Abstract
Ts3 is an alpha scorpion toxin from the venom of the Brazilian scorpion Tityus serrulatus. Ts3 binds to the domain IV voltage sensor of voltage-gated sodium channels (Nav) and slows dow n their fast inactivation. The covalent structure of Ts3 toxin is uncertain, and the structure of the folded protein molecule is unknown. Here we report the total chemical synthesis of four candidate Ts3 toxin protein molecules and the results of structure-activity studies that enabled us to establish the covalent structure of biologically active Ts3 protein molecule. We also report synthesis of the mirror image form of Ts3 toxin protein, and the use of racemic protein crystallography to determine the folded (tertiary) structure of biologically active Ts3 toxin by X-ray diffraction.
Keywords: Ts3 toxin, chemical protein synthesis, racemic protein crystallography
Ts3 is an alpha scorpion toxin protein natural product isolated from the venom of the Brazilian scorpion Tityus serrulatus [1]. Alpha-scorpion toxins belong to the family of Nav channel gating modifier proteins, and they mainly bind to the loop connecting S3 and S4 in Nav domain IV.[2,3] Upon binding to the Nav channels, these toxins inhibit the fast inactivation of Nav channels without dramatically affecting the activation of the channels.[4-8]
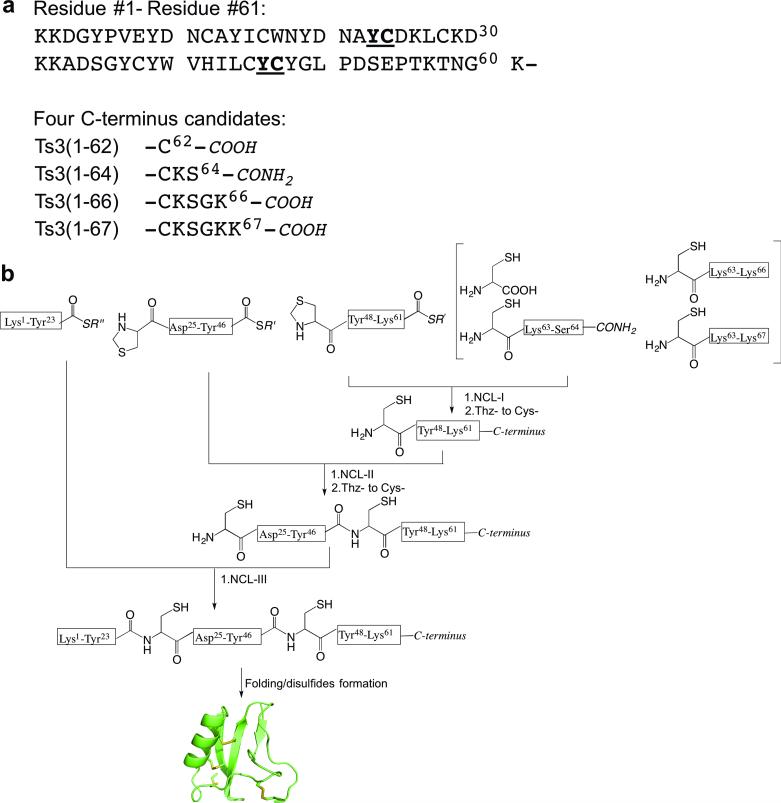
Prior to the work in this paper, there was no X-ray or NMR structure of the Ts3 protein. Furthermore, there is confusion in the literature about the C-terminus of the Ts3 polypeptide chain: some papers stated that the active Ts3 toxin had 62 amino acid residues,[9] while others believed it had 64 amino acid residues.[10,11] From analysis of the gene sequence, Martin-Eauclaire et al. proposed that the 64 residue Ts3 toxin polypeptide chain should terminate as the caroboxamide.[10] In the same paper, they also suggested there could be three different C-terminal candidates for the correct sequence of the Ts3 toxin protein.[10] To resolve this confusion and to determine the amino acid sequence and covalent structure of active Ts3 toxin, our first task was to prepare the three proposed Ts3 polypeptide chains together with the precursor polypeptide chain Ts3(1-67) (Scheme 1a), in order to determine which if any of these polypeptides folds to form biologically active Ts3 toxin protein.
