-72 his5-2 leu2-1 lys1-1 met8-1 trp5-48 ura4-1) and IS110-18A ([PSI] − MATa aro7-1 leu2-2 ilv1-2 his4-166 lys2-101 met8-1 trp5-48 ura4-1) were purchased from ATCC. To confirm the genotype of these strains we performed next generation sequencing. In brief, genomic DNA from these strains was extracted and libraries were generated using the Nextera XT Library Preparation Kit according to manufacturer's instructions (Illumina). Libraries were pooled and sequenced on the Illumina MiSeq platform, generating 150 bp paired-end reads. Sequence reads were mapped to the yeast reference genome S288C version R64 (Saccharomyces Genome Database, SGD, http://www.yeastgenome.org) using the short-read aligner BWA (31). Single-nucleotide variants (SNVs) were identified and filtered with the help of the SAMtools toolbox (32). Each SNV was annotated with a custom-made Perl script and gene information downloaded from SGD on 21 January 2014. The read alignments in the regions of interesting candidate SNVs were visually inspected with the IGV viewer (33,34). Using standard homologous recombination technique we deleted two major efflux pumps PDR1 and PDR3 from both yeast strains and generated B0133-3B-AB13 ([PSI]+ MATa can1-100 cyc1-72 his5-2 leu2-1 lys1-1 met8-1 trp5-48 ura4-1 PDR1Δ::URA4 PDR3Δ::HIS5) and IS110-18A-AB13 ([PSI] − MATa aro7-1 leu2-2 ilv1-2 his4-166 lys2-101 met8-1 trp5-48 ura4-1 PDR1Δ::URA4 PDR3Δ::HIS4) strains. Primers used to amplify PCR products for homologous recombination and primers to confirm deletion of PDR1 and PDR3 are listed in Supplementary Table S3.
Yeast PTC readthrough screening assay
Exponentially growing B0133-3B-AB13 cells were seeded in 384-well plates at A600 of 0.05 in Synthetic Complete medium lacking methionine and supplemented with 2.5 μM paromomycin. 157,000 chemicals from Prestwick, Sigma LOPAC, Microsource Spectrum, Biomol, Chembridge DiverSet and Lankenau Chemical Genomics Center were added to the wells using a Platemate Plus automated liquid handler (Matrix Technologies) equipped with a FP3 384 pin tool (0.7 mm diameter), at a final concentration of ∼7 μM. Plates were incubated at 30°C for 48 h and yeast growth was determined by A600 measurement using a PowerWave HT Microplate Spectrophotometer (BioTek Instruments). Compounds that induced >2.5-fold increase in cell growth compared with 2.5 μM paromomycin alone were retested at various concentrations in the presence or absence of paromomycin. Fresh samples of the active compounds were acquired from the respective suppliers and tested against various nonsense mutations including met8-1, trp5-48 and lys2-101 in B0133-3B-AB13 and IS110-18A-AB13 strains.
Human cells
GM16485 primary fibroblasts were purchased from the Coriell Biorepository. hTERT immortalized SD123 fibroblasts were provided by Dr Cornelius Boerkoel (University of British Columbia). C25CI48 and HSK001 immortalized myoblasts from an unaffected individual and a DMD patient, were generated as previously described (35). HDQ-P1 and ESS-1 cell lines were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). EKVX cells were from the NCI Developmental Therapeutics Program (DTP). MDA-MB-361, Caov-3, SW900, NCI-H1688, Calu-6, SK-MES-1, HCC1937, UACC-893, NCI-H1299 and HCT116 cells were purchased from ATCC. GM16485, SD123, HDQ-P1, Caov-3, SK-MES-1 and HCT116 cells were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM, Sigma-Aldrich) supplemented with 10% or 15% fetal bovine serum (FBS, Sigma-Aldrich) and 1× antibiotic-antimycotic (Gibco/Thermo Fisher Scientific) at 37°C and 5% CO2. Human myoblasts were cultured in Skeletal Muscle Cell Growth Medium (PromoCell) supplemented with 20% FBS and 1× antibiotic-antimycotic at 37°C and 5% CO2. Myoblasts were differentiated into myotubes in differentiation medium consisting of DMEM (Gibco/Thermo Fisher Scientific) supplemented with 10 μg/ml insulin (Sigma) and 1× antibiotic-antimycotic. All other cell lines (ESS-1, MDA-MB-361 and UACC-893) were cultured in RPMI-1640 medium supplemented with 10% or 20% FBS and 1× antibiotic–antimycotic at 37°C and 5% CO2. GM16485 fibroblasts are derived from a patient with a late infantile neuronal ceroid lipofuscinosis harboring compound heterozygous nonsense mutations in the TPP1 gene (NM_000391.3:c.379C>T/c.622C>T; NP_000382.3:p.127X/p.R208X). SD123 fibroblasts are derived from an SIOD patient harboring homozygous nonsense mutations in the SMARCAL1 gene (NM_014140.3:c.49C>T; NP_054859.2:p.R17X). HSK001 myoblasts are derived from a patient with DMD harboring nonsense mutation in the DMD gene (NM_004006.2:c.6103G>T;NP_003997.1:p.E2035X). The HCT116 cell line has wild-type TP53 while other cancer cell lines harbor the following homozygous mutations in their TP53 gene: MDA-MB-361 (NM_000546.5:c.166G>T; NP_000537.3:p.E56X), Caov-3 (NM_000546.5:c.406C>T; NP_000537.3:p.Q136X), SW900 (NM_000546.5:c.499C>T; NP_000537.3:p.Q167X), NCI-H1688 (NM_000546.5:c.574C>T; NP_000537.3:p.Q192X), Calu-6 (NM_000546.5:c.586C>T; NP_000537.3:p.R196X), EKVX (NM_000546.5:c.610G>T; NP_000537.3:p.E204X), HDQ-P1 (NM_000546.5:c.637C>T; NP_000537.3:p.R213X), ESS-1 (NM_000546.5:c.637C>T; NP_000537.3:p.R213X), SK-MES-1 (NM_000546.5:c. 892G>T; NP_000537.3:p.E298X), HCC1937 (NM_000546.5:c.916C>T; NP_000537.3:p.R306X), UACC-893 (NM_000546.5:c.1024C>T; NP_000537.3:p.E342X), NCI-H1299 (NM_000546.5:c.1_954>AAG; NP_000537.3:p.?) (COSMIC).
Automated p53 immunofluorescence 96-well plate assay
HDQ-P1 cells cultured in DMEM containing 10% FBS and 1× antibiotic-antimycotic were seeded at 4000 cells per well in PerkinElmer View 96-well plates. The next day, the medium was replaced with fresh culture medium containing the compounds to be tested. After 72 h, the culture medium was removed by aspiration, the cells were fixed with 3% paraformaldehyde, 0.3% Triton X-100 and 1.5 μg/ml Hoechst 33323 in phosphate-buffered saline (PBS) pH 7.2 for 20 min. The cells were rinsed once with PBS and incubated for 2 h with a blocking solution of 3% bovine serum albumin (BSA) in PBS. The blocking solution was removed by aspiration and cells were incubated with 0.1 μg/ml DO-1 p53 mouse monoclonal antibody (Santa Cruz) in blocking solution for 90 min. The wells were washed once with PBS for 5 min and then incubated with Alexa 488-conjugated goat anti-mouse antibody (Thermo Fisher Scientific) in blocking buffer for 90 min. The wells were washed once with PBS for 5 min, 75 μl PBS was added, the plates were covered with a black adherent membrane and stored at 4°C overnight. Nuclear p53 immunofluorescence intensity was measured using a Cellomics ArrayScan VTI automated fluorescence imager. Briefly, images were acquired with a 20× objective in the Hoechst and GFP (XF53) channels. Images of 15 fields were acquired for each well, corresponding to ∼2000 cells. The Compartment Analysis bioapplication was used to identify the nuclei and define their border. The nuclear Alexa 488 fluorescence intensity was then measured and expressed as % positive nuclei, using as a threshold the fluorescence intensity of nuclei from untreated cells, 50–75, depending on experiment.
Automated electrophoresis western analysis assay
Cells were seeded at 1–2 × 105 cells per well in tissue culture-treated six-well plates. The next day, the medium was replaced with fresh medium containing compounds to be tested and cells were incubated for 48–96 h. The wells were replenished with fresh medium and compounds every 48 h. The medium was removed by aspiration, and cell monolayers were rinsed with 1 ml ice-cold PBS. Cells were lysed in 80 μl lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate) supplemented with fresh 1 mM Na3VO4, 1 mM dithiothreitol and 1× complete protease inhibitor cocktail (Roche Molecular Biochemicals). Lysates were pre-cleared by centrifugation at 18 000 g for 15 min at 4°C. Supernatants were collected, protein was quantitated using the Bradford assay and lysates were adjusted to 0.5–1 mg/ml protein. Capillary electrophoresis western analysis was carried out with manufacturer's reagents according to the user manual (ProteinSimple WES). Briefly, 5.6 μl cell lysate was mixed with 1.4 μl fluorescent master mix and heated at 95°C for 5 min. The samples, blocking reagent, wash buffer, DO-1 p53 antibody (0.5 μg/ml), mouse α-vinculin antibody (1:800 or 1:2000, clone 728526, R&D Systems), secondary antibody and chemiluminescent substrate were dispensed into the microplate provided by the manufacturer. The electrophoretic separation and immunodetection was performed automatically using default settings. The data was analyzed with inbuilt Compass software (ProteinSimple). The truncated and full-length p53 peak intensities (area under the curve) were normalized to that of the vinculin peak, used as a loading control. In most figures, electropherograms are represented as pseudo-blots, generated using Compass software. TPP1 western analysis was carried out similarly, using the cell lysates prepared for TPP1 activity measurements and Abcam ab54685 α-TPP1 antibody (0.8 μg/ml). Dystrophin western analysis was carried out similarly using cell lysates prepared from myotubes and Abcam ab15277 α-dystrophin antibody (1:200).
