Abstract
Blood coagulation and platelet adhesion remain major impediments to the use of biomaterials in implantable medical devices. There is still significant controversy and question in the field regarding the role that surfaces play in this process. This manuscript addresses this topic area and reports on state of the art in the field. Particular emphasis is placed on the subject of surface engineering and surface measurements that allow for control and observation of surface-mediated biological responses in blood and test solutions. Appropriate use of surface texturing and chemical patterning methodologies allow for reduction of both blood coagulation and platelet adhesion, and new methods of surface interrogation at high resolution allow for measurement of the relevant biological factors.
1. Introduction
Biomaterials have been routinely used in all fields of medicine and surgery for a range of medical applications from short term dressing to long term implants, and play a critical role in the treatment of disease and the improvement of health care1,2. Despite the wide use of biomaterials in clinical environments, the biocompatibility of the materials is still far from ideal and a variety of adverse reactions such as inflammation, fibrosis, infection, and thrombosis, may be triggered 3–5. The biocompatibility of implants and successful application of these medical devices are largely dependent on the biological events occurring at the surfaces. Probing these specific and direct interactions between biomaterials and tissue components will markedly improve the field’s understanding of the mechanisms of biocompatibility and thereby to improve the development of new biomaterials 6.
Blood is often the first body fluid that comes into contact with blood-contacting devices. Blood-material interactions trigger a complex series of events including protein adsorption, platelet adhesion and activation, coagulation, and thrombosis. Rapid adsorption of plasma proteins is the first event occurring on biomaterial surface during blood/material interactions, and leads to activated, adsorbed proteins that can catalyze, mediate, or moderate the subsequent biological responses to biomaterials 7–9. Surface-induced thrombosis is the main problem impeding development of long-term blood contacting devices 10,11. Thrombus formation on device surfaces is partially due to platelet-mediated reactions12 and partially due to coagulation of blood plasma13.
Biological responses are a complex process governed by many factors, but it is widely accepted that surface properties of a biomaterial dictate the biological response. Surface properties such as chemistry, topography, surface free energy, elasticity, and charge may moderate protein and cell interactions, and ultimately the host response. For example, platelet adhesion and activation on biomaterial surfaces is influenced by surface properties such as energy14, charge15, and composition16. However, platelet adhesion and activation are largely mediated by proteins such as fibrinogen 17,18. As plasma proteins can rapidly adsorb onto the material surface to form a “conditioning film” following blood contact, this adsorbed protein layer may minimize the direct effect of biomaterial surface properties on cell responses, and the surface-biology interactions are mediated by proteins (Figure 1).
Figure 1.
The interaction between biomaterials and biological entities at the interface is influenced by surface property and proteins.
The aim of this work is to outline the current understanding of the phenomena of thrombosis with particular emphasis placed on cardiovascular biomaterials. As surface chemistry and topography of a biomaterial are known to be the important parameters that influence the biological responses to surfaces, this manuscript focuses on the protein adsorption, platelet adhesion, and blood coagulation at biomaterial interfaces with different surface properties.
2. Protein adsorption and platelet adhesion on biomaterial surfaces
2.1 Plasma proteins and platelet adhesion
When a biomaterial comes into contact with blood, a layer of plasma proteins typically adsorbs to the surface and mediate biological responses to biomaterials. Blood contains many hundreds of proteins with a wide range of biological functions and activity, and present in vastly different concentrations. Albumin, immunoglobulins and fibrinogen are the most abundant proteins in plasma and represent more than 50% of all plasma proteins. In considering surface-induced thrombosis, albumin is generally considered to be inert towards platelet adhesion and activation 19, while fibrinogen is a central protein in the process of biomaterial-induced thrombosis 20–22. Other plasma proteins such as fibronectin23,24, vitronectin25,26, and von Willebrand factor (vWF)27,28, have been shown to be capable of mediating platelet adhesion to materials when pre-adsorbed to the surface. These adsorbed proteins can bind to platelets via cell membrane receptors such as GPIIb/IIIa (known as integrin αIIbβ3), GPIb, and other receptors, and such binding will induce platelet adhesion and aggregation. Adherent platelets become activated and mediate clotting events such as platelet aggregation and formation of thrombus.
The ability of proteins to promote platelet adhesion varies with substrates and flow condition. Fibrinogen has been identified as the major protein mediating platelet adhesion biomaterials at low shear stress29 and plays a prominent role in development of surface-induced thrombosis due to its multi-functional role serving as a ligand for platelet adhesion, linking platelet aggregates together, and stabilizing thrombi as fibrin polymer20,30,31. vWF circulates in plasma as a series of heterogeneous multimers, mediating platelet tethering, translocation and finally adhesion to areas of injured endothelium under higher shear stress conditions. Immobilization and shear induce conformational changes in vWF, and enhance the binding of vWF to glycoprotein Ib alpha on platelet cell membrane, resulting in platelet adhesion 32,33. Fibronectin plays little or no role in mediating platelet adhesion on material surfaces pre-adsorbed with normal plasma or fibrinogen, but it does support platelet adhesion in fibrinogen-depleted plasma34. Vitronectin is a multifunctional 75-kD glycoprotein that is present in plasma and the extracellular matrix, and functions as a complement regulatory promoting growth and attachment of cells. Vitronectin can bind to platelets via membrane protein GPIIbIIIa, and plays a role in platelet adhesion and formation of stable platelet aggregates 35,36.
2.2 Surface properties influence protein adsorption
A number of different factors have been shown to influence the composition and function of the adsorbed proteins on implanted biomaterials37. Among these are the solution concentration of the proteins, the properties of the surface and the intrinsic “surface activity” of the protein38–40. Other studies show time dependent factors involved in protein adsorption and function on biomaterials41–44. A fundamental premise in the field of biomaterials is that the surface properties of a biomaterial affect the adsorption, the structure and the function of proteins 45,46. Surface properties that have been shown to be important (1) surface free energy and wettability, (2) surface chemistry and functional group, and (3) surface topography and roughness 37,47,48.
2.2.1 Surface energy and wettability
The introduction of a biomaterial surface in blood creates a new interface between cellular and fluid components of blood and the material. The thermodynamic quantity of surface free energy of both solid and liquid phases leads to the interfacial free energy at the interface, which can be used to assess the biocompatibility of biomaterials49. In the case of blood plasma-biomaterial interface, it has been suggested that the interfacial free energy should be in the order of 1–3 dyne/cm for long-term compatibility with blood50. Protein adsorption is an energy-driven process that involves the properties of both the substrate surface and the proteins themselves. Calculation of the interfacial energy of protein interaction with surface from the surface free energy of a biomaterial is helpful to quantitatively understand the mechanisms of protein adsorption on varied surfaces51–53. It has been reported that there is a correlation between the amount of protein adsorbed on the polymeric materials such as poly(methylmethacrylate) (PMMA), polystyrene (PS), and poly(dimethylsiloxane) (PDMS) and the total surface energy of the substrate, i.e., adsorption is higher on low energy substrates54.
Surface wettability driven by surface energy is a key factor in determining adsorption kinetics and the amount of proteins adsorbed on the surface55,56. Wettability can be characterized by the Dupré work of water adhesion to surfaces, W =γLV (1+cosθ), where the interfacial energy of water γLV = 71.97 mJ/m2 at 25°C 57,58. Generally water contact angle θ is measured and represented by the water adhesion tension τ, (τ =γLV cosθ), to estimate the surface wettability (hydrophilic or hydrophobic). Vogler et al. defined the terms “hydrophilic” and “hydrophobic” from experimental observation, with hydrophilic surfaces exhibiting advancing water contact angle < 65°, and hydrophobic surfaces exhbiting water contact angle > 65° 59,60. A more precise definition of the relative terms hydrophobic and hydrophilic for use in biomaterials was further derived from calculation of wetting energetics, where hydrophilic surface wetting expends >1.3 kJ/mole-of-surface-sites, and hydrophobic surface wetting expends <1.3 kJ/mole-of- surface-sites. Considering the hydrogen bond of surrounding water involved in surface wetting, hydrophilic surfaces wet with >1 hydrogen bond per water molecule whereas hydrophobic surfaces wet with <1 hydrogen bond per water molecule 61.