Scheme 1.
Candidate amino acid sequences for the Ts3 polypeptide chain and synthetic strategy for their preparation. a) Sequences of candidate Ts3 polypeptide chains.[9,10] b) Modular synthetic strategy based on native chemical ligation for preparation of the four candidate Ts3 polypeptide chains. ‘C-terminus’ is one of the four candidates listed in scheme 1a.
We have previously reported total synthesis of the Ts1 toxin protein molecule,[12] but that experience was of limited use for the current work. Although isolated from the same scorpion venom, Ts3 toxin and Ts1 toxin are very different protein molecules: they have distinct biological activities - Ts1 is a beta-scorpion toxin while Ts3 is an alpha-scorpion toxin; there is little sequence homology between the two polypeptide chains (only 16/53 conserved non-Cys residues [Figure 1]); and, Ts3 has much greater structural ambiguity at its C-terminus than was the case with Ts1 toxin. Although Ts3 toxin and Ts1 toxin do share a conserved pattern of eight Cys residues in the amino acid sequence of their polypeptide chains (Figure 1), it has been shown that, even where the pattern of Cys residues is conserved scorpion venom proteins can have distinct tertiary structures.[13]
Figure 1.

Comparison of the amino acid sequences of Ts3 and Ts1 toxins. Alignment based on Cys residues (bold); conserved amino acids underlined. Unitl now, the presence in mature biologically active Ts3 protein of the C-terminal residues enclosed in parentheses was in dispute.
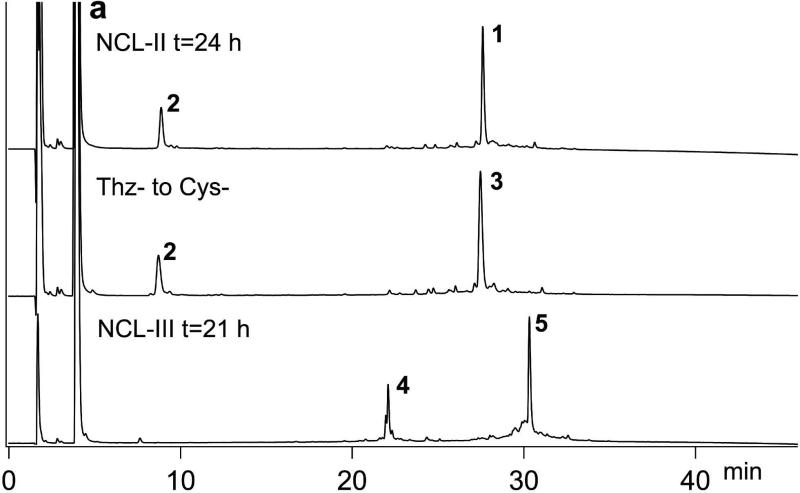
Total chemical synthesis of the candidate Ts3 polypeptide chains was carried out using a modular approach as shown in Scheme 1b. Native chemical ligation sites were chosen at -Tyr23- Cys24- and -Tyr46-Cys47- so that, after preparation of the C-terminal peptide variants, the peptide segments used were of similar length (viz. 23; 23; and 16, 18, 20, or 21 amino acids), Peptide segments were synthesized as described in the SI (Figures S1-9). Four distinct C-terminal peptide segments were prepared by native chemical ligation of Thz47-Lys61-thioester to the corresponding C-terminal Cys-peptide, or cysteine (Scheme 1b). Analytical data for the preparation by native chemical ligation of the full-length Ts3(1-64)-CONH2 polypeptide chain are shown in Figure 2a. Synthetic manipulations (see SI) for making each of the four full-length candidate Ts3 polypeptide chains were essentailly similar (Figures S10-12).
Figure 2.