SDS-PAGE and immunoblotting
SIOD fibroblasts were seeded, treated and lysed as above, protein was quantified, and western blotting was assayed as previously described (36). In brief, 20 μg protein from each lysate was separated on an 8% polyacrylamide gel, electroblotted onto a nitrocellulose membrane and blocked overnight in 5% (w/v) nonfat milk supplemented with 2% (v/v) goat serum. Membranes were incubated with primary antibodies, rabbit α-SMARCAL1 (1:2000, gift from Dr Cornelius Boerkoel) and rabbit α-β-actin (1:20 000, Novus Biologicals), washed three times with PBS + 0.1% (v/v) Tween-20 (PBS-T), and incubated with HRP-conjugated goat α-rabbit IgG secondary antibody, washed again with PBS-T and developed using enhanced chemiluminescence substrate (Millipore).
Aminoglycosides
G418 disulfate was purchased from Toku-E, gentamicin sulfate and hygromycin B from MicroCombiChem, netilmicin sulfate from AK Scientific and all other aminoglycosides from Sigma.
Synthesis of CDX5 compounds
CDX5-1: A mixture of 4-aminophthalimide (500 mg, 3.1 mmol; TCI) and 2,6-dichlorobenzyl bromide (740 mg, 3.1 mmol; Aldrich) in N,N-dimethylformamide (0.5 M) was stirred at 60°C for 6 h then at room temperature overnight. The reaction mixture was quenched with saturated NaHCO3 and extracted with ethyl acetate. The combined organic layers were dried over Na2SO4 and concentrated under vacuum. The crude material was purified either by automated flash chromatography (10–50% ethyl acetate in hexanes) or by preparative HPLC to give the desired compound in 30% yield.
CDX5-35 and CDX5-52: A mixture of appropriate benzyl halide (1 mmol) and potassium phthalimide (1 mmol) in N,N-dimethylformamide (1.5 ml) was heated at 120°C under nitrogen for 2–3 h. The product was then precipitated out by addition of water (5 ml) to the mixture. The solids were filtered and washed with water and air dried to give the desired product in 30–80% yield.
CDX5-4, CDX5-53, CDX5-109 and CDX5-186: To a solution of the appropriate phthalic anhydride (1 mmol) in acetic acid (2 ml) was added the appropriate benzyl amine (1 mmol). The mixture was heated at 100°C under nitrogen for 3–4 h then cooled to room temperature. The resultant crystals were collected by filtration and washed with ethanol to give the desired product in 20–80% yield.
CDX5-13 was obtained from the Lankenau Chemical Genomics Center.
Characterization of CDX5 compounds
CDX5-1: 2-(2,6-dichlorobenzyl)isoindoline-1,3-dione. 1H NMR (400 MHz, DMSO-d6) δ 10.80 (s, 1H), 7.55 (dd, J = 13.2, 8.2 Hz, 3H), 7.48 – 7.40 (m, 1H), 7.15 (t, J = 4.7 Hz, 1H), 7.03 (d, J = 2.1 Hz, 1H), 6.97 (dd, J = 8.4, 2.1 Hz, 1H), 4.53 (d, J = 4.6 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 170.07, 169.81, 154.14, 136.18, 135.86, 133.44, 131.11, 129.23, 125.00, 118.91, 115.96, 105.49, 43.32.
CDX5-4: N-(2-(2,5-Dichlorobenzyl)-1,3-dioxoisoindolin-4-yl)acetamide. 1H NMR (400 MHz, Chloroform-d) δ 9.50 (s, 1H), 8.85 (d, J = 8.5 Hz, 1H), 7.74 (dd, J = 8.6, 7.3 Hz, 1H), 7.64–7.55 (m, 1H), 7.36 (d, J = 8.3 Hz, 1H), 7.27–7.18 (m, 2H), 4.95 (s, 2H), 2.30 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 169.61, 169.28, 167.20, 137.69, 136.27, 134.90, 133.03, 131.35, 131.29, 130.88, 129.21, 128.73, 125.09, 118.38, 115.38, 38.94, 25.00.
CDX5-35: 2-(Naphthalen-1-ylmethyl)isoindoline-1,3-dione. 1H NMR (400 MHz, CDCl3) δ 8.37 (d, J = 8.4 Hz, 1H), 7.88–7.82 (m, 3H), 7.80 (d, J = 8.2 Hz, 1H), 7.74–7.69 (m, 2H), 7.64–7.56 (m, 2H), 7.54–7.47 (m, 1H), 7.43 (dd, J = 8.0, 7.3 Hz, 1H), 5.34 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 168.40 (s), 134.19 (s), 133.90 (s), 132.20 (s), 131.48 (s), 131.31 (s), 128.87 (s), 128.81 (s), 127.44 (s), 126.67 (s), 125.96 (s), 125.42 (s), 123.61 (s), 123.53 (s), 39.67 (s).
CDX5-52: 2-(3,4-dichlorobenzyl)isoindoline-1,3-dione. 1H NMR (400 MHz, CDCl3) δ 7.86 (dd, J = 5.4, 3.1 Hz, 2H), 7.73 (dd, J = 5.5, 3.1 Hz, 2H), 7.52 (d, J = 2.0 Hz, 1H), 7.39 (d, J = 8.2 Hz, 1H), 7.28 (dd, J = 8.5, 2.2 Hz, 1H), 4.79 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 167.95 (s), 136.52 (s), 134.37 (s), 132.08 (s), 130.81 (s), 130.76 (s), 128.21 (s), 123.68 (s), 40.65 (s).
CDX5-53: 2-(2,6-Dichlorobenzyl)-5-fluoroisoindoline-1,3-dione. 1H NMR (400 MHz, chloroform-d) δ 7.84 (dd, J = 8.2, 4.5 Hz, 1H), 7.50 (dd, J = 7.0, 2.3 Hz, 1H), 7.44 – 7.33 (m, 3H), 7.22 (dd, J = 8.6, 7.4 Hz, 1H), 5.16 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 167.69, 166.45, 165.14, 136.49, 134.69, 134.59, 130.48, 129.71, 128.50, 127.63, 127.60, 125.75, 125.66, 121.17, 120.93, 111.30, 111.06, 38.47.
CDX5-109: 2-(2-Chloro-5-(trifluoromethyl)benzyl)isoindoline-1,3-dione. 1H NMR (400 MHz, CDCl3) δ 7.91 (dd, J = 5.3, 3.2 Hz, 2H), 7.78 (dd, J = 4.1, 3.0 Hz, 2H), 7.51 (t, J = 9.4 Hz, 3H), 5.02 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 167.90 (s), 137.10 (s), 134.70 (s), 134.50 (s), 131.97 (s), 130.41 (s), 129.61 (q, J = 33.0 Hz), 125.97 (p, J = 3.8 Hz), 123.82 (s), 39.34 (s).
CDX5-186: 2-(2,6-Dichlorobenzyl)-4,7-difluoroisoindoline-1,3-dione. 1H NMR (400 MHz, Chloroform-d) δ 7.48–7.31 (m, 4H), 7.22 (dd, J = 8.7, 7.4 Hz, 1H), 5.15 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 163.07, 155.00, 154.96, 152.37, 152.33, 136.52, 130.01, 129.85, 128.52, 125.06, 124.93, 124.88, 124.75, 38.50.
RNA extraction and RT-PCR
RNA was extracted from HDQ-P1 cells using the RNeasy Plus Mini Kit (Qiagen). cDNA synthesis was performed using High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). Quantitative real-time RT-PCR for detection of TP53 mRNA was performed using the ABI StepOnePlus Real-Time PCR system, using the primers listed in Supplementary Table S3.
Site directed mutagenesis
The three TP53 mutations c.637C>T, c.637-639delCGAinsTAG, and c.637-638delCGinsTA resulting in p53 R213X with all three possible stop codons (TGA-C, TAG-C, TAA-C, respectively) were engineered into pDONR223-p53 (hORFeome V8.1) using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and primers listed in Supplementary Table S3. The mutant strand synthesis reaction was performed by PCR using 10x kit reaction buffer, pDONR223-p53 (60ng), forward and reverse primers (125 ng), dNTP mix, Quick Solution reagent and lightning enzyme. Eighteen PCR reaction cycles were performed with DNA denaturing step at 95°C for 20 s, primer annealing at 60°C for 10 s, and polymerase extension at 68°C for 3.5 min. Following digestion of the parental DNA with Dpn I restriction enzyme, DNA for each sample was chemically transformed into XL10-Gold Ultracompetent cells and spread on spectinomycin-containing (100 μg/ml) LB agar plates. DNA was isolated from spectinomycin resistant clones and the TP53 cDNA was Sanger-sequenced in its entirety to confirm the introduction of the desired nonsense mutations and to ensure that mutations were not introduced elsewhere in the cDNA. Finally, to generate expression clones we used the LR recombination reaction to recombine the mutated cDNA samples from the Entry vectors into the pcDNA-6.2/V5-DEST vector (Thermo Fisher Scientific). Samples were chemically transformed into Top10 competent cells and spread on ampicillin-containing (100 μg/ml) LB agar plates. DNA from resistant clones was isolated and the p53 R213X was confirmed by Sanger sequencing.