In general, hydrophobic surfaces tend to adsorb larger amount of proteins than hydrophilic62 as the hydrophilic surface is strongly bounded with water molecules that are difficult to be displaced by adsorbing proteins, while protein readily displaces water from the hydrophobic surface to become adsorbed51. The above experimental and theoretical observations of surface hydrophilic/hydrophobic transiting at θ = 65° may also explain the sharp changes in the protein adsorption to material surfaces with water contact angles centered around 65° in diverse circumstances53,63–65. Cha et al. measured the difference in the amount of protein adsorbed at solid-vapor and solid-liquid interfaces and found it decreased with the adhesion tension τ from maximal values measured on the methyl-terminated self-assembled monolayers surface through zero (no protein adsorption in excess of bulk solution concentration) near τ = 30 mN/m (θ = 65°)53. Other independent experiments show that the protein adsorption capacity decreases monotonically with increasing adsorbent hydrophilicity to a limit of detection near τ = 30 mN/m (θ = 65°) for all protein/surface combinations studied 64,65. All these results suggest that the structured interfacial water (with thickness ~ 2–3 nm) plays an important role in protein adsorption66.
Protein adsorption on surfaces can be understood from the aspect of interfacial energetics and measurement of adsorbed amount, but also from the direct measurement of the strength of the molecular interaction between protein and the material surface. The force measurement techniques of an atomic force microscopy (AFM) using a protein-modified probe tip have been powerful tools to analyze the actual interaction forces between protein and material surfaces67. Since the normal AFM tip is very sharp with radii of end in the range of 10–20 nm, force measurement using a protein-modified model probe is generally regarded as individual proteins interacting with surface. Such characteristics of individual proteins in interaction with surface offer much insight into the environment presented by biomaterial interfaces.
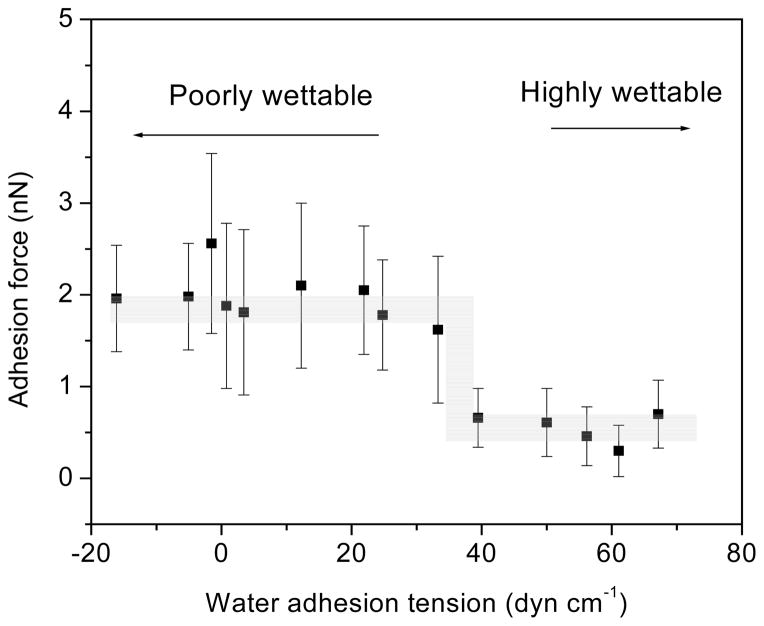
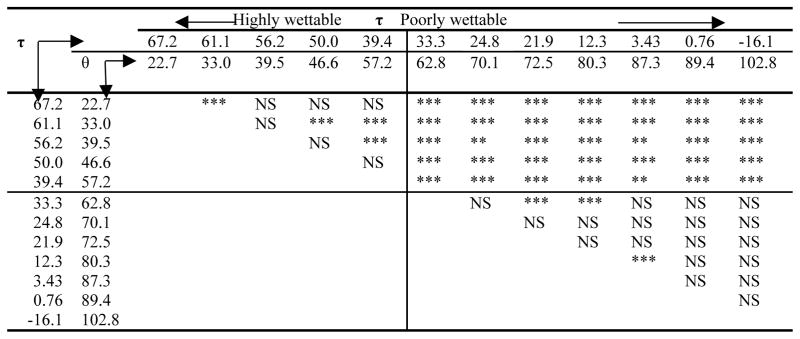
Table 1 shows the adhesion forces between model self-assemble monolayer (SAM) surfaces and plasma proteins albumin, coagulation factor XII, and coagulation factor XIIa. (FXII and FXIIa are involved in contact activation of the coagulation cascade). All proteins exhibit strong adhesion to the hydrophobic methyl-terminated octadecyltrichlorsilane (OTS) surface and weak adhesion to water-wettable glass surfaces, despite FXII supposedly being uniquely attracted to glass surfaces. The force measurements were extended to look at a range of water wettabilities in a polymer by treating low density polyethylene (LDPE) in a glow-discharge plasma68. The contact angle and adhesion forces between albumin, FXII, and fibrinogen modified tips with the substrate were measured. All of the proteins studied exhibited a step increase in adhesion force as the contact angles of the surface increased above θ~60° (τ < 36.4 dyn/cm). The step occurred within a narrow range of wettabilities over ~ 60–65°. Figure 2 illustrates the trend of adhesion force of fibrinogen interaction with LDPE surface. ANOVA shows that adhesive forces between the poorly wettable surfaces are not different from each other, adhesive forces between the wettable surfaces are not different from each other, but there are substantial differences between the two groups, as illustrated in Table 2. The presence of the step increase in adhesion forces was also observed in interactions of proteins and a series of SAM-surfaces. For SAMs with θ < 66° (-OH, -CONH2, and -EG3OH), weak adhesion was observed, while for 66° < θ < 104° (-CH3, -OPh, -CF3, -CN, -OCH3), strong adhesion was observed and increased with the molecular weight of the proteins69. Results suggested that the effect of surface wettability on protein adhesion is actually quite straightforward and that subtle changes in wettability not affect protein adhesion unless that change yields a transition across this θ=60–65° region68.
Table 1.
Contact angle and adhesive force values for protein modified AFM probes
| Surface | OTS | APTES | Glass | |
|---|---|---|---|---|
| Advancing Water Contact Angle (°) | 110 | 59 | <10 | |
| Adhesion force (nN) (mean ± std. dev.) | Standard Si3N4 AFM Probe | 6.5 ± 4.8 | 1.3 ± 1.1 | 0.8 ± 0.2 |
| FXII-Modified AFM Probe | 4.4 ± 2.1 | 2.0 ± 0.9 | 0.5 ± 0.6 | |
| FXIIa-Modified AFM Probe | 3.4 ± 1.1 | 0.5 ± 0.4 | 0.5 ± 0.4 | |
| Albumin-Modified AFM Probe | 5.6 ± 4.2 | 1.1 ± 0.7 | 0.5 ± 0.4 | |
Figure 2.
Average adhesion forces of human fibrinogen coated AFM tips to LDPE surfaces with different water adhesion tensions. (error bar: standard deviation) (Reprint from Xu and Siedlecki68 with permission from Elsevier)
Table 2.
ANOVA analysis of adhesion forces for fibrinogen and LDPE surfaces with different water adhesion tensions (τ, dyn.cm−1) and water contact angle (θ, degree). τ =γLV cosθ (Reprint from Xu and Siedlecki68 with permission from Elsevier).