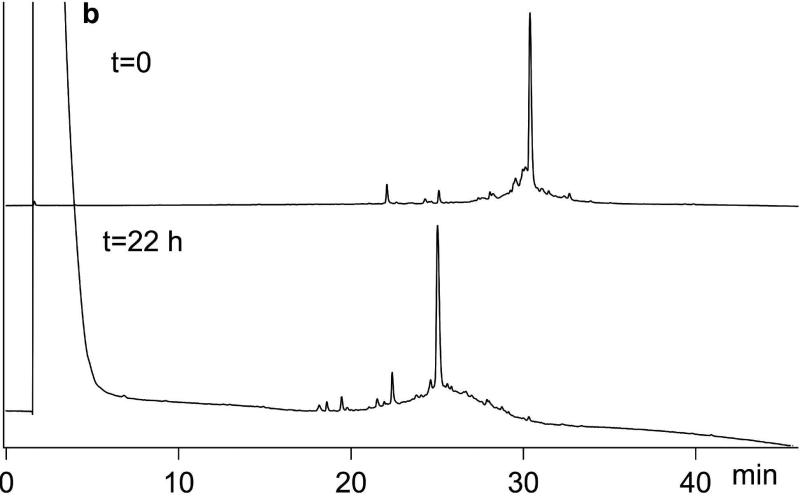
Chemial synthesis of Ts3 (1-64)-CONH2 protein. a) Analytical LCMS data for the synthesis of polypeptide Lys1-Ser64-CONH2. 1 Thz24-Ser64-CONH2; 2 Cys47-Ser64-CONH2; 3 Cys24-Ser64-CONH2; 4 Lys1-Tyr23-COS thiolactone; 5 Desired product Lys1-Ser64-CONH2. b) Analytical HPLC data for folding/disulfides formation of crude Ts3(1-64)-CONH2 polypeptide. Folding conditions: [polypeptide] 0.02 mg/mL; 2.0 mM GSH/1.0 mM GSSG; 0.5 M L-arginine·HCl; 1 mM EDTA; 0.1 M Tris, 20% DMSO pH 8.5; room temperature. Analytical LCMS conditions are described in the SI.
Folding the synthetic Ts3 polypeptide chains turned out to be extremely challenging. Conventional conditions that used a thiol-disulfide redox couple, together with a low concentration of the chaotrope Gu· HCl to keep misfolded/mispaired disulfide intermediates in solution, did not produce any discrete folded product. Nor did the folding conditions we had previously developed for Ts1.[12] Addition of 20% v/v DMSO to the redox buffer gave acceptable yields of discrete folded products, of mass 8 Da less than that of the corresponding polypeptide chains, indicating the formation of 4 disulfide bonds. After careful optimization, the final folding conditions used were: [polypeptide] 0.02 mg/mL; 2.0 mM GSH/ 1.0 mM GSSG; 0.5 M L-arginine· HCl; 1.0 mM EDTA; 0.1 M Tris, 20% DMSO, pH 8.5; room temperature. Data for the folding of Ts3(1-64)-CONH2 is shown in Figure 2b. Using these conditions, we were able to fold all four Ts3 candidate polypeptide chains, with concomitant formation of 4 disulfides in each case as shown by the loss of 8 Da. Analytical LCMS data for the four variant synthetic candidate Ts3 proteins are shown in Figure S13.