Transient transfection
H1299 cells were transiently transfected with pcDNA-6.2/V5-DEST vector expressing either WT p53 or one of the three p53 R213X mutants with different stop codons using Lipofectamine 2000 (Thermo Fisher Scientific). Immunofluorescence microscopy using anti-V5 antibody revealed more than 80% transfection efficiency 48 h after transfection. Twenty four hours after transfection, each sample was split into four wells of a six-well plate (105 cells/well) and 24 h later either left untreated or treated with 20 μM G418, 10 μM CDX5-1, or 20 μM G418 and 10 μM CDX5-1. After 48 h, the cells were lysed and subjected to automated capillary electrophoresis western analysis (ProteinSimple WES).
In vitro transcription and translation assay
pcDNA-6.2/V5-DEST vectors expressing either WT p53 or one of the three p53 R213X mutants were linearized with FastDigest MssI restriction enzyme (Thermo Fisher Scientific). Linear DNA fragments were subjected to RNA synthesis using mMESSAGE mMACHINE T7 ULTRA Transcription Kit (Thermo Fisher Scientific) to generate 5′ capped and poly(A) tailed RNA. The in vitro translation assay was carried out using the 1-Step Human Coupled IVT Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, 500 ng RNA was added to the Hela cell lysate-based protein expression system and incubated at 30°C for 90 min without or with 0.5 μM G418, 1 μM CDX5-1, or a combination of 0.5 μM G418 and 1 μM CDX5-1. Samples were diluted in water and subjected to automated electrophoresis western analysis for p53 detection.
TPP1 activity assay
GM16485 primary fibroblasts seeded into six-well plates were exposed to compounds for up to 9 days. The wells were replenished with fresh medium and compounds every 3 days. At the end of the experiment, the cell monolayers were washed with ice-cold PBS and scraped using a rubber policeman into 0.1 ml lysis buffer without Na3VO4. The lysates were clarified by centrifugation at 15 000 × g for 10 min at 4°C. TPP1 enzyme activity was determined as in (37) with modifications. Lysates were diluted 1:5 in 50 mM sodium acetate pH 4.0 and pre-incubated at 37°C for 1 h. After pre-incubation, 20 μg of total protein from GM16485 lysates or 5 μg of total protein from lysates of fibroblasts from unaffected individuals was incubated in 150 μl of 50 mM sodium acetate pH 4.0 containing a final concentration of 62.5 μM Ala-Ala-Phe-7-amido-4-methylcoumarin for 2 h at 37°C. Fluorescence was measured using a TECAN Infinite M200 spectrophotometer with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Assays were carried out under conditions where product formation was linear with respect to protein concentration and time.
Statistical analysis
Data are presented as mean ± SD unless otherwise stated. Comparisons were made using the two-tailed Student's t-test and differences were considered significant at a P-value of <0.05. Combination index (CI) was calculated using the CompuSyn software (38), with very strong synergistic activity defined as CI < 0.1.
RESULTS
Development of a yeast PTC readthrough screening assay
We wished to design a stringent high-throughput screening assay that reports readthrough of endogenous genes rather than artificial constructs. We developed a yeast cell-based assay that links readthrough of endogenous PTC-containing genes to increased cell growth. Saccharomyces cerevisiae cells carrying a PTC allele of a biosynthetic gene (e.g. met8-1) do not grow in the absence of the relevant product metabolite (e.g. methionine). Exposure to a compound that stimulates PTC readthrough allows cells to grow in the absence of the metabolite, i.e. acts as a chemical suppressor of the nonsense mutation. We selected strains B0133-3B and IS110-18A, reported to harbor PTCs in several biosynthetic genes (39,40). The strains were subjected to whole genome sequencing to verify the PTC sequences and identify their location within each gene (Supplementary Table S1). To maximize the likelihood of identifying active chemicals, the pleiotropic drug resistance genes PDR1 and PDR3 were deleted in both strains, redesignated B0133-3B-AB13 and IS110-18A-AB13. B0133-3B-AB13 was selected for the high-throughput screening assay because it carries the [PSI+] prion form of translation termination factor erf3p which weakly suppresses nonsense mutations by ∼1% (41). The [PSI−] strain IS110-18A-AB13 was used for further testing of active compounds.
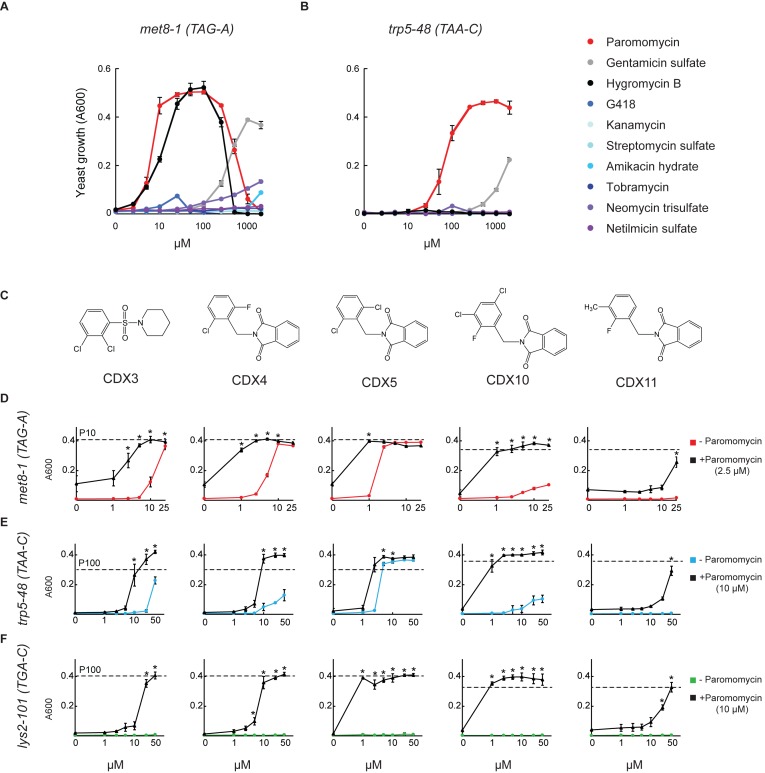
A number of aminoglycoside antibiotics have been shown to suppress certain nonsense alleles in yeast (5,6). To validate the screening assay we tested 10 different aminoglycosides for suppression of met8-1 (TAG-A) and trp5-48 (TAA-C) in the [PSI+] strain. Paromomycin was the most potent suppressor, enabling growth in the absence of methionine or tryptophan; hygromycin B suppressed met8-1 only; and gentamicin suppressed both, but only at very high concentrations (Figure 1A and B) consistent with previous reports (5,6). The seven other aminoglycosides showed negligible activity (Figure 1A and B).
Figure 1.
Chemical suppression of yeast nonsense alleles. (A, B) Suppression of met8-1 and trp5-48 nonsense alleles by aminoglycosides. B0133-3B-AB13 yeast cells were grown in 96-well plates without methionine (A) or without tryptophan (B) and with the indicated concentrations of aminoglycosides. Yeast growth (A600) was measured at 40 h. Shown are mean ± S.D. (n = 3). (C–F) Synergistic suppression of met8-1, trp5-48 and lys2-101 nonsense alleles by CDX compounds and paromomycin. (C) Structures of the CDX compounds identified in the high throughput screen. B0133-3B-AB13 (D and E) or IS110-18A-AB13 (F) yeast cells were grown in 96-well plates without methionine (D) tryptophan (E) or lysine (F) and with different concentrations of the CDX compounds shown above each column, without (colored curves) or with paromomycin (black curves). The selected paromomycin concentration of 2.5 μM (D) or 10 μM (E and F) was subactive for these alleles. The dashed lines labelled P10 or P100 indicate growth with paromomycin alone at 10 or 100 μM, respectively. The +4 nucleotide is indicated for each codon. The asterisks indicate synergistic combinations (CI <0.1). Shown are mean ± S.D. (n = 3).
In the [PSI−] strain, which also carries a TGA-C PTC (lys2-101), paromomycin suppressed lys2-101, as well as met8-1 and trp5-48 (Supplementary Figure S1). Therefore, the assay provides a sensitive and quantitative high-throughput means of examining PTC readthrough at any of the three termination codons (TGA, TAG and TAA) in different biosynthetic genes and genetic backgrounds. The data also illustrate the earlier observations that aminoglycosides do not uniformly suppress all nonsense mutations (5,6).
Identification of small molecules that potentiate the activity of paromomycin
A first screen of 30,000 compounds tested for suppression of met8-1 in the [PSI+] strain yielded no active compound. This disappointing result led us to consider the hypothesis that efficient PTC readthrough might require the concerted action of two compounds. We therefore screened for small molecules that potentiate the PTC readthrough activity of paromomycin by testing 157,000 compounds for suppression of met8-1 in the presence of 2.5 μM paromomycin, a sub-active concentration in this assay (Figure 1A). This screen yielded ten compounds capable of suppressing met8-1 (TAG). Five of these compounds also suppressed trp5-48 (TAA) in the same [PSI+] strain in the presence of a low paromomycin concentration (Figure 1C–E).
The five compounds which demonstrated PTC readthrough activity against both met8-1 and trp5-48 were further evaluated at different concentrations, with and without various concentrations of paromomycin for suppression of the three genes with PTC in the [PSI+] and the [PSI−] strains. Four showed readthrough activity as single agents on at least one of the three genes (Figure 1D–F). Interestingly, all five compounds showed increased activity in the presence of a sub-active concentration of paromomycin and all combinations suppressed all three genes (Figure 1D–F and Supplementary Figure S1 for complete data). Remarkably, several combinations demonstrated very strong synergistic activity, with combination indices (CI) below 0.1 (42). As a striking example, compound CDX5 showed no activity as a single agent on lys2-101 even at a high concentration of 50 μM and paromomycin showed no activity at 10 μM, but the combination of 1 μM CDX5 and 10 μM paromomycin showed optimal activity, comparable to that of paromomycin alone at 100 μM (Figure 1F). Four of the five active compounds (CDX4, 5, 10 and 11) are closely related members of the same N-substituted phthalimide class while CDX3 is structurally distinct. These findings show that combining two compounds can efficiently suppress different nonsense alleles with all three possible termination codons in yeast.