NS : Not significant
: significant p <0.001
: significant p<0.01
: significant p<0.05
2.2.2 Surface chemical structure and functional group
Surface chemistry (elemental constituents and functional group) gives rise to surface properties such as wettability and charge, and influences the extent of protein adsorption, denaturation, and functional activity. It plays an important role in biocompatibility of implants8. To reduce unwanted-protein adsorption on surfaces, intensive research efforts has gone towards engineering surfaces with functional groups to repel proteins. Ostuni et al. described that typical characteristics of protein-resistant surface should be hydrophilic, neutral charge, containing hydrogen-bond acceptors, but without hydrogen-bond donors70. Although there appears to be exceptions, it is believed that the identification of protein resistant chemical structure and/or functional group of surface can help to control protein adsorption and biological events following adsorption, and thus enhance the biocompatibility of biomaterials8.
Surface chemical structure and functional groups affect the extent of protein adsorption, denaturation, and epitope exposure8,71–74. Compared to the complexity of polymer surface chemistry, SAMs have precisely controlled density and conformation of single or multiple functional groups, and can be easily prepared with different terminal groups. Therefore, SAMs applied on model surfaces are used to study surface chemistry effects on protein adsorption and biological responses75,76. Hydrophobic functionalized surfaces tend towards more protein adsorption than the hydrophilic surfaces77. Aminated surfaces are able to form hydrogen bonds with proteins while negative charged groups may react less with proteins compared to other surface71,74. Kidoaki and Matsuda78 measured the adhesion forces of blood plasma proteins and above SAMs modified surfaces by AFM and revealed that the hydrophobic methyl functional group exhibited the highest forces for all measured proteins (fibrinogen, albumin, and IgG),while carboxyl functional group was lowest, with fibrinogen exhibiting higher adhesion than albumin and IgG. SAMs also allow production of surfaces containing different functional groups with varying for studies of protein adsorption and biological responses79,80. For example, binary mixing of alkanethiol SAMs terminated with -COOH and one of the end groups –CH3, -OH or -NH2 on gold surfaces influences the surface charge, and alters the secondary structure of covalently bound fibronectin81. Mixing longer chain methyl- and shorter chain hydroxyl-terminated alkanethiols at different ratios produced a series of surfaces containing different percentages of components. Fibrinogen adsorption, platelet adhesion and activation were found to decrease with increasing –OH groups (increasing hydrophilicity), and there is good correlation between fibrinogen adsorption and platelet adhesion 14.
In contrast to SAMs, surface polymerization can be used to create polymer brushes, where an array of macromolecular chains (polymer) are bonded to a substrate surface at one end, while in an extended conformation to minimize segment–segment overlaps82. The polymer chains that extend to the grafting surface can exhibit properties distinctly different from chains in solution so that the substrate surface grafted such polymers can possess novel properties, and have been applied to many areas including medical applications83,84. One of most effective polymer brushes is poly(ethyleneoxide) (PEO) or poly(ethyleneglycol) (PEG), which are considered to possess excellent blood compatibility. PEG is widely used in the production of biomaterials because of a resistance to protein adsorption, weak immunogenicity, and good compatibility with living cells 85. In vitro and in vivo experiments show that PEG coatings on PVC drain tubes significantly suppressed plasma protein adsorption and platelet adhesion, leading to reduced risk of thrombus formation, tissue damage, and cytotoxic effects, suggesting PEG grafted surfaces have great potential for clinical applications86. Protein resistance of PEG is related to grafting density, chain length, molecular weight, protein size, polymer-protein interactions and protein-surface interactions87–91. Generally, it is believed that PEG protein-resistance arises from steric repulsion, chain mobility, and excluded volume effects92–95. Jin et al. 96 recently described the protein adsorption mechanism on PEG-grafted polyethylene surface, stating that dense PEG brush can release more trapped water molecules to resist protein adsorption. However, other responses to PEG-modified materials still remain unclear. It should be noted that unanticipated responses such as hypersensitivity reactions caused by PEG-modified surfaces have been reported. There is also evidence that PEG surface may activate the complement system involved in the foreign body reaction to these biomaterials97,98 so it is not clear that resisting protein adsorption does not mean that protein interactions cannot still occur.
2.2.3 Surface topography and roughness
Surface topography and roughness are important factors in determining the biological responses to a foreign material. Numerous studies have demonstrated that micro- and nanoscale structured surfaces influence cell behaviors99–101 such as orientation102, morphology103, adhesion104, proliferation105, function regulation106, and gene expression107. Topographical surfaces also influence adhesion of bacteria108,109 and platelets110,111, and may affect protein adsorption112. The engineering of surfaces with well-defined micro- and nanoscale topographies has become relevant as a strategy to control biological responses113,114.
There is evidence that surface topography might influence the amount of protein adsorbed on surface and alter the adsorption ratio spatial distribution, conformation, and surface binding affinity of different proteins114. As the dimensions of proteins are the nanometer range, nanoscale topographies are thought to influence proteins, whereas micrometer sized topographic surfaces would appear smooth at the protein scale, minimizing the effect of these topographies on protein behavior. However, there is some evidence that even micro-sized topographies can influence protein interactions with surfaces, and greater amounts of proteins were found on surfaces bearing micrometer scale roughness112,115. In fact, the adsorption of proteins on topographical surfaces appears to be a result of complex processes influenced by surface wettability, surface chemistry, and surface topography, including dimension and shape of features and roughness. However, when assessing the field as a whole, there appears to be no general trend in the amount of protein adsorbed onto topographically-defined surfaces, even when analyzing just a single type of protein.
Surface texturing affects the surface wettability and surface contact area without changing the bulk properties of the material or the surface chemistry. Surface texturing of hydrophobic materials can increase the surface hydrophobicity as evidenced by water contact angles up to >150°, termed superhydrophobic, due to enhancing water repellency by trapped air between structures, whereas hydrophilic materials can be made more wettable with texturing (so-called superhydrophilic) due to the increased contact area of the liquid with surface114. Superhydrophobic or superhydrophilic surfaces prepared by addition of surface topography can decrease or increase protein adsorption on surfaces depending on size, shape, and composition of surface feature, and type of proteins. Interactions of proteins or cells with surfaces exhibiting extreme wettability have been reviewed elsewhere116.
The size of topographical feature on hydrophobic surfaces affects protein adsorption. It has been reported that superhydrophobic material surfaces bearing nanoscale topography suppress protein adsorption, while surfaces having micrometer-sized roughness increase protein adsorption112,117. Koh et al.117 suggested that topographical parameters for reducing protein fibrinogen adsorption and platelet responses would be high aspect ratio (>3:1) with reduced interspacing (<200nm) or high density. This possible mechanism of reduction in protein adsorption is that nanoscale topography limits the contact of the protein solution to the top of the structures, reducing the surface area for protein adsorption due to the Cassie-Baxter effect, with liquid sitting on top of the rough surface and air in the hollows112. Indeed, the trapped air between the structures plays an important role in reducing protein adsorption on superhydrophobic structured surfaces. Leibner et al.118 showed that air trapped within the interstices of fibular structure of ePTFE used in vascular grafts prevented intimate contact with protein solutions. Moreover, air trapped in nanoscale topography blocks the interactions of proteins with surface, and lower protein adsorption. For TiO2 nanotube surfaces bearing either superhydrophobic or superhydrophilic microtemplated domains, less protein was found on the superhydrophobic domains in the presence of trapped air, and more was bound to superhydrophilic domains with trapped air. However, more proteins were found on superhydrophobic domains than hydrophilic domains after the air was removed by ultra-sonication119. These superhydrophobic nanoscale surfaces reduce protein adsorption, but also promote desorption of proteins under flow conditions112. In fact, almost all proteins could be removed from some nanoscale surfaces, which might be very useful in microfluidic devices116.