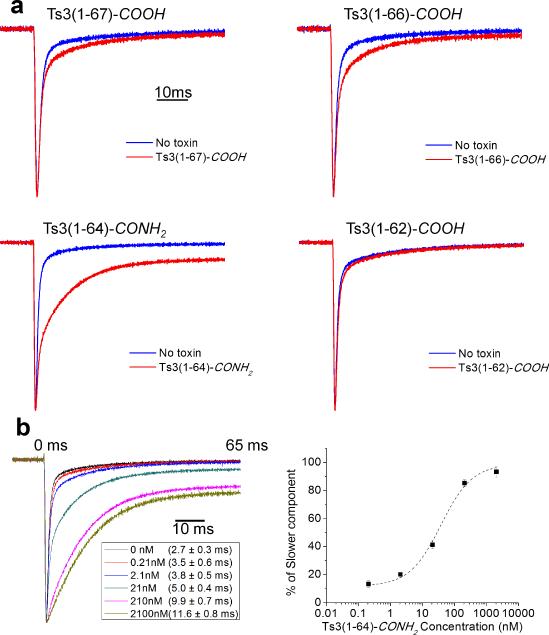
Structure-activity profiles of the four candidate Ts3 protein molecules were evaluated by the fast inactivation decay rate of sodium currents recorded from heterologously expressed at skeletal muscle voltage-gated sodium channel (rNav1.4) in the presence of 210 nM of each protein (5 min incubation at a holding potential of −90 mV) (Figure 3). Ts3(1-64)-CONH2 protein showed obvious disruption the rNav1.4 fast inactivation (Figure 3a), while the other three protein molecules showed minimal effects. on treatement with Ts3(1-64)-CONH2 protein, fast inactivation decay of rNav1.4 became slower in a dose-dependent manner (Figure 3b). The effect saturated at concentrations between 210-2100 nM, consistent with previous reports using Ts3 toxin isolated from scorpion venom.[8] Ts3(1-64)-CONH2 protein can thus be presumed to be the principal toxic component of natural Ts3 toxin isolated from scorpion venom.
Figure 3.
Biological activities of Ts3 protein candidates. a) Disruption of rNav1.4 fast inactivation compared among the 4 different candidate Ts3 proteins. Na+ ionic current, elicited by a step-pulse to −10 mV from a holding potential of −90 mV, was measured without (blue traces) and with (red traces) 210 nM of each candidate protein. b) Dose-response assay of rNav1.4 fast inactivation modification by Ts3(1-64)-CONH2 protein. The right panel shows the proportion of the slower component of the fit as a function of toxin concentration. Error bars indicate standard error of the mean (number of cells = 3-6 cells).
With the covalent structure of biologically active Ts3 protein in hand, we set out to determine its folded structure by X-ray crystallography. Crystallization of L-Ts3(1-64)-CONH2 toxin protein was problematic, so racemic protein crystallization was used.[14-16] Preparation of the D-Ts3(1-64)-CONH2 polypeptide chain and its folding were as described for the L-protein enantiomer. (Figures S14-16). Crystallization trials using the Hamption Index (HR2-144) screen and a racemic mixture at 10 mg/mL (5 mg/mL of L-Ts3(1-64)-CONH2 toxin plus 5 mg/mL of D-Ts3(1-64)-CONH2 protein) produced diffraction-quality crystals from a single condition [0.1 M BIS-TRIS pH 6.5, 28% w/v Polyethylene glycol monomethyl ether 2,000 at 19 °C] after three weeks.