PTC readthrough in a human cell line
We next examined whether the five active compounds identified in the yeast screen could also induce PTC readthrough in human cells. Mammary carcinoma HDQ-P1 cells homozygous for R213X (TGA-C, where C is the 3′-flanking nucleotide) in exon 6 of the TP53 gene (43) were selected for testing based on convincing evidence of readthrough by the aminoglycoside G418 (44), confirmed in this study (Supplementary Figure S2A).
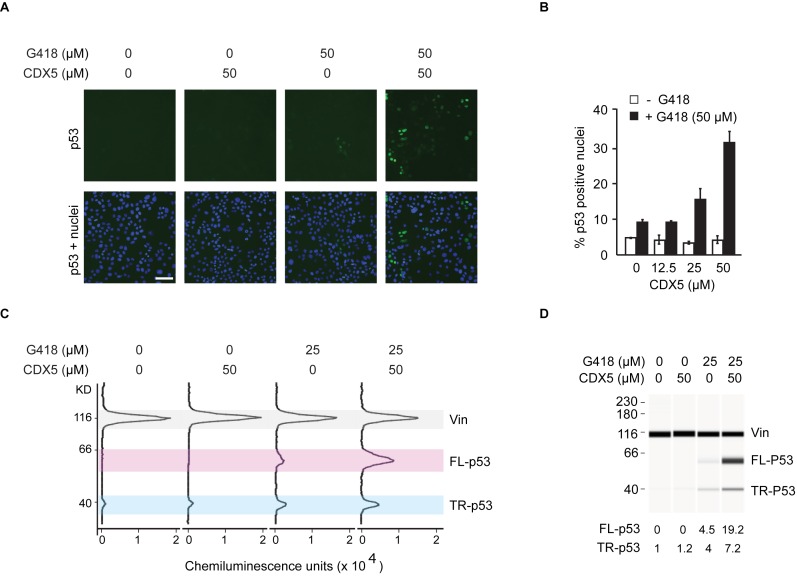
Nuclear localization sequences and a tetramerization domain located in the C-terminus of p53 contribute to its retention in the nucleus (45,46). p53 truncated at R213 lacks these sequences. To assess p53 R213X readthrough with relatively high throughput, we established an automated 96-well fluorescence microscopy assay to detect and quantitate nuclear p53, reasoning that PTC readthrough would result in the nuclear accumulation of full-length p53. HDQ-P1 cells showed little or no nuclear p53 signal (Figure 2A). Among several aminoglycosides tested at concentrations of 50, 100 and 200 μM only G418 induced a concentration-dependent increase in cells showing nuclear p53, consistent with the formation of full-length p53 due to readthrough. Paromomycin, which is active in yeast, and other approved aminoglycosides displayed no PTC readthrough activity at these concentrations. After 72 h exposure, 50 μM G418 induced nuclear p53 expression in 9% of cells (Figure 2A and B) while 200 μM G418 induced nuclear p53 expression in nearly all cells (Supplementary Figure S2B). None of the CDX compounds showed detectable PTC readthrough activity when used alone and only CDX5 in combination with 50 μM G418 induced a concentration-dependent increase in cells with nuclear p53 (Figure 2A and B).
Figure 2.
PTC readthrough at p53 R213X in HDQ-P1 human breast carcinoma cells. (A and B) Automated p53 immunofluorescence microscopy assay. Cells grown in 96-well plates were exposed for 72 h to different concentrations of CDX5 without or with 50 μM G418. The proportion of cells showing nuclear p53 immunofluorescence was determined as an indirect but high throughput measure of PTC readthrough. Representative images are shown in (A), with p53 immunofluorescence shown in green and nuclei in blue (bar, 100 μm). Quantitative data are shown in (B) (mean ± S.D., n = 3). Additional images are shown in Supplementary Figure S2. (C) Automated capillary electrophoresis western analysis. Cells were exposed for 96 h to the indicated compounds. The results are electropherograms of the chemiluminescence detection of bound antibodies. TR-p53: truncated p53, FL-p53: full-length p53, Vin: vinculin loading control. (D) The results from panel C are displayed as ‘pseudo blots’ for ease of visualization. The area under the truncated and full-length p53 peaks was first normalized to the vinculin loading control to account for variations in protein loading. To provide lane-to-lane comparison, the amount of truncated and full-length p53 was further divided by the amount of truncated p53 found in untreated cells. These numbers are displayed under the lanes. The data show that CDX5 does not induce the formation of full-length p53 as a single agent but that it strongly potentiates the PTC readthrough activity of G418.
Western analysis using an automated capillary electrophoresis system was used to measure p53 R213X readthrough quantitatively. HDQ-P1 cells expressed very low but measurable levels of truncated p53 and no detectable full-length p53 (Figure 2C and D). Alone, CDX5 caused no increase in truncated or full-length p53. G418 alone caused a 4-fold increase in both truncated and full-length p53 relative to the amount of truncated p53 present in untreated cells (Figure 2C and D). The combination of CDX5 and G418 caused a 7-fold increase in truncated p53 and a 19-fold increase in full-length p53 (Figure 2C and D). Therefore, CDX5 potentiates the PTC readthrough activity of G418 at p53 R213X in human HDQ-P1 cells.
CDX5 structure-activity relationship (SAR)
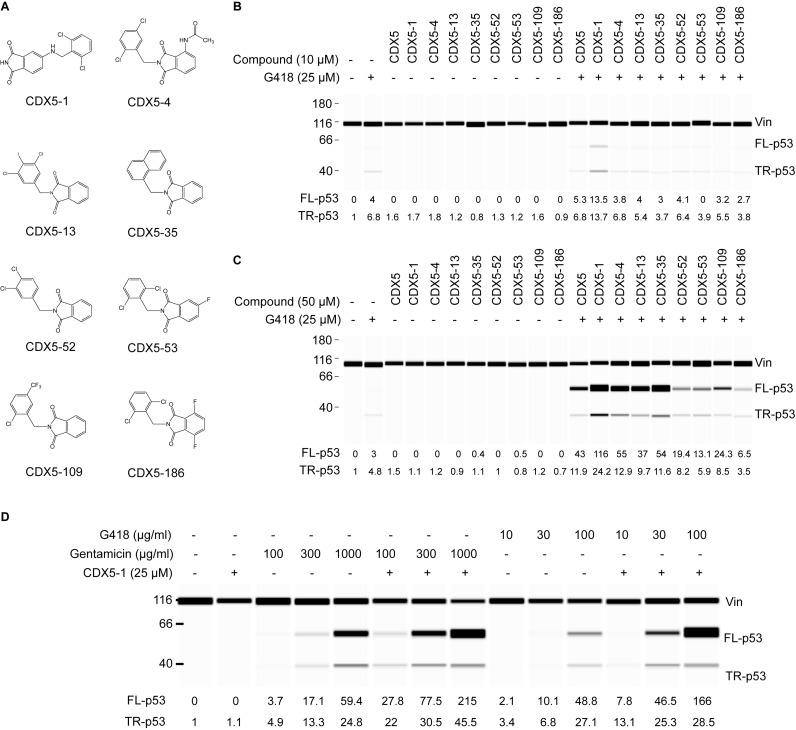
To better understand the structural requirements for PTC readthrough potentiation by CDX5 and to identify more potent analogs, structural derivatives were prepared wherein both aromatic rings were systematically functionalized with a wide range of polar and non-polar functional groups. Additionally, template hopping was explored to provide a more diverse family of core structures. Over 170 compounds were prepared and their effect on p53 R213X readthrough in HDQ-P1 cells was analyzed by automated fluorescence microscopy at different concentrations, with and without 50 μM G418. None of these compounds increased the number of cells with nuclear p53 when used alone but eight compounds, when combined with G418, increased nuclear p53 over 2-fold compared to G418 alone (Supplementary Table S2). Western analysis confirmed that the eight compounds did not induce PTC readthrough when used alone (Figure 3). When combined with 25 μM G418, CDX5-1, CDX5-4 and CDX5-35 induced more readthrough than the parent compound CDX5 (Figure 3). CDX5-1 was the most potent, inducing PTC readthrough at a lower concentration of 10 μM in combination with G418 (Figure 3). CDX5-1 also potentiated PTC readthrough by high concentrations of gentamicin (Figure 3D). CDX5-1 was therefore chosen for further study.
Figure 3.
PTC readthrough by CDX5 analogs in combination with aminoglycosides. HDQ-P1 cells were exposed for 96 h to the analogs shown in (A) at 10 μM (B) or 50 μM (C), without or with 25 μM G418 and were analysed for formation of full-length p53, quantified relative to the amount of truncated p53 in untreated cells. (D) HDQ-P1 cells were exposed for 72 h to CDX5-1 and different concentrations of gentamicin or G418. Concentrations are shown in μg/ml to enable comparison with gentamicin, which is a mix of related aminoglycosides.