Increasing amounts of protein are found on textured surfaces with micro- or nano-sized features. Liu120 fabricated a three-dimensional microfluidic chip for DNA and protein analysis using the lotus leaf as a template, and found the device to have 5 times higher sensitivity than flat surface after the superhydrophobic surface was changed to hydrophilic by a simple oxygen plasma treatment. This increase in protein uptake is due to the increase in surface area by topography. Luong-Van et al.114 reported that protein uptake linearly increased with surface area when textured surface structures were wetted by protein solution. However, it is not always sufficient to explain observed increase in protein adsorption strictly by surface area. Chen et al.121 fabricated dot-like patterns in PDMS with protrusion diameter bottom of 12 μm, and intervals of 7.5 μm between, and found that adsorbed fibrinogen increased 46% but surface area increased only about 8%, suggesting that the surface area factor was not solely responsible for increases in protein adsorption. Increases in protein adsorption are also found with nanoscale roughness. Rechendorff et al.122 found fibrinogen adsorption on tantalum films increased with increasing root-mean-square roughness (2.0 to 32.9 nm) beyond the accompanying increase in surface area. Vlachopoulou et al.115 observed that BSA adsorption was enhanced up to 10 times on high-aspect-ratio plasma-induced nanotextured PDMS surface. Scopelliti et al.123 quantified the protein adsorption on titanium with surface roughness ranging from 14 to 32 nm, and found fibrinogen and BSA adsorption 500% higher than the theoretical monolayer coverage on rough surfaces. The suggested possible mechanism for these findings is that surface nanoscale pores promote protein nucleation and aggregation so that proteins are trapped in pores, contributing to protein multilayer formation.
2.3 Fibrinogen adsorption and functional activity and platelet adhesion
Fibrinogen is a central protein in the process of biomaterial-induced thrombosis, promoting thrombus formation through binding of the platelet integrin receptor αIIbβ3 (GPIIb/IIIa), leading to platelet immobilization, activation and aggregation124, and also being the precursor to fibrin, the predominant structural component in blood clotting/coagulation. Fibrinogen is a symmetric molecule, composed of two sets of three intertwined polypeptide chains termed the Aα, Bβ and γ chains, with a total molecular weight of 340 kDa. The six chains fold together in a trinodular structure with a center globular knot (termed the E domain) containing portions of all six polypeptide chains, and flanked on each side by a larger globular structure (the D domain) containing one of the sets of three polypeptide chains. Each fibrinogen molecule possesses three pairs of potential platelet-binding peptide sequences, two RGD sequences in each of the Aα chains (RGDF and RGDS) and a dodecapeptide sequence (HHLGGAKQAGDV) in each of the γ chains; the γ chain dodecapeptide sequence being the primary ligand for platelet adhesion to adsorbed fibrinogen 22,125. It is now becoming accepted that the conformation and availability of platelet-binding sites in fibrinogen are more important in mediating platelet adhesion than is the amount of adsorbed protein 126,127. The discussion of conformational structure and functional activity of adsorbed proteins in this manuscript is mainly based on AFM measurement. This is not to discount the other techniques and methods have been used to study the conformation of proteins adsorbed on biomaterial surface, such as time-of-flight secondary ion mass spectrometry with principal component analysis128, and improved circular dichroism methods129.
2.3.1 Time-dependent conformational change and energy profiles involved in fibrinogen adsorption on surfaces
Fibrinogen undergoes conformational changes following adsorption to biomaterial substrates41,130,131. Conformational changes are both time- and surface-dependent. It was observed that structural deformation (spreading) of fibrinogen increased with material surface hydrophobicity; quantitative analysis of the different domains showed that the overall molecular length and widths of the individual D and E domains increased while the heights decreased when the material surface is more hydrophobic132. The time dependence of these changes in fibrinogen upon adsorption was directly observed in our lab133.
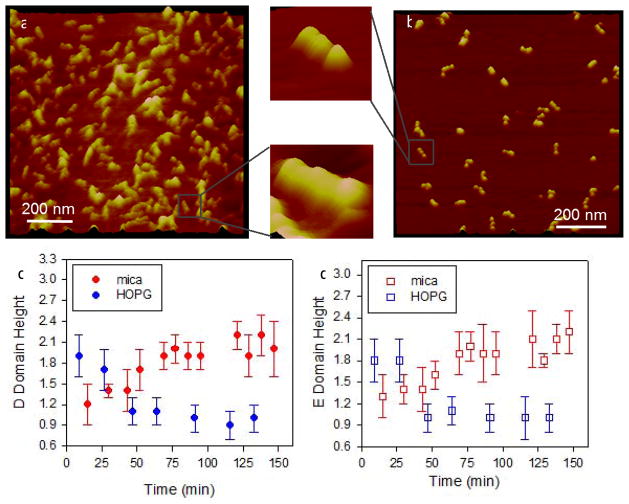
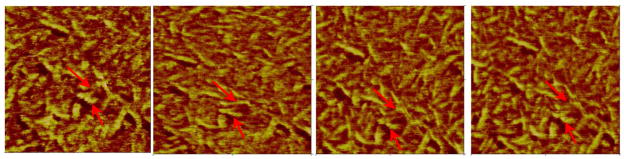
Figure 3 illustrates the time-dependent conformational changes in fibrinogen on a hydrophobic highly ordered pyrolytic graphite (HOPG) material (Fig. 3a) and a hydrophilic muscovite mica substrate (Fig. 3b). Fibrinogen was continuously imaged by AFM for ~ 2 hours. Spreading curves were generated by plotting heights of D and E domains of individual fibrinogen molecules. On HOPG, a clear decrease in height was observed with time, indicating protein spreading (Fig. 3c). On mica, the height of the D and E domains increased with time (up to 60 min) eventually reaching a plateau height of 2.1 nm (Fig. 3d). Together with other studies, these data demonstrate that proteins undergo conformational changes following contact with material surfaces and these changes are dependent on the properties of the underlying substrate.
Figure 3.
Time dependent spreading of fibrinogen on model substrates, (a) HOPG and (b) muscovite mica. Individual molecules could clearly be observed as seen in the higher magnification images. Spreading of the molecules was followed by measuring heights of the (c) D domains and (d) E domains for ~ 2 hrs following adsorption. (error bar: standard deviation) (Reproduced from Agnihotri and Siedlecki133 with permission from ACS)
Time-dependent unfolding of protein on surface causes variations in surface-protein interaction. AFM force mode makes it possible to measure the interaction strength of protein-surface interactions by bringing a protein into contact with a material surface and then separating. Assessing the effects of contact times on protein adhesion forces reveals information about the unfolding energy involved in protein-surface interactions. Low-density polyethylene (LDPE) was modified by glow discharge plasma to yield surfaces spanning a range of water wettability. Adhesion forces between protein and substrate increased with increasing protein contact time up to 50sec, and with smaller adhesion forces measured on wettable surfaces. Changes in adhesion forces with contact time were modeled by a simple exponential equation and energy barriers for protein unfolding were calculated with the Arrhenius equation134. The analysis yielded activation energies of 18.0–22.6 kT for human fibrinogen on hydrophobic surfaces, and 18.6–23.2 kT for hydrophilic surfaces68. Based on the observations of changes in adhesion force at early time points and the changes in height of D and E domains in fibrinogen at ~2 hours, a two-step spreading model was proposed, with the first step being very rapid (on the order of seconds to minutes) and involving rearrangement of protein surface amino acids, and the second step taking much longer and involving rearrangement of the internal amino acids to the hydrophobic surface68.
The energy profiles in interactions of protein/surface can be further explored through examining the interaction forces between protein and surface as a function of the loading rate using AFM measurement, where loading rate (rf) is defined as the product of probe retraction velocity and spring constant of the cantilever. In measuring the strength of bonds between biological receptor molecules and their ligands, the bond strengths measured under a variety of loading rates are governed by the barriers in the energy landscape along the force-driven, bond dissociation pathway135–137. Using the Bell model138, dissociation rates and energy barriers in biological molecule reactions can be quantified. These measurements can be extended to probe protein-surface interactions and reveal the energy profiles in interactions of fibrinogen with surfaces139.