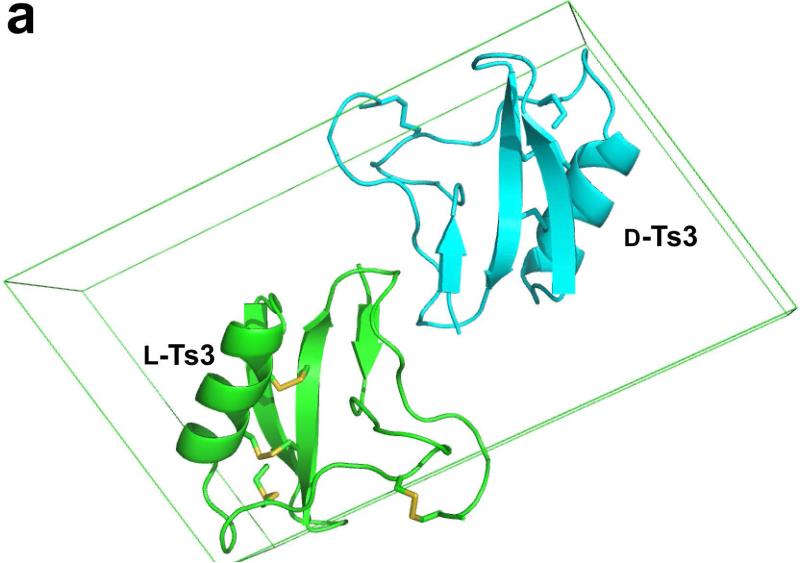
Diffraction data were collected from the racemic D/L-protein crystal at an energy level of 12.6 keV using the NE-CAT beamline 24-ID-E at the Advanced Photon Source at Argonne National Laboratory. Reasonably complete diffraction data were collected to a resolution of 1.93 Å. Diffraction intensity statistics revealed that the race mic D/L-protein crystallized in space group P1<bar>, with one enantiomer in the asymmetric Unit. The X-ray structure of racemic D/L-Ts3(1-64)-CONH2 protein was solved by molecular replacement[17] using the scorpion toxin LQH-alpha-IT (PDB ID: 2ASC) as a search model. The final model was refined to a crystallographic R-factor of 0.26 (R-free 0.30) using Phenix.[18] Note that this centrosymmetric R factor corresponds to a conventional R factor that is ~33% smaller (i.e. R-factor ~0.20).[19] The packing of D-Ts3(1-64)-CONH2 and L-Ts3(1-64)-CONH2 proteins in the unit cell is shown in Figure 4a, with the two enantiomeric molecules in the unit cell.