Time course of p53 PTC readthrough and mRNA accumulation
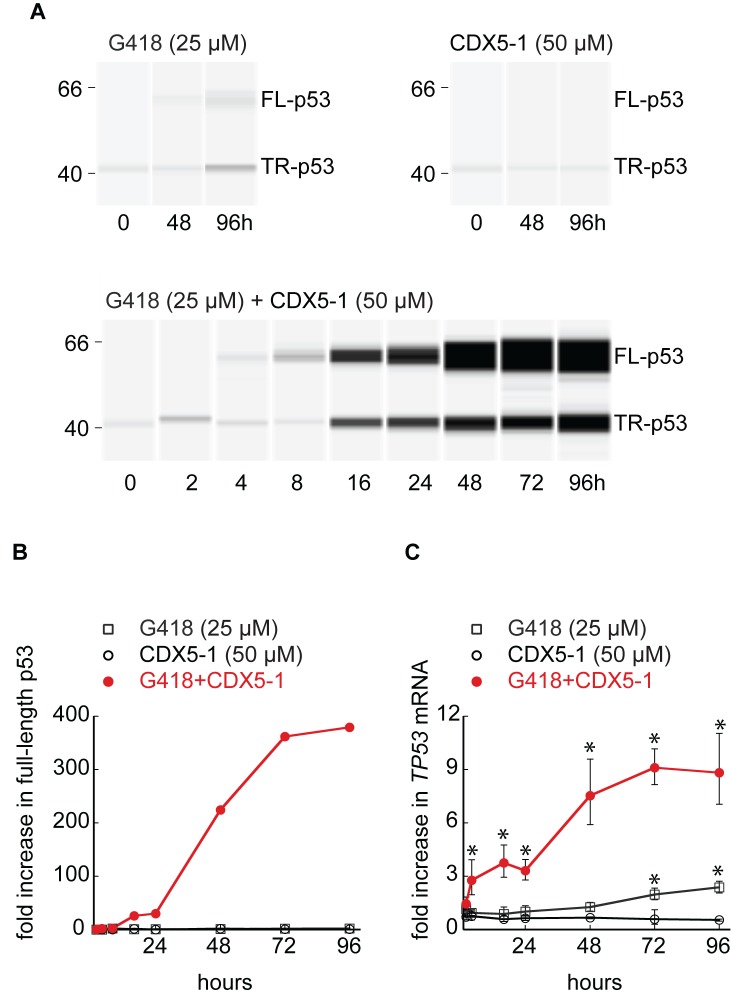
To gain a better understanding of readthrough potentiation by CDX5-1, we measured the accumulation of full-length p53 and truncated p53 over the course of 96 h exposure of HDQ-P1 cells to the compounds (Figure 4). G418 at a low concentration of 25 μM induced a very small amount of full-length p53, first detectable at 48 h and increasing to 2.3-fold at 96 h (Figure 4A and B). 50 μM CDX5-1 induced no full-length p53 at any time tested (Figure 4A and B). When G418 was combined with CDX5-1, full-length p53 levels increased dramatically to nearly 400 times the level of p53 present in untreated cells (Figure 4A and B). Furthermore, full-length p53 became first detectable after only 4 h exposure to the combination and increased steeply from 30- to 361-fold from 24 to 72 h, after which it reached a plateau. This time course illustrates the magnitude by which CDX5-1 potentiates G418 PTC readthrough activity.
Figure 4.
Time course of PTC readthrough. HDQ-P1 were exposed to compounds for different times and analysed for formation of full-length p53 (A and B), and TP53 mRNA (C). Panel A shows representative time points for G418 and CDX5-1 and the entire time course for the combination of G418 and CDX5-1. Panel B shows quantitation of full-length p53 relative to the amount of truncated p53 found in untreated cells for the entire time course for all three treatments. Panel C shows triplicate measurements of TP53 mRNA (± S.D., n = 3) from the same samples as panels A and B. * indicates statistically significant differences between different treatment conditions and untreated samples (P < 0.05).
We also measured TP53 mRNA during the time course experiment. The level of TP53 mRNA in untreated HDQ-P1 cells was ∼12% of that found in HCT116, a cancer cell line with WT TP53, consistent with its recognition as a PTC-containing mRNA followed by degradation by nonsense mediated RNA decay (NMD). HDQ-P1 cells exposed to G418 showed a ∼2-fold increase in TP53 mRNA at 72 and 96 h (Figure 4C). CDX5-1 caused no increase in TP53 mRNA at any time point. Exposure to both G418 and CDX5-1 resulted in a much larger, time-dependent increase in TP53 mRNA (Figure 4C); TP53 mRNA increased ∼3-fold by 4 h, coinciding with the first detection of readthrough by western analysis, and it increased further to a maximum of 9-fold at 72 h. Therefore, CDX5-1 also potentiates the stabilization of TP53 mRNA by G418. The lack of increase in TP53 mRNA during exposure to CDX5-1 alone shows that it does not transcriptionally activate the TP53 gene. This result also indicates that exposure of HDQ-P1 cells to CDX5-1 alone does not inhibit TP53 mRNA degradation and that it does not potentiate the PTC readthrough activity of G418 by inhibiting NMD. CDX5-1 and G418 alone did not change the expression of p53 downstream target genes P21, PUMA, and miRNA34a but the combination of CDX5-1 and G418 significantly increased their expression (Supplementary Figure S3), indicating that p53 produced by PTC readthrough is active as a transcription factor.
Potentiation of readthrough at all three termination codons
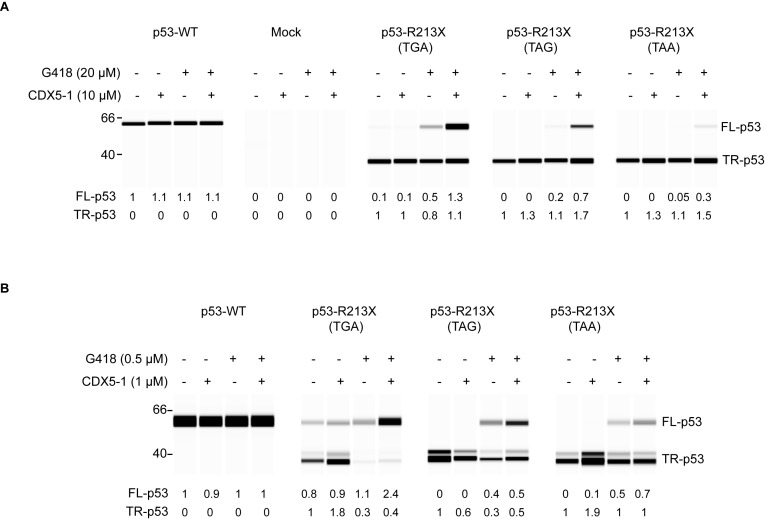
Previous work showed that basal and gentamicin-induced PTC readthrough efficiency differs for the three termination sequences, being highest for TGA, intermediate for TAG and lowest for TAA (47). To determine whether the identity of the termination codon influences readthrough by the compound combination, we generated four TP53 cDNA expression plasmids containing the WT arginine codon CGA or the termination codons TGA, TAG or TAA at position 213. Following transient transfection of p53-null H1299 cells with each construct, the cells were exposed to CDX5-1, G418, or both for 48 h. Untreated cells transfected with WT TP53 cDNA expressed only full-length p53, as expected, and exposure to the compounds did not affect WT p53 levels (Figure 5A). Untreated cells transfected with p53 R213X (TGA, TAG or TAA) constructs all expressed a prominent truncated p53 band and only p53 R213X (TGA) showed a small amount of full-length p53 indicating a low level of spontaneous readthrough at this PTC (Figure 5A). G418 induced full-length p53 production more efficiently at TGA than TAG, and no detectable readthrough at TAA. CDX5-1 alone at 10 μM did not increase PTC readthrough in any of the cells. However, the combination of CDX5-1 and G418 increased full-length p53 production at the TGA, TAG and TAA codons compared to G418 alone (Figure 5A). These findings indicate that the combination of CDX5-1 and G418 is able to induce full-length p53 from all three different PTC sequences at position R213, with highest efficiency for TGA, followed by TAG and TAA codons.
Figure 5.
PTC readthrough at p53 R213X (TGA, TAG and TAA) sequences in cells and in vitro. (A) H1299 cells transiently transfected with the indicated constructs were exposed for 48 h to CDX5-1 and G418, alone or in combination and full-length and truncated p53 was quantified relative to the amount of p53 in untreated cells. p53-WT: R213; Mock: transfection reagents only. (B) PTC readthrough in a cell-free translation extract. The indicated 5′ capped and 3′ poly(A) tailed TP53 mRNAs were subjected to in vitro translation in the presence of the indicated compounds. Full-length p53 and truncated p53 were quantified relative to full-length p53 (in WT) or truncated p53 (in mutants) in untreated reactions.
PTC readthrough in vitro
Aminoglycosides, which bind to ribosomes and directly target the translational machinery, can induce PTC readthrough in cell-free in vitro translation assays (48–51). To gain insight into the mechanism by which CDX5-1 potentiates readthrough by G418, we examined its activity in a HeLa cell-free translation assay. TP53 mRNA bearing the R213X (UGA-C) PTC was generated by in vitro transcription and further 5′-capping and 3′ poly(A)-tail addition. This mRNA was translated efficiently in vitro, forming a prominent truncated p53 band (Figure 5B). Some full-length p53 was also detected, indicating spontaneous readthrough (Figure 5B). The relative amount of spontaneous readthrough in the cell-free extract (Figure 5B) was higher than in the transfected cells (Figure 5A). In the presence of G418, in vitro translation of TP53 R213X (UGA) mRNA resulted in the production of more full-length p53 and less truncated p53 than without G418, indicating increased readthrough in this system (Figure 5B). CDX5-1 alone caused a slight increase in truncated p53 (Figure 5B). In a separate experiment carried out with a different batch of the in vitro translation kit, CDX5-1 increased both truncated p53 and full-length p53 (Supplementary Figure S4). Addition of both CDX5-1 and G418 caused a larger increase in full-length p53 than either compound alone (Figure 5B and Supplementary Figure S4).