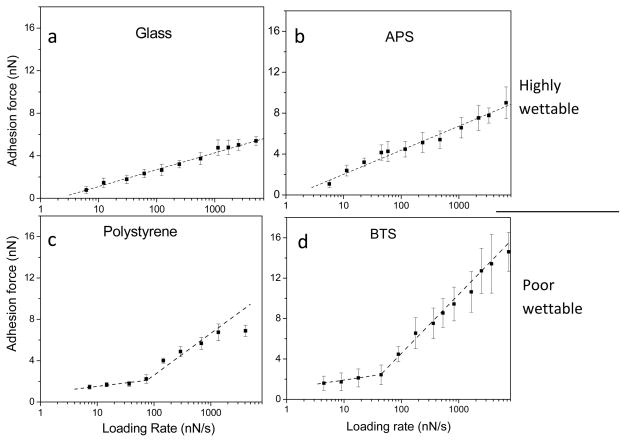
Dynamic forces were measured between colloid materials modified with SAMs of different wettability and fibrinogen. Figure 4 illustrates the adhesive forces between fibrinogen and these model surfaces as a function of loading rate. The linear relationship between adhesion forces and the ln(rf) for wettable surfaces (bare glass and 3-aminopropyltrichlorosilane (APS)-coated) suggests a single energy barrier for interactions of fibrinogen with these hydrophilic surfaces (Figs. 4a and b), while two line segments with differing slopes between hydrophobic surfaces and fibrinogen (Figs. 4c and d) suggests that there are multiple energy barriers and multiple transition states during interactions of fibrinogen with hydrophobic surfaces139. Dynamic force spectroscopy analysis was performed to fit the plots of adhesion force against ln(rf) by following equation:
where k is the Boltzmans constant, T is the absolute temperature, koff is the thermal off-rate at zero force, and χβ is the effective distance (Å) between the bound and transition states along the direction of applied force that measures the width of the energy well that traps the interacting molecules in the bond state. The results show that the off-rate (koff) for protein interactions with hydrophilic surfaces lie in the range of 1.35–2.92 s−1, about 3–10 fold greater than those for protein interactions with hydrophobic surfaces at the lower loading rates (Table 3). The bond lifetimes on the hydrophobic surfaces are longer than they are on hydrophilic surfaces, consistent with increased adhesion forces observed between proteins and hydrophobic surfaces. This analysis of dynamic forces reveals energy profiles in interactions of protein and surfaces, providing insights into the role of surface wettability on protein interaction with biomaterials.
Figure 4.
Adhesion forces of fibrinogen and colloid surfaces under different loading rates. High wettable surfaces: (a) glass and (b) 3-aminopropyltrichlorosilane (APS), and poorly wettable surfaces: (c) polystyrene and (d) n-butyltrichlorosilane (BTS). Multiple energy barriers are present between fibrinogen and hydrophobic surfaces while a single energy barrier is present between fibrinogen and hydrophilic surfaces. (error bar: standard deviation) (Reprint from Xu and Siedlecki139 with permission from ACS)
Table 3.
Parameters for dynamic model of fibrinogen interaction with surface. (Reprint from Xu and Siedlecki139 with permission from ACS)
| Wettability | Surface | Water contact angle (°) | Loading rate (nN/s) | χβ | koff (s−1) | τ (s) | |
|---|---|---|---|---|---|---|---|
| Highly wettable |
|
Glass | 11.3±1.7 | 6–5100 | 0.06 | 2.92 | 0.34 |
| Polystyrene (plasma treated) | 22.3±1.6 | 6–3100 | 0.02 | 1.99 | 0.50 | ||
| APS | 57.2±1.7 | 5–6500 | 0.04 | 1.35 | 0.74 | ||
|
| |||||||
| Poorly wettable |
|
Polystyrene | 82.2±1.5 | 6–100 100–4100 |
0.13 0.04 |
0.23 4.98 |
4.35 0.20 |
| BTS | 90.4±1.2 | 4–100 100–7400 |
0.11 0.02 |
0.19 5.50 |
5.12 0.18 |
||
| OTS | 103.0±1.9 | 5–100 100–3200 |
0.07 0.01 |
0.42 13.8 |
2.38 0.07 |
||
2.3.2 Time-dependent fibrinogen functional activity and platelet adhesion
Conformational change following protein adsorption results in transient exposure of functional epitopes and time-dependent platelet adhesion. Studies of the availability of functional epitopes in adsorbed proteins help to understand platelet responses to surfaces. Although many techniques have been applied to analyze the protein structure, immunological methods using antibodies directly recognize the functional/activity changes in proteins41,140. AFM with antibody-functionalized probes offers a unique method to recognize and identify adsorbed proteins141–143. It allows for examination of multiple proteins at the molecular scale144,145, and also probes the conformation, orientation, and activity of adsorbed proteins with appropriate choice of antibodies146. As the γ-chain dodecapeptide sequence in fibrinogen is the primary ligand for platelet adhesion to adsorbed fibrinogen, the availability of the dodecapeptide can be viewed as an indicator of the functional activity of fibrinogen as related to the conformation and/or orientation of the adsorbed protein18,41,147.
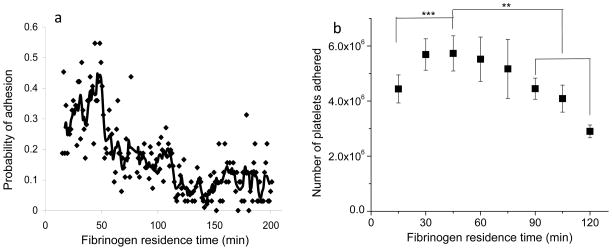
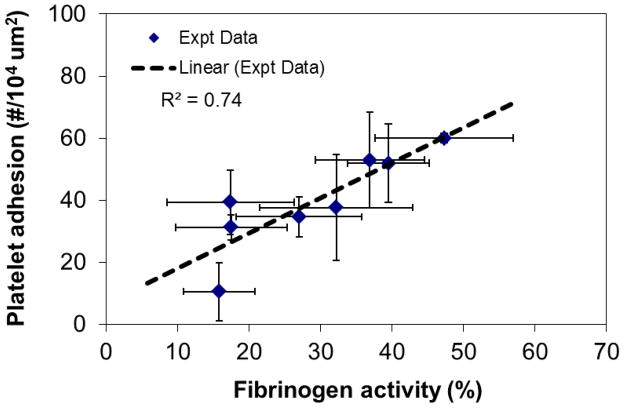
The force measurement mode of AFM provides an approach to study fibrinogen functional activity as recognized by monoclonal antibodies (mAb) at differing residence times. An AFM probe coupled with mAb that recognizes the last 20 amino acids of the γchain of fibrinogen (γ392–411) was used to measure the interaction forces between mAb and adsorbed fibrinogen. Figure 5(a) illustrates the probability of this mAb encountering it’s antigen in the platelet-binding region as a function of fibrinogen residence time after adsorption to a hydrophilic muscovite mica surface. Data demonstrate that the activity of fibrinogen is time-dependent, with the maximum likelihood of recognition occurring at around 45 min and decreasing at longer residence times148. Macro-scale assessments of platelet adhesion using the LDH assay showed that platelet adhesion varied with residence time of fibrinogen, and also reached a peak at ~45 min (Fig. 5b), correlating well with the molecular scale AFM results.
Figure 5.