Figure 4.
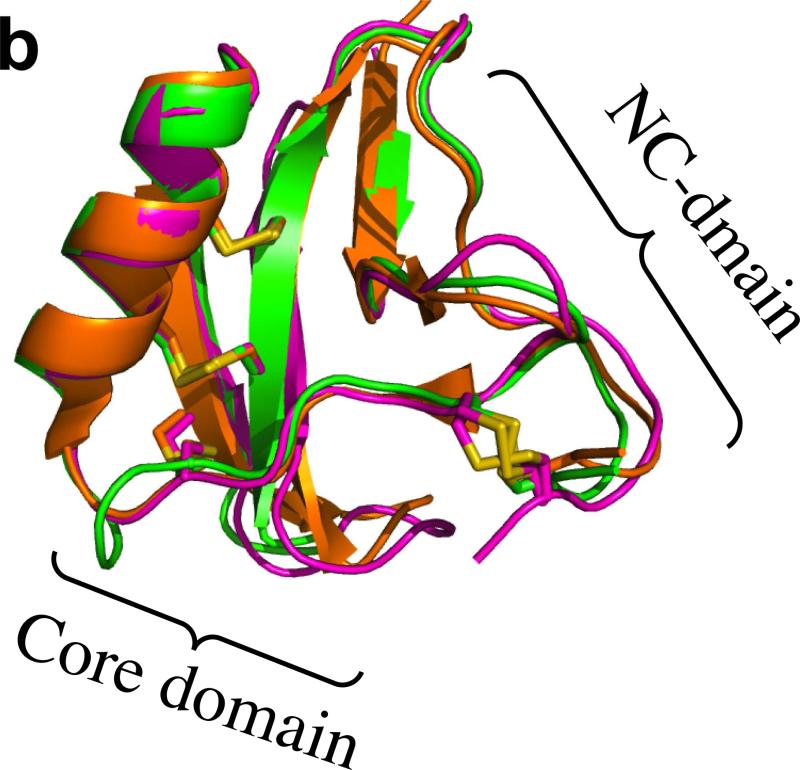
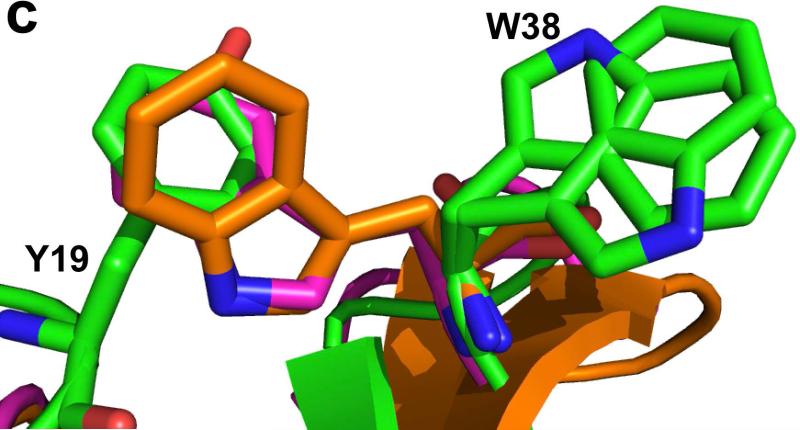
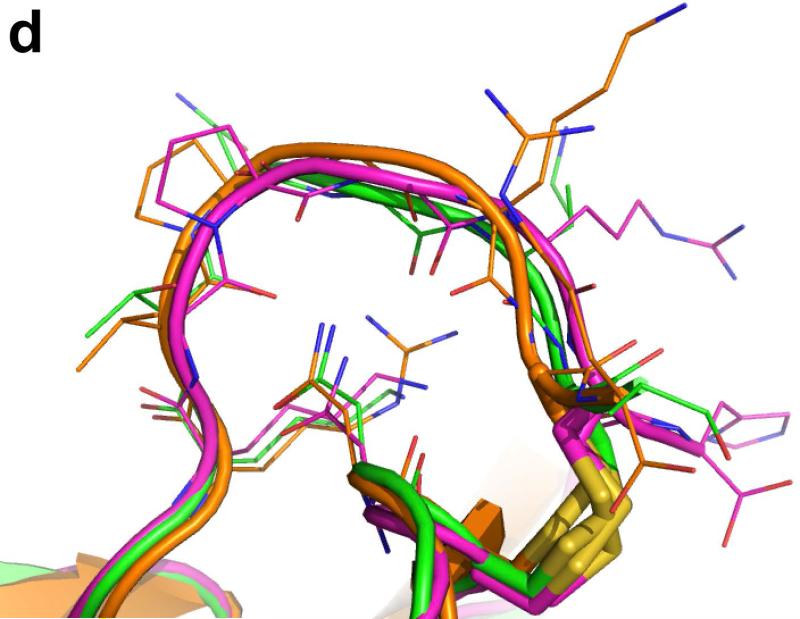
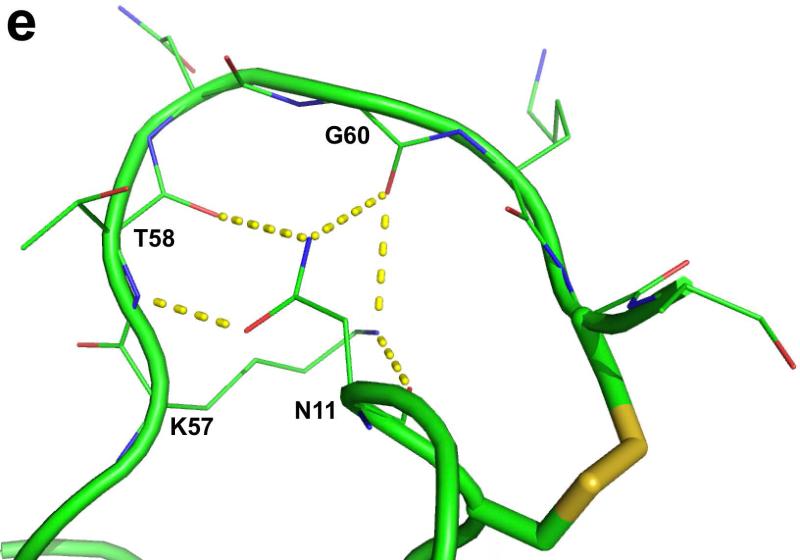
Crystal structure of Ts3(1-64)-CONH2 toxin protein. a) Crystal structre of the D/L racemate of Ts3(1-64)-CONH2 protein, (PDB 5CY0). Cartoon representation of the packing of d-Ts3(1-64)-CONH2 (Cyan) and l-Ts3(1-64)-CONH2 (Green) proteins in the unit cell in P1<bar>. b) Structure alignment of Ts3 toxin (green) and alpha scorpion toxin LQH-α-IT (golden) and Aah2 (magenta).[20] c) Core domain hydrophobic surface comparison between Ts3 and other alpha scorpion toxins. d) NC-domain comparison between Ts3 and other alpha scorpion toxins. e) Hydrogen bonding of Asn11 and Lys57 with residues in the C-terminal region of Ts3.