We also examined the effect of the compounds on in vitro translation of TP53 mRNA bearing a UAG or a UAA instead of UGA at codon 213. Little or no spontaneous readthrough was observed with these nonsense codons (Figure 5B). G418 induced readthrough less efficiently at UAG, as evident from the lower ratio of full-length p53 to truncated p53 and even less efficiently at UAA (Figure 5B). Combining CDX5-1 with G418 caused a slight increase in readthrough at both UAG and UAA compared to G418 alone (Figure 5B). Additionally, the compounds, alone or in combination, had no effect on the translation of WT TP53 mRNA (Figure 5B). Therefore, CDX5-1 increased readthrough by G418 in vitro, but the enhancement observed was less pronounced than in cells.
PTC readthrough at different positions in the TP53 gene
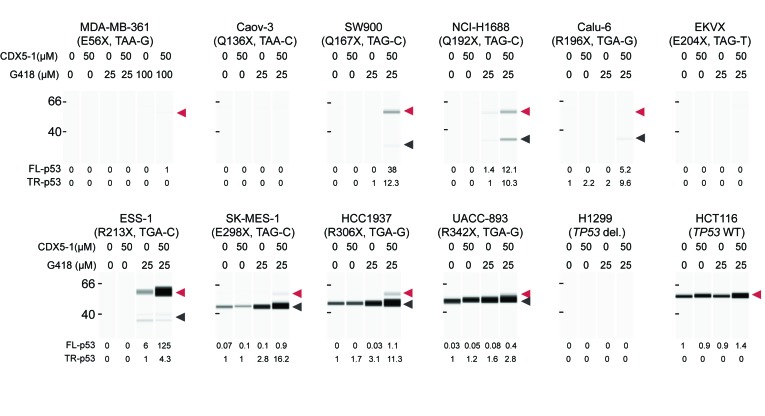
To determine whether the compound combination can induce p53 PTC readthrough at positions other than R213X and in cancer cell lines from different tissue origins we selected ten human cancer cell lines with homozygous TP53 nonsense mutations at different positions in the TP53 coding region (Figure 6). These cell lines originate from breast, lung, ovary and endometrium and all contain a different spectrum of somatic alterations according to the COSMIC Cell Line Project. Exposure to 50 μM CDX5-1 alone did not induce p53 PTC readthrough in any of the cell lines, as was the case for HDQ-P1 cells. 25 μM G418 induced measurable PTC readthrough in only two of the cell lines, ESS-1 (R213X, TGA) and NCI-H1688 (Q192X, TAG). The combination of 25 μM G418 and 50 μM CDX5-1 induced measurable readthrough in 7 of 8 cell lines with TGA (Calu-6, ESS1, HCC1937, UACC-893) or TAG (SW900, NCI-H1688, SK-MES-1) PTCs (Figure 6), and very weakly in one of the two cell lines with a TAA termination codon, MDA-MB-361 (E56X). Neither compound alone nor in combination induced p53 production in H1299 p53-null cells or increased p53 levels in HCT116 cells with WT TP53. These findings show that the combination of CDX5-1 and G418 is effective against a considerably wider range of TP53 nonsense mutations than G418 alone.
Figure 6.
PTC readthrough in different human cancer cell lines with homozygous nonsense mutations at different positions in the TP53 gene. The cell lines (mutations shown in parentheses), were exposed for 96 h to CDX5-1 and G418, alone or in combination, and full-length p53 (red arrowhead) and truncated p53 (black arrowhead) was quantified relative to truncated p53 found in untreated cells, or to truncated p53 found in treated cells if no p53 signal was detected in untreated cells. The +4 nucleotide is indicated for each codon.
PTC readthrough in rare genetic disease models
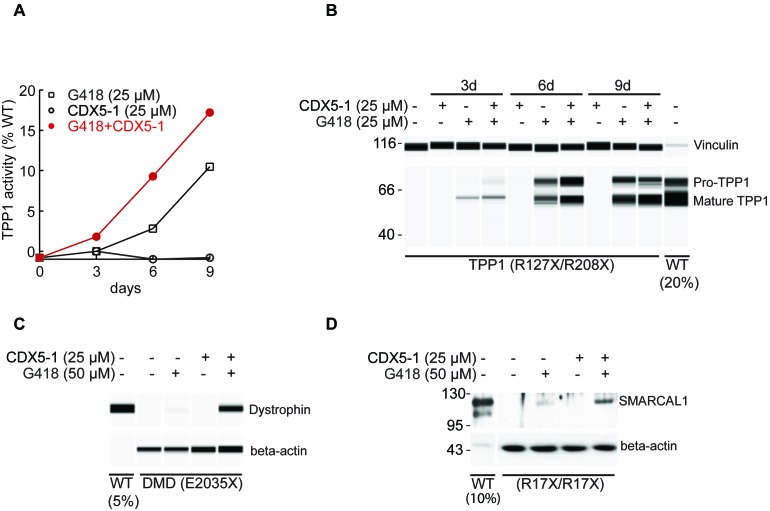
Compounds that induce PTC readthrough could in principle be used to treat inherited diseases caused by nonsense mutations. To exemplify whether the combination of compounds identified in this study can also induce readthrough of PTCs causing genetic diseases, primary fibroblasts from a late infantile neuronal ceroid lipofuscinosis (CLN2) patient, a myoblast line established from a patient with DMD, and a fibroblast line derived from a patient with Schimke immuno-osseous dysplasia (SIOD) were selected. The CLN2 cells have compound heterozygous nonsense mutations (R127X/R208X) in the TPP1 gene encoding the lysosomal enzyme tripeptidyl peptidase 1. The DMD myoblasts have nonsense mutation (E2035X) in the DMD gene that encodes dystrophin, and the SIOD cells have a homozygous nonsense mutation (R17X/R17X) in the SMARCAL1 gene encoding the DNA annealing helicase SMARCAL1. All patient-derived cells were exposed to 25 or 50 μM G418, 25 μM CDX5-1 or the combination for up to 9 days. TPP1 enzyme activity was measured using a fluorometric assay and expression of full-length TPP1, dystrophin and SMARCAL1 proteins were assessed by immunoblotting. Untreated CLN2 cells displayed no detectable TPP1 enzyme activity (Figure 7A). Exposure to CDX5-1 alone induced no enzyme activity. G418 induced a time-dependent increase in activity first detectable at day 3. The combination induced greater TPP1 activity at all time points tested (Figure 7A). The maximum TPP1 activity observed was 17% of the average activity obtained from fibroblasts of two unaffected individuals. The extracts used for enzyme activity measurement were also subjected to western analysis to examine formation of full-length TPP1. Fibroblasts from unaffected individuals showed two immunoreactive bands corresponding to the proenzyme and mature form of TPP1 (37) and patient fibroblasts showed no detectable bands (Figure 7B). As expected from the TPP1 enzyme activity results, CDX5-1 alone induced no TPP1 protein. G418 alone caused a time-dependent increase in both TPP1 proenzyme and mature form and the combination of G418 and CDX5-1 induced more TPP1 protein than G418 alone at all time points tested (Figure 7B). Similarly, untreated or CDX5-1-treated cells from DMD and SIOD patients did not show any detectable full-length dystrophin or SMARCAL1, respectively. Treatment with G418 alone induced a small amount of full-length dystrophin in DMD cells and some full-length SMARCAL1 in SD123. The combination of G418 and CDX5-1 induced more full-length dystrophin and SMARCAL1 in DMD and SIOD cells, respectively (Figure 7C and D).
Figure 7.
TPP1, DMD and SMARCAL1 PTC readthrough in patient-derived cells. (A) GM16485 fibroblasts (TPP1: R127X, TGA-C; R208X, TGA-T) were exposed to the indicated concentrations of compounds for different times and TPP1 enzyme activity was determined and expressed relative to the average activity of untreated fibroblasts from two unaffected individuals (WT). (B) The same cell extracts were analysed for formation of TPP1 by automated capillary electrophoresis western analysis. Extracts from WT fibroblasts were also analysed, using 20% of the amount of protein used for GM16485. Vinculin was used as a loading control. (C) HSK001 myoblasts derived from a DMD patient with nonsense mutation (DMD: E2035X, TAG-G) were differentiated into myotubes and exposed to the indicated concentrations of compounds for 3 days and dystrophin expression level was determined by automated capillary electrophoresis western analysis. Extracts from WT myotubes were also analyzed, using 5% of the amount of protein used for DMD cells. Beta-actin was used as a loading control. (D) SD123 fibroblasts with a homozygous SMARCAL1 nonsense mutation (R17X, TGA-C) were exposed to the indicated concentrations of compounds for 6 days and SMARCAL1 levels were determined by western blotting. Extracts from WT fibroblasts were also analyzed, using 10% of the amount of protein used for SIOD cells. Beta-actin was used as a loading control.
We also tested the effect of G418 and CDX5-1, alone and in combination, on the viability of HDQ-P1, HCT116, fibroblast and myoblast cell lines (Supplementary Figure S5). During three-day exposure to 25 μM CDX5-1, a concentration that effectively potentiated readthrough in rare disease cells, the rate of cell proliferation decreased and proliferation resumed when the compound was washed away (Supplementary Figure S5). 50 μM CDX5-1, which maximally potentiated p53 readthrough in HDQ-P1 cells, caused a more pronounced slowing of cell proliferation, which was also reversible upon removal of the compound. G418 at 25 or 50 μM had little or no effect on cell proliferation. Combining G418 and CDX5-1 had little additional effect on proliferation compared with CDX5-1 alone, except for fibroblasts, which appeared to become stably arrested.
DISCUSSION
Currently available PTC readthrough compounds show only weak activity in vitro and in animals, and those tested in clinical trials show response only in a fraction of patients (52), which restricts their usefulness as therapies. Our identification of small molecules that strongly potentiate PTC readthrough by paromomycin at multiple nonsense alleles in yeast was encouraging because it raised the possibility that combining an aminoglycoside with a potentiator might also induce more efficient and broad PTC readthrough in human cells. The phthalimide class of small molecules represented by CDX5 exerted similar effects on TP53 R213X in human cells similar to those observed for alleles like lys2-101 in yeast; CDX5 was inactive as a single agent but, in combination with paromomycin in yeast or G418 in human cells, it strongly potentiated PTC readthrough.