(a) Probability of interaction between a mAb against γchain 392–411 and its antigen on fibrinogen measured by AFM. The solid line, the average of 5 points, is included for visualization purposes. (b) Platelet adhesion data from multiple experiments (n ≥ 6 for each time point) showing changes in platelet adhesion as a function of fibrinogen residence time on mica substrates. (**: p<0.01, ***: p<0.001, error bar: standard deviation) (Reprint from Soman, Rice and Siedlecki148 with permission from ACS)
Protein adsorption to biomaterials occurs from protein mixtures in most biomedical applications. The presence of other proteins results in adsorption competition149, and also alters the conformational structure and dynamics of adsorbed proteins. Our studies show that inclusion of albumin into a fibrinogen solution moves the peak biological activity of fibrinogen to earlier residence time points150. With a protein concentration ratio of 50:50 (Fibrinogen:BSA) in binary protein solutions, the peak biological activity of adsorbed fibrinogen shifts to ~15 min and activity decreases with increasing residence time. In the case of 10% fibrinogen in a binary protein solution, no peaks in fibrinogen activity were observed, and the activity remained essentially steady at a lower value. Measurement of platelet adhesion carried out under the same conditions show correlation between changes in activity of fibrinogen and platelet adhesion.
It should be noted that the studies of protein conformation were often carried on the model substrates such as mica and gold while the conformation of fibrinogen response to biomedical related polymeric surface is less reported. There is evidence that conformation of protein is related to chemistry and surface properties of polymeric biomaterials. Berglin et al. 151 reported that the conformation of fibrinogen adsorbed to acrylic polymeric substrate is related to polymer chain flexibility and fibrinogen maintained a more native conformation on the flexible polymer. More work is needed to focus on protein conformational response and platelet adhesion to individual biomedical polymeric surfaces.
2.4 Protein adsorption and platelet adhesion on polyurethane biomaterial surface
2.4.1 Microphase separation structure and protein adsorption
The surface chemical structure of polymeric biomaterials is often complicated compared to model materials. Block copolymers are composed of different segments having different properties, and are widely used in biomedical applications. One of the most important structures formed in block copolymers is found in polyurethane materials, and is termed microphase separation. This microphase separation creates a surface microenvironment where chemical composition and physicochemical properties are distributed spatially, and therefore have the potential to induce different levels of protein adsorption/activity across the material surface. Polyurethane (PU) and poly(urethane urea) (PUU) block copolymers are used in a variety of blood-contacting medical devices due to their broad range of mechanical properties, fatigue resistance and relatively-good hemocompatibility152–154, and these properties are believed to arise from the presence of separated microphases in the copolymers. Microphase separation is a result of thermodynamic immiscibility of the polar hard segments (made from diisocyanate and either a diol or a diamine) and the relatively non-polar soft segments (often a polyether but increasingly PU materials are being made from polycarbonates or mixed PTMO/PDMS precursors), where hard domains act as thermoplastic crosslinks and reinforcing agents dispersed in a soft segment rich background matrix155.
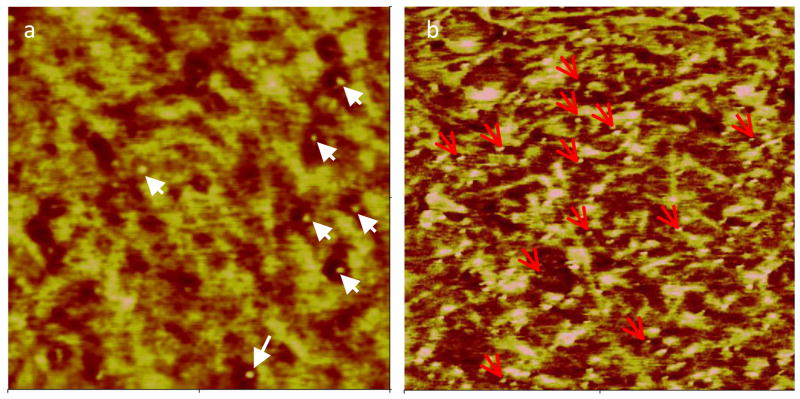
Numerous studies have shown that the Microphase separation structure influences biological responses to surfaces, including protein adsorption156–159, platelet adhesion/ activation160,161, and cell attachment162,163. We measured adhesion forces of a protein modified AFM probe with a PUU surface, and correlated mechanical properties with local adhesion forces. Results showed that low adhesion forces were associated with hard domain regions164. A nanogold-labeled protein conjugate was further used to visualize individual protein adsorption to the separate microstructures on PUU surfaces (Figure 6). Analysis of the image shows an ~ 50% increase in the number of proteins per unit area on the soft segment material compared to the hard segment regions, suggesting preferential adsorption to the less polar, more hydrophobic, soft segment region164.
Figure 6.
AFM images of nanogold conjugated BSA adsorbed on PUU surface imaged under (a) ambient condition and (b) in PBS buffer. Gold beads indicated by arrows are seen in phase image where lighter color represents hard domains. Quantification reveals more than 2 fold increase in number of labeled proteins per unit area on the soft segment regions compared to the hard segment. (Scan size: 500 ×500 nm2, reprint from Xu and Siedlecki164 with permission from Wiley)
2.4.2 Effect of microphase chemistry of polyurethane on protein adsorption and platelet adhesion
A variety of polyurethanes have been developed and synthesized for meeting varied requirements in medical device applications155. Principle candidate PU biomaterials generally consist of similar hard segments, but with different hard segment content and varying soft segment chemistry. The soft segments used in PU include polyether, aliphatic polycarbonate (PC), and polydimethylsiloxan (PDMS). These different soft segments produce different phase separation structures and affect material properties165,166, resulting in different biological responses including fibrinogen functional activity and platelet adhesion167.
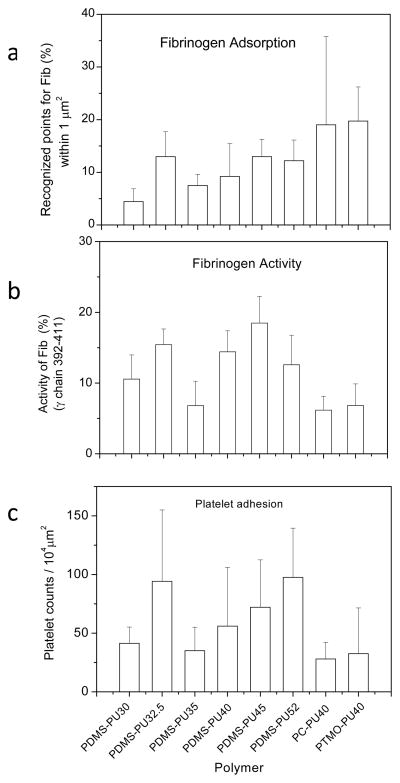
As a first effort at relating polymer microphase chemistry and structure, protein activity and subsequent platelet adhesion, we assessed the properties of a series of polyurethane materials derived from polycarbonate (PC-PU40), polytetramethylene oxide(PTMO-PU40), and PDMS (PDMS-PU series) chemistries. The number following the polymer chemistry description is the hard segment content of the material. Figure 7 summarizes the fibrinogen adsorption as recognized by polyclonal antibody, functional activity as recognized by mAb, and platelet adhesion on these materials. The amount of adsorbed fibrinogen recognized generally increased with hard segment content except for one material, the PDMS-PU32.5 (Fig. 7a). This trend was roughly consistent with platelet adhesion results (Fig. 7c). However, larger amounts of fibrinogen were measured on the PC-PU40 and PTMO-PU40 surfaces despite the fact that fewer platelets adhered on these materials. This inconsistency between platelet adhesion and fibrinogen adsorption suggests that the amouns of fibrinogen adsorbed may not be the key factor determining platelet adhesion. Rather, the availability of γchain dodecapeptide in fibrinogen is more important. Results showed that fewer specific binding events recognized by the mAb against fibrinogen γ392–411 were measured with proteins adsorbed on PC-PU40 and PTMO-PU40 materials (Fig. 7b). Generally, the activity of fibrinogen measured by the mAb was consistent with platelet adhesion on the three types of polymers tested (Fig. 8). However, it should be noted that there are two other RGD sites in the α chain that may also be involved in mediating platelet adhesion, acting perhaps separately or synergistically with the C-terminus of the γchain126.
Figure 7.