As shown by comparison with the structures of the prototypical alpha scorpion toxins LQH-α-IT and Aah2 (PDB ID: 1AHO) the Ts3(1-64)-CONH2 protein molecule has a typical alpha scorpion toxin fold made up of an α-helix (residues 21-31) packed against a three-stranded anti-parallel β-sheet (residues 2-4, 35-39, 44-50) (Figure 4b). Alpha scorpion toxins have two surfaces that interact with the Nav channel: a “Core domain”, formed by loop residues that connect the secondary structure elements, and an “NC-domain”, formed by a five-residue turn near the N-terminus (residues 8-12) and the C-terminal region.[21] In the Core domain of Ts3, the Tyr19 side chain was found to overlap with the side chain of the functionally important Trp38 of other alpha scorpion toxins (Figure 4c). The NC-domain of Ts3 is quite similar to the NC-domain of the other members of the alpha scorpion toxin family in terms of both structure (Figure 4d), and surface electrostatics. The C-terminal region of Ts3 and similar alpha-scorpion toxins is heavily populated with multiple positively charged residues. Side chains of conserved Asn11 and Lys57 of Ts3 form a hydrogen bonding network with the main chain amide at the C-terminus which, together with a disulfide bond at Cys62, holds the C-terminus in position for interacting with the sodium channel (Figure 4e).
A robust total synthesis of the Ts3 protein molecule using a modular synthetic strategy has enabled us to prepare the four suggested covalent structures for Ts3 toxin. Correlating biological activity measurements with the known structures of these synthetic proteins verified the covalent structure of biologically active Ts3 toxin. Using racemic protein crystallization, diffraction quality crystals of racemic Ts3 protein were obtained, and from the resulting X-ray diffraction data we determined the three-dimensional structure of biologically active Ts3 toxin. The covalent and tertiary structures of biologically active Ts3 toxin reported here constitute essential information for future work aimed at developing Ts3 protein as a biophysical probe of the Nav channel,[22] in order to complement and extend the information obtained using labeled Ts1 neurotoxin.[23]
Supplementary Material
Acknowledgements
This research was supported in part by funds from NIH Grants U54 GM087519, and R01-GM030376 to F. Bezanilla and by the American Heart Association (13POST14800031) to T. Kubota. Use of NE-CAT beamline 24-ID at the Advanced Photon Source is supported by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Bobo Dang, Department of Chemistry, Department of Biochemistry & Molecular Biology, Institute for Biophysical Dynamics, University of Chicago Chicago, IL 60637
Dr. Tomoya Kubota, Department of Biochemistry & Molecular Biology, University of Chicago, Chicago, IL 60637
Dr. Kalyaneswar Mandal, Department of Chemistry, Department of Biochemistry & Molecular Biology, Institute for Biophysical Dynamics, University of Chicago Chicago, IL 60637
Prof. Dr. Ana M. Correa, Department of Biochemistry & Molecular Biology, University of Chicago, Chicago, IL 60637
Prof. Dr. Francisco Bezanilla, Department of Biochemistry & Molecular Biology, Institute for Biophysical Dynamics, University of Chicago Chicago, IL 60637
Prof. Dr. Stephen B. H. Kent, Department of Chemistry, Department of Biochemistry & Molecular Biology, Institute for Biophysical Dynamics, University of Chicago Chicago, IL 60637.