A range of modifications to the CDX5 benzyl ring and phthalimide rings, including polar and non-polar substitutions, heteroaromatics and altered ring size, mostly caused loss of activity and yielded no compounds with higher potency than the parent screening hit. A template hopping approach produced CDX5-1 as the single more potent compound. The extent to which CDX5-1 potentiated the readthrough activity of G418 was remarkable, as illustrated in Figure 3 and particularly in Figure 4B, where CDX5-1 increased G418-induced readthrough at p53 R213X up to 180-fold compared to G418 alone. Equally striking was that CDX5-1 accelerated the earliest detection of readthrough from 48 h for G418 alone to 4 h for the combination.
Studies using reporter constructs in vitro and in cell culture have shown that the relative level of basal or aminoglycoside-induced PTC readthrough is TGA>TAG>TAA (47,53–54). Our results show that CDX5-1 can potentiate the readthrough activity of G418 at all three PTCs and that it does not alter this sequence preference. The presence of a uracil just before and of a cytosine just after a PTC in mRNA increases readthrough by gentamicin (47). In HDQP-1 cells TP53 R213X is flanked by these two nucleotides, likely providing an optimal sequence context for G418-induced readthrough. Substitution nonsense mutations account for 8% of TP53 mutations in cancer. R213X is the most common, affecting 1.3% of patients (COSMIC), but there are several others, most of which are not in an optimal sequence context. Our analysis of ten additional human cancer cell lines with homozygous TP53 nonsense mutations at ten different positions provides a comprehensive description of PTC readthrough in an endogenous gene. In two of these cell lines the PTC was flanked by 5′ T and 3′ C, one with the same R213X mutation as HDQ-P1, and the other with Q192X. Both showed readthrough with G418 alone and increased readthrough with the combination. The eight others had either one or none of these flanking nucleotides and showed no readthrough with G418 alone. Six had some level of response with the combination while two showed no response. Therefore, combining G418 with CDX5-1 can elicit PTC readthrough even in non-optimal sequence contexts.
Of the 10 cell lines examined, three showed a detectable level of truncated protein. These cell lines had PTCs towards the 3′ end of the TP53 coding sequence and the level of truncated protein was higher the closer the PTC was to the normal termination codon (Figure 6). Interestingly, these three cell lines showed no p53 PTC readthrough with G418 alone and relatively little with the combination, compared with the high levels of PTC readthrough observed at R213X and Q192X in cells with no detectable truncated protein. Therefore, the presence of truncated protein does not appear to predict response to PTC readthrough compounds.
The mechanism by which CDX5-1 potentiates readthrough by G418 remains to be elucidated but some possibilities can be ruled out. CDX5-1 does not stimulate TP53 gene expression because it does not increase the cellular level of TP53 mRNA in HDQ-P1 cells with TP53 nonsense mutations or in HCT116 cells with WT TP53 when used alone (Figure 4 and Supplementary Figure S3). The fact that CDX5-1 does not increase TP53 mRNA in HDQ-P1 cells also argues against it being an NMD inhibitor because inhibition of NMD by downregulation of UPF proteins or with small molecules like NMDI-1 generally increases the cellular level of mRNAs containing nonsense mutations (14,55–58). Additionally, CDX5-1 enhanced the PTC readthrough activity of G418 in a cell-free in vitro translation system indicating that it does not act by modulating the expression of other genes such as genes controlling translational termination or NMD.
The time course of accumulation of full-length p53 protein and of mRNA during exposure to G418 and CDX5-1 is also informative. During the first 12 h, full-length p53 protein rose slowly, followed by a period of rapid full-length p53 formation that reached a half-maximal point after 45 h (Figure 4). The p53 mRNA increased slowly and by less than 3-fold during exposure to G418 alone but it increased more rapidly and to nearly 9-fold with the combination before reaching a plateau. The sigmoidal curve of protein accumulation is compatible with a self-amplification mechanism whereby TP53 mRNA is normally kept at a very low level because translation of any PTC-containing mRNA molecule leads to its recognition and degradation by NMD. Stalling of a translating ribosome at a PTC and inefficient termination of translation are believed to be triggers for NMD (59,60). The combination of G418 and CDX5-1 may facilitate rapid pairing of a near-cognate aminoacyl-tRNA at the PTC-bound ribosome and incorporation of an amino acid residue, leading not only to formation of full-length p53 but also to escape from recognition by NMD in a fraction of these mRNAs. Further translation of the PTC-containing mRNAs that escaped degradation would then lead to cycles of additional readthrough and further mRNA stabilization. In this model, TP53 mRNA initially increases as a consequence of readthrough but the increased mRNA levels in turn enable additional readthrough events. Such a self-amplification loop could explain the sigmoidal increase in full-length p53 over time.
The combination of CDX5-1 and G418 appears to induce PTC readthrough less efficiently at TP53 nonsense mutations situated closer to the 3′ end of the transcript and showing high levels of truncated p53 (Figure 6). It is likely that translation termination is more efficient at these 3′-proximal PTCs and consequently that the ribosomes do not stall for as long as they do at R213X, thus lowering the probability that a near-cognate aminoacyl-tRNA will pair before translation termination, and consequently reducing readthrough. Therefore, more efficient drug-induced readthrough would occur at PTCs with favorable flanking nucleotides and positions that do not permit efficient translation termination.
The finding that combining CDX5-1 with G418 induces more readthrough and at a broader number of PTCs in cancer cells than G418 alone raised the possibility that such combinations might also broadly suppress nonsense mutations in untransformed cells for treatment of genetic diseases. We found that the combination efficiently induced readthrough in cells from CLN2, DMD and SIOD patients bearing nonsense mutations in the TPP1 (R127X/R208X), DMD (E2035X) and SMARCAL1 (R17X/R17X) genes. Of note, TPP1 is the most commonly mutated gene in late infantile neuronal ceroid lipofuscinosis. DMD is also the most common and the most lethal of muscular dystrophies. PTC readthrough has not been observed before in SIOD. Gentamicin has been found previously to increase TPP1 activity in lymphoblast cells with the L104X and R208X mutations (61) and in fibroblasts with the R127X and R208X mutations (62). The restoration of 17% of WT TPP1 activity by the combination might be sufficient for therapeutic activity since genetic studies have shown that mouse hypomorphs expressing 6% of WT TPP1 activity in the brain had relatively mild pathology and locomotor dysfunction and a near normal 20-month median lifespan, compared to 4.5 months for mice lacking TPP1 activity (63). CDX5-1 reversibly slowed down the proliferation of immortalized fibroblasts and myoblasts. It is not yet known whether this effect on cell proliferation may be separated from the PTC readthrough potentiating activity by further chemical modification or whether they are mechanistically linked and not separable. It will be important to further optimize the compounds and test them in animal models to determine whether therapeutically relevant PTC readthrough can be achieved with a good therapeutic window in vivo.
Overall, these findings raise hope that combinations of an aminoglycoside and a potentiator might enable use of lower aminoglycoside doses to minimize adverse effects such as nephrotoxicity and ototoxicity, as well as induce PTC readthrough more broadly than current drugs to benefit a larger number of patients.
Supplementary Material
Acknowledgments
We thank Mel Reichman and Scott Donover (Lankenau Institute for Medical Research) for screening data analysis and provision of compounds, Peter Stirling for technical advice and Cornelius Boerkoel for cell lines, antibodies, encouragement and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Microgrant from the Rare Disease Foundation (to M.R.); Canadian Cancer Society [702681 to M.R.]; Genome BC [POC027 to M.R.]; Mitacs [IT03752 to A.B.H.]. Funding for open access charge: Canadian Cancer Society.
Conflict of interest statement. M.R. and A.B.H. have an ownership interest in Codon-X Therapeutics. A patent application pertaining to the results presented in the paper has been filed with the United States Patent and Trademark Office. The authors declare no additional competing financial interests.