(a) Fibrinogen adsorption on PUs as recognized by a polyclonal antibody; (b) Functional activity of adsorbed fibrinogen detected by a monoclonal antibody against the fibrinogen γchain 392–411; (c) platelets adhesion on PU surfaces. Note: The number following the type of PU is the percentage hard segment content. (mean ± standard deviation) (Reprint with permission from 167)
Figure 8.
Relationship of platelet adhesion and fibrinogen bioactivity on polyurethane surfaces. (mean ± standard deviation)
2.4.3 Protein adsorption and platelet adhesion with dynamic restructuring polyurethane biomaterial surfaces
Microphase separation structure in segmented polyurethanes is the result of thermodynamic immiscibility of the hard and soft segments. The structure is influenced by block length and chemical composition of segments, but is also affected by the environmental conditions. Studies show that polyurethane materials undergo reorganization and reorientation in aqueous environments for minimizing surface free energy, resulting in enhancement of hard domains at the surface168–170. Figure 9 illustrates sequential in-situ AFM phase images of a PUU film following hydration in phosphate buffered saline (PBS) for 21 hrs and shows that hard domains in PUU undergo reorientation. The analysis of phase angle distribution showed that the percentage of areas having high phase angle shift increased with hydration time, suggesting enrichment of hard domains at the surface164. Hydration-induced surface enrichment of hard domains was also confirmed by measurements of local mechanical properties using a nano-indentation technique171. Water contact angle measurements showed that the surface becomes more wettable with hydration time due to the migration of hydrophilic domains to surface, consistent with the observation of others172. Dynamic restructuring of hard domains increases surface wettability and reduces the interaction forces between proteins and surfaces. For example, the water contact angle at the initial stage of PUU was near 90° and adhesion force with BSA was 1.5±0.3 nN, while the contact angle decreased to 70° and adhesion force with BSA dropped to 0.5 ± 0.3 nN at 53 hrs hydration164.
Figure 9.
Sequential phase images of PUU under PBS buffer hydrated for 21 hrs, showing hard domain reorientation and reorganization due to hydration. The images were captured at approximately 9 min intervals and high phase angle shifts (lighter colors) represent the “harder” regions. The lines point to landmarks that can be used for reference in monitoring the dynamic changes.
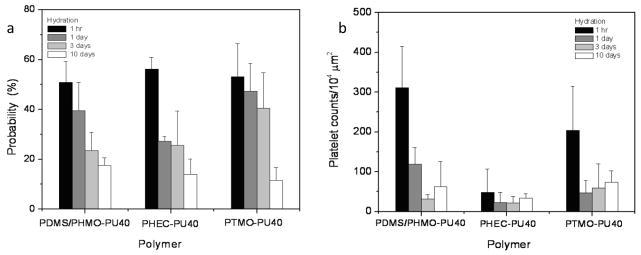
Dynamic restructuring of PUU hard domains of PUU also decreases the functional activity of adsorbed fibrinogen and subsequent platelet adhesion. On a series of PU biomaterials with different soft segments, higher fibrinogen activity was observed on surfaces at hydration time of 1 hr compared to samples with longer hydration times (Fig. 10a). Immuno-AFM force measurements identified the platelet binding epitope in adsorbed fibrinogen 51%, 56% and 53% of the measures for the PDMS/PHMO-PU40, PHEC-PU40 and PTMO-PU40 surfaces, respectively, when samples were hydrated in PBS for 1 hr. Increasing the hydration time of the polymers decreased this measure of activity for fibrinogen on these materials. When hydrated for 10 days prior to a 10 minute incubation with fibrinogen, only 18% (PDMS/PHMO-PU40), 14% (PHEC-PU40) and 12% (PTMO-PU40) of the force measures showed an interaction for the mAb against γ392–411. Corresponding platelet adhesion is shown in Figure 10b, and shows that regardless of the soft segment chemistry, PU materials show the highest platelet adhesion at 1 hr hydration time. This result is consistent with the higher activities of fibrinogen found on the surfaces, again showing that the availability of the γ-chain dodecapeptide in fibrinogen correlates with platelet adhesion.
Figure 10.
(a) Probability of recognition by mAb shows the functional activity of fibrinogen on polyurethane surfaces following hydration for 1 hr, 1 day, 3 days and 10 days, (b) platelet adhesion on polyurethane surfaces following hydration with same duration. (reprint from reference171). (error bar: standard deviation) (reprint from Xu, Runt, and Siedlecki171 with permission from Elsevier)
Taken together, phase restructuring during hydration changes the local microenvironments and the physicochemical/mechanical properties at the interface of polyurethane. As a result, the interactions of blood with biomaterial surfaces, including platelet adhesion/activation, thrombus formation, and cell attachment/growth, is expected to change during this process. Fibrinogen plays the dual roles of serving as a ligand for platelet adhesion to the surface and also for linking platelets together into aggregates20. The decrease in platelet adhesion on PU surfaces after hydration, consistent with the measured decrease in fibrinogen bioactivity on the surface, strongly suggests that phase rearrangements in PU could affect thrombus formation. The varying degrees of phase separation/restructuring in different PU polymers would therefore be expected to affect protein adsorption, platelet adhesion/activation, cell attachment and proliferation, as well as bacterial adhesion.
2.5 Protein adsorption and platelet adhesion on textured biomaterial surface
2.5.1 Surface topography controlling biological responses to surface
Physical approaches to biomaterial design have been a promising route for controlling biological responses113. Design and control material surface topography has had a profound impact on cell and tissue behaviors including platelet adhesion and bacterial adhesion, as well as cell growth. The motivation to use physical properties to control biological response comes from nature, for example, the skin of sharks, the mussel, lotus leaf, and the inner surface of blood vessels173–177 have all provided templates for the fabrication of materials. These natural surfaces are micro-topographically structured and possess intrinsic non-adherence function. These materials have inspired novel biointerface materials 178. Materials with topographical features mimicking shark skin show resistance to marine biofouling108,179–181. This topography applied into biomedical-related polymeric materials was found to disrupt biofilm formation of Staphylococcal aureus182 and inhibit platelet adhesion117, demonstrating applicability for medical devices. We developed textured surfaces having ordered array of pillars and demonstrated that these materials reduced the adhesion of Staphylococcal strains and inhibit biofilm formation183,184. Previous studies showed these materials inhibit platelet adhesion and activation110,111.
Surface features influencing biological responses include feature shape and size, distance between features, and organization of features185. Submicron textured PUU surfaces (dimension/separation of 400/400 nm and 500/500 nm) consisting of ordered arrays of pillars with dimension and separation of pillars less than 1 μm produce a dramatic reduction in accessible contact area for bacteria or platelet interaction, and minimize opportunities for bacteria and platelet to be trapped in inter-pillar spaces111,183. This design is consistent with the finding of other researchers that show that surface structures are most effective when in the range of 50 to 90% of the diameter or length of settling organisms180 and the height of nanocylinders on structured surface should be >160 nm186.