References
- 1.Possani LD, Martin BM, Mochcamorales J, Svendsen I. Carlsberg Res. Commun. 1981;46:195. [Google Scholar]
- 2.Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. J. Biol. Chem. 1996;271:15950. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 3.Benzinger GR, Kyle JW, Blumenthal KM, Hanck DA. J. Biol. Chem. 1998;273:80. doi: 10.1074/jbc.273.1.80. [DOI] [PubMed] [Google Scholar]
- 4.Catterall WA. J. Biol. Chem. 1977;252:8660. [PubMed] [Google Scholar]
- 5.Couraud F, Rochat H, Lissitzky S. Biochem. Biophys. Res. Commun. 1978;83:1525. doi: 10.1016/0006-291x(78)91394-3. [DOI] [PubMed] [Google Scholar]
- 6.Kirsch GE, Skattebol A, Possani LD, Brown AM. J. Gen. Physiol. 1989;93:67. doi: 10.1085/jgp.93.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos FV, Coronas FI, Beirão PSL. Br. J. Pharmacol. 2004;142:1115. doi: 10.1038/sj.bjp.0705793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos FV, Chanda B, Beirao PS, Bezanilla F. J. Gen. Physiol. 2008;132:251. doi: 10.1085/jgp.200809995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cologna CT, Marcussi S, Giglio JR, Soares AM, Arantes EC. Protein Pept. Lett. 2009;16:920. doi: 10.2174/092986609788923329. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Eauclaire MF, Céard B, Ribeiro AM, Diniz CR, Rochat H, Bougis PE. FEBS Lett. 1994;342:181. doi: 10.1016/0014-5793(94)80496-6. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira CE, Ifa DR, Corso G, Santagada V, Caliendo G, Antunes E, De Nucci G. FASEB J. 2003;17:485. doi: 10.1096/fj.02-0635fje. [DOI] [PubMed] [Google Scholar]
- 12.Dang B, Kubota T, Correa AM, Bezanilla F, Kent SBH. Angew. Chem. Int. Ed. Engl. 2014;53:8970–8974. doi: 10.1002/anie.201404438. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:9116. [Google Scholar]
- 13.Saucedo AL, Flores-Solis D, Rodríguez de la Vega RC, Ramírez-Cordero B, Hernández-López R, Cano-Sánchez P, Noriega Navarro R, García-Valdés J, Coronas-Valderrama F, de Roodt A, Brieba LG, Domingos Possani L, del Río-Portilla F. J. Biol. Chem. 2012;287:12321. doi: 10.1074/jbc.M111.329607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SBH. J. Am. Chem. Soc. 2008;130:9695. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang B, Kubota T, Mandal K, Bezanilla F, Kent SBH. J. Am. Chem. Soc. 2013;135:11911. doi: 10.1021/ja4046795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeates TO, Kent SBH. Annu. Rev. Biophys. 2012;41:41. doi: 10.1146/annurev-biophys-050511-102333. [DOI] [PubMed] [Google Scholar]
- 17.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J. Appl. Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Acta Crystallogr. D. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luzzati PV. Acta Crystllogr. 1952;5:802. [Google Scholar]
- 20.Smith GD, Blessing RH, Ealick SE, Fontecilla-Camps JC, Hauptman HA, Housset D, Langs DA, Miller R. Acta Crystallogr. D. 1997;53:551. doi: 10.1107/S0907444997005386. [DOI] [PubMed] [Google Scholar]
- 21.Karbat I, Frolow F, Froy O, Gilles N, Cohen L, Turkov M, Gordon D, Gurevitz M. J. Biol. Chem. 2004;279:31679. doi: 10.1074/jbc.M402048200. [DOI] [PubMed] [Google Scholar]
- 22.Selvin PR. Annu. Rev. Biophys. Biomol. Struct. 2002;31:275. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 23.Kubota T, Brugarolas P, Dang B, Finolurdaneta RK, Frezza L, French RJ, Kent SBH, Bezanilla F, Correa AM. Biophysical Journal. 2014;106:133a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.