REFERENCES
- 1.Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 2.Benzer S., Champe S.P. A change from nonsense to sense in the genetic code. Proc. Natl. Acad. Sci. U.S.A. 1962;48:1114–1121. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawthorne D.C., Mortimer R.K. Super-suppressors in yeast. Genetics. 1963;48:617–620. doi: 10.1093/genetics/48.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manney T.R. Action of a super-suppressor in yeast in relation to allelic mapping and complementation. Genetics. 1964;50:109–121. doi: 10.1093/genetics/50.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh A., Ursic D., Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 6.Palmer E., Wilhelm J.M., Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 7.Burke J.F., Mogg A.E. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G418 and paromomycin. Nucleic Acids Res. 1985;13:6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.François B., Russell R.J.M., Murray J.B., Aboul-ela F., Masquida B., Vicens Q., Westhof E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Loubresse N.G., Prokhorova I., Holtkamp W., Rodnina M. V., Yusupova G., Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 10.Bedwell D.M., Kaenjak A., Benos D.J., Bebok Z., Bubien J.K., Hong J., Tousson A., Clancy J.P., Sorscher E.J. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat. Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 11.Du M., Jones J.R., Lanier J., Keeling K.M., Lindsey J.R., Tousson A., Bebök Z., Whitsett J.a, Dey C.R., Colledge W.H., et al. Aminoglycoside suppression of a premature stop mutation in a Cftr-/- mouse carrying a human CFTR-G542X transgene. J. Mol. Med. 2002;80:595–604. doi: 10.1007/s00109-002-0363-1. [DOI] [PubMed] [Google Scholar]
- 12.Clancy J.P., Bebök Z., Ruiz F., King C., Jones J., Walker L., Greer H., Hong J., Wing L., Macaluso M., et al. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2001;163:1683–1692. doi: 10.1164/ajrccm.163.7.2004001. [DOI] [PubMed] [Google Scholar]
- 13.Wilschanski M., Yahav Y., Yaacov Y., Blau H., Bentur L., Rivlin J., Aviram M., Bdolah-Abram T., Bebok Z., Shushi L., et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N. Engl. J. Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 14.Linde L., Boelz S., Nissim-Rafinia M., Oren Y.S., Wilschanski M., Yaacov Y., Virgilis D., Neu-Yilik G., Kulozik A.E., Kerem E., et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton-Davis E.R., Cordier L., Shoturma D.I., Leland S.E., Sweeney H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik V., Rodino-Klapac L.R., Viollet L., Wall C., King W., Al-Dahhak R., Lewis S., Shilling C.J., Kota J., Serrano-Munuera C., et al. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann. Neurol. 2010;67:771–780. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 17.Shulman E., Belakhov V., Wei G., Kendall A., Meyron-Holtz E.G., Ben-Shachar D., Schacht J., Baasov T. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: a strategy for the treatment of genetic diseases. J. Biol. Chem. 2014;289:2318–2330. doi: 10.1074/jbc.M113.533588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue X., Mutyam V., Tang L., Biswas S., Du M., Jackson L.A., Dai Y., Belakhov V., Shalev M., Chen F., et al. Synthetic aminoglycosides efficiently suppress cystic fibrosis transmembrane conductance regulator nonsense mutations and are enhanced by ivacaftor. Am. J. Respir. Cell Mol. Biol. 2014;50:805–816. doi: 10.1165/rcmb.2013-0282OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatti R.A. SMRT compounds correct nonsense mutations in primary immunodeficiency and other genetic models. Ann. N. Y. Acad. Sci. 2012;1250:33–40. doi: 10.1111/j.1749-6632.2012.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du M., Liu X., Welch E.M., Hirawat S., Peltz S.W., Bedwell D.M. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L., Jung M.E., Damoiseaux R., Completo G., Fike F., Ku J.-M., Nahas S., Piao C., Hu H., Gatti R.A. A new series of small molecular weight compounds induce read through of all three types of nonsense mutations in the ATM gene. Mol. Ther. 2013;21:1653–1660. doi: 10.1038/mt.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilberberg A., Lahav L., Rosin-Arbesfeld R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut. 2010;59:496–507. doi: 10.1136/gut.2008.169805. [DOI] [PubMed] [Google Scholar]
- 23.Arakawa M., Shiozuka M., Nakayama Y., Hara T., Hamada M., Kondo S., Ikeda D., Takahashi Y., Sawa R., Nonomura Y., et al. Negamycin restores dystrophin expression in skeletal and cardiac muscles of mdx mice. J. Biochem. 2003;134:751–758. doi: 10.1093/jb/mvg203. [DOI] [PubMed] [Google Scholar]
- 24.Auld D.S., Lovell S., Thorne N., Lea W.a, Maloney D.J., Shen M., Rai G., Battaile K.P., Thomas C.J., Simeonov A., et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auld D.S., Thorne N., Maguire W.F., Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElroy S.P., Nomura T., Torrie L.S., Warbrick E., Gartner U., Wood G., McLean W.H.I. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol. 2013;11:e1001593. doi: 10.1371/journal.pbio.1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 28.Kerem E. Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir. Med. 2014;18:11–12. doi: 10.1016/S2213-2600(14)70100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bushby K., Finkel R., Wong B., Barohn R., Campbell C., Comi G.P., Connolly A.M., Day J.W., Flanigan K.M., Goemans N., et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014;50:477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan N.J. Ataluren: first global approval. Drugs. 2014;74:1709–1714. doi: 10.1007/s40265-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L., et al. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donohue E., Tovey A., Vogl A.W., Arns S., Sternberg E., Young R.N., Roberge M. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J. Biol. Chem. 2011;286:7290–7300. doi: 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lojewski X., Staropoli J.F., Biswas-Legrand S., Simas A.M., Haliw L., Selig M.K., Coppel S.H., Goss K.A., Petcherski A., Chandrachud U., et al. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum. Mol. Genet. 2014;23:2005–2022. doi: 10.1093/hmg/ddt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou T.-C., Martin N. CompuSyn software for drug combinations and for general dose effect analysis. 2007 [Google Scholar]
- 39.Ono B.I., Fujimoto R., Ohno Y., Maeda N., Tsuchiya Y., Usui T., Ishino-Arao Y. UGA suppressors in Saccharomyces cerevisiae: allelism, action spectra and map positions. Genetics. 1988;118:41–47. doi: 10.1093/genetics/118.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono B.I., Stewart J.W., Sherman F. Yeast UAA suppressors effective in psi+ strains serine-inserting suppressors. J. Mol. Biol. 1979;128:81–100. doi: 10.1016/0022-2836(79)90309-7. [DOI] [PubMed] [Google Scholar]
- 41.Liebman S.W., Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J. Bacteriol. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou T. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 43.Wang C.S., Goulet F., Lavoie J., Drouin R., Auger F., Champetier S., Germain L., Têtu B. Establishment and characterization of a new cell line derived from a human primary breast carcinoma. Cancer Genet. Cytogenet. 2000;120:58–72. doi: 10.1016/s0165-4608(99)00253-8. [DOI] [PubMed] [Google Scholar]
- 44.Floquet C., Deforges J., Rousset J.-P., Bidou L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011;39:3350–3362. doi: 10.1093/nar/gkq1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaulsky G., Goldfinger N., Ben-Ze'ev A., Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol. Cell. Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang S.H., Clarke M.F. Regulation of p53 localization. Eur. J. Biochem. 2001;268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- 47.Floquet C., Hatin I., Rousset J.-P., Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2012;8:e1002608. doi: 10.1371/journal.pgen.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebibo-Sabbah A., Nudelman I., Ahmed Z.M., Baasov T., Ben-Yosef T. In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome. Hum. Genet. 2007;122:373–381. doi: 10.1007/s00439-007-0410-7. [DOI] [PubMed] [Google Scholar]
- 49.Goldmann T., Rebibo-Sabbah A., Overlack N., Nudelman I., Belakhov V., Baasov T., Ben-Yosef T., Wolfrum U., Nagel-Wolfrum K. Beneficial read-through of a USH1C nonsense mutation by designed aminoglycoside NB30 in the retina. Invest. Ophthalmol. Vis. Sci. 2010;51:6671–6680. doi: 10.1167/iovs.10-5741. [DOI] [PubMed] [Google Scholar]
- 50.Sánchez-Alcudia R., Pérez B., Ugarte M., Desviat L.R. Feasibility of nonsense mutation readthrough as a novel therapeutical approach in propionic acidemia. Hum. Mutat. 2012;33:973–980. doi: 10.1002/humu.22047. [DOI] [PubMed] [Google Scholar]
- 51.Gómez-Grau M., Garrido E., Cozar M., Rodriguez-Sureda V., Domínguez C., Arenas C., Gatti R.A., Cormand B., Grinberg D., Vilageliu L. Evaluation of aminoglycoside and non-aminoglycoside compounds for stop-codon readthrough therapy in four lysosomal storage diseases. PLoS One. 2015;10:e0135873. doi: 10.1371/journal.pone.0135873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keeling K.M., Xue X., Gunn G., Bedwell D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manuvakhova M., Keeling K., Bedwell D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard M.T., Anderson C.B., Fass U., Khatri S., Gesteland R.F., Atkins J.F., Flanigan K.M. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann. Neurol. 2004;55:422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- 55.Usuki F., Yamashita A., Higuchi I., Ohnishi T., Shiraishi T., Osame M., Ohno S. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol. 2004;55:740–744. doi: 10.1002/ana.20107. [DOI] [PubMed] [Google Scholar]
- 56.Usuki F., Yamashita A., Kashima I., Higuchi I., Osame M., Ohno S. Specific inhibition of nonsense-mediated mRNA decay components, SMG-1 or Upf1, rescues the phenotype of ullrich disease fibroblasts. Mol. Ther. 2006;14:351–360. doi: 10.1016/j.ymthe.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez-Hilarion S., Beghyn T., Jia J., Debreuck N., Berte G., Mamchaoui K., Mouly V., Gruenert D.C., Déprez B., Lejeune F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J. Rare Dis. 2012;7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keeling K.M., Wang D., Dai Y., Murugesan S., Chenna B., Clark J., Belakhov V., Kandasamy J., Velu S.E., Baasov T., et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fatscher T., Boehm V., Gehring N.H. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015;72:4523–4544. doi: 10.1007/s00018-015-2017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celik A., Kervestin S., Jacobson A. NMD: At the crossroads between translation termination and ribosome recycling. Biochimie. 2015;114:2–9. doi: 10.1016/j.biochi.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller J.N., Chan C.H., Pearce D.A. The role of nonsense-mediated decay in neuronal ceroid lipofuscinosis. Hum. Mol. Genet. 2013;22:2723–2734. doi: 10.1093/hmg/ddt120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sleat D.E., Sohar I., Gin R.M., Lobel P. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur. J. Paediatr. Neurol. 2001;5:57–62. doi: 10.1053/ejpn.2000.0436. [DOI] [PubMed] [Google Scholar]
- 63.Sleat D.E., El-Banna M., Sohar I., Kim K.H., Dobrenis K., Walkley S.U., Lobel P. Residual levels of tripeptidyl-peptidase I activity dramatically ameliorate disease in late-infantile neuronal ceroid lipofuscinosis. Mol. Genet. Metab. 2008;94:222–233. doi: 10.1016/j.ymgme.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.