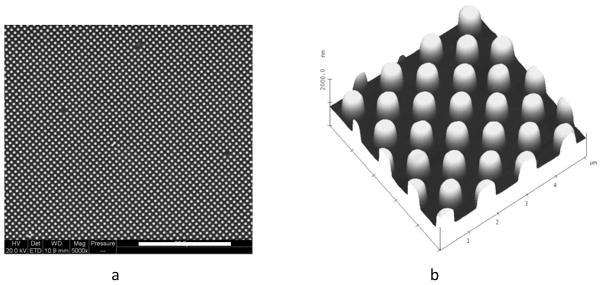
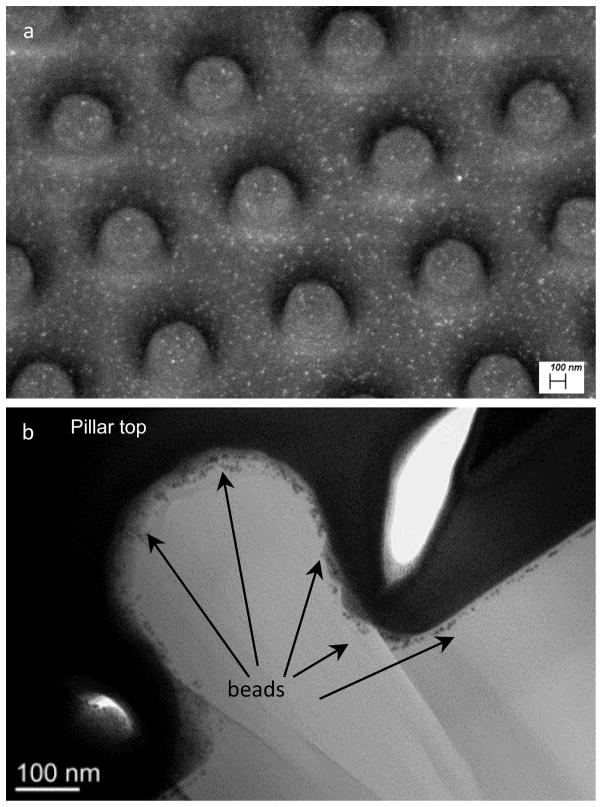
PUU textured films can be physically prepared by soft lithography, two-stage replication molding techniques where a master pattern is fabricated in silicon, a silicone mold is cast and PUU replicas created from the silicone negative111. A variety of techniques have been applied to characterize the surfaces. Figure 11 illustrates representative SEM and AFM images of a textured PUU surface having a 500/500 nm pattern. Analysis of these surfaces revealed the top surface of this 500/500 nm pattern is only about 24.5% of the nominal surface area. Reductions in surface contact area reduce opportunity for platelet or bacteria to adhere to the surface. Another characteristic of textured surfaces can be an increase in surface hydrophobicity. Water contact angles of submicron patterned PUU surface reach nearly 145°, near to the “superhydrophobic”, while the water contact angle for a smooth surface PUU surface was just 93°183. The increase in hydrophobicity of textured surfaces is believed to be due to air captured in the spaces between pillars187. Evidence suggests that both reduction in surface contact area and increase in hydrophobicity influence the biological events on surfaces.
Figure 11.
(a) SEM and (b) AFM images showing topography of textured PUU film surface with 500/500 nm pattern. (Reprint from Xu and Siedlecki 183 with permission from Elsevier)
2.5.2 Protein adsorption on textured biomaterial surfaces
Protein adsorption on topographically-modified surfaces depends on both the surface features and the proteins, as described in Section 2.2.3. The amount of total protein or of a selective protein is quantified to indicate the behavior of protein adsorption, often by using traditional colorimetric protein assays188, the enzyme-linked immunosorbent assay (ELISA)117, radiolabeling189, electrophoresis118, quartz crystal microbalance (QCM)122, or any of the myriad other techniques available. The main weakness to these methods is that it only provides the overall quantification of protein adsorption and often misses one of the most important characteristics of topographic surfaces – the increase in surface area. It has been suggested that protein adsorption on topographic surfaces may be non-uniform and vary locally.
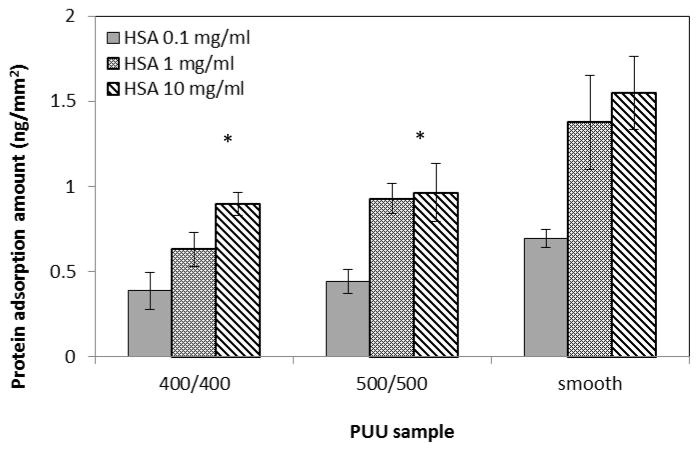
Directly imaging or detecting single molecular proteins on surface is difficult, especially on polymeric biomaterial surfaces with textured topographies. With the development of nanotechnology in biology and medicine, nanoparticles conjugated with proteins have numerous applications in sensing, imaging, delivery, catalysis, therapy and control of protein structure and activity190. Among nanoparticles, gold nanoparticles are receiving significant attention because their unique physical, chemical, and biological properties are quite different from the bulk of their counterparts191 and can be easily detected by microscopy techniques such as electron microscopy and AFM. We used nanogold particles conjugated to an anti-fibrinogen antibody and identified fibrinogen from dual-component protein solutions following adsorption and imaging by by AFM144. We have also characterized the adsorption of proteins coupled with 6 nm nanogold beads on textured surface by field emission scanning electron microscopy (FESEM) and transmission electron microscope (TEM) (Fig. 12). The bright spots in SEM images illustrate the nanogold beads, indicating the presence of adsorbed protein. FESEM image demonstrates that the beads distribute around the top and bottom surfaces, and more beads (i.e., proteins) were detected on the bottom surface than on the top surface of pillars. The side view of a single pillar (TEM, Fig 12b) shows that the beads (i.e., proteins) are distributed on pillar top, edge, sidewall, and bottom areas. These results suggested that protein adsorption on textured surface with submicron pillars occurred not only on the top surface of pillars, but also on the sidewall and bottom areas. As a result, the quantification of protein adsorption by other methods should be normalized to the entire surface area including sidewall of pillars and other topographic features. Figure 13 illustrates the trends of adsorption of human albumin on textured PUU films with smooth, 400/400 nm, and 500/500 nm patterns, measured by ELISA. Results demonstrated that less protein was adsorbed on textured surfaces, suggesting that submicron textured surface may produce the heterogeneous local microenvironments at pillar surface area and alters the protein adsorption behavior, resulting in different densities of protein adsorbed.
Figure 12.
(a) FESEM with 30 degree angle and (b) TEM images of 500/500 nm patterned PUU surfaces adsorbed with nano-gold conjugated BSA, showing the distribution of protein adsorbed on textured pillar surface.
Figure 13.
Human albumin adsorption on textured PUU surfaces at different concentrations. (*: p<0.05, error bar: standard deviation)
2.5.3 Platelet adhesion on textured biomaterial surface
Platelet response to topographical surface is influenced by the surface physical features and chemical composition of substrate. Koh et al.192 prepared poly(lactic-co-glycolic-acid) films with multi-walled carbon nanotubes having two different nanotube orientations, either random or vertically aligned, and found the film with vertically aligned nanotubes exhibited very low levels of fibrinogen adsorption and platelet adhesion. Reduction of platelet adhesion was regarded to be attributed to both chemical and topographical effects. They further studied the effect of topographical features on platelet adhesion and concluded that the optimal range of topographical dimensions of the pillars with height of 300–800 nm, width of 100–200 nm, and interspacing distances <200 nm117. Other researchers using polymer demixing methods created structured polymer films with typical feature sizes ranging from the nanometer to the micrometer, and found that increasing the feature size of the surface encourages von Willebrand factor adsorption and thus platelet adhesion and consequent thrombus formation193. Ye et al.194 used dual scale structures with nano and micro dimensions as model substrates to investigate the impact of surface topography on platelet response, and found fewer platelets adhesion and no activation on such surfaces. We measured the platelet adhesion on smooth and patterned PUU film surfaces at low shear stresses across a 0–10 dyn/cm2 range. Significant reduction in platelet adhesion was found on 400/400 nm and 700/700 nm PUUs for shear stress <5 dyn/cm2 compared to smooth. Additionally, non-adherent platelets did not demonstrate increased activation after transient exposure to textured surface. These results suggest that using surface topography can reduce platelet adhesion and activation111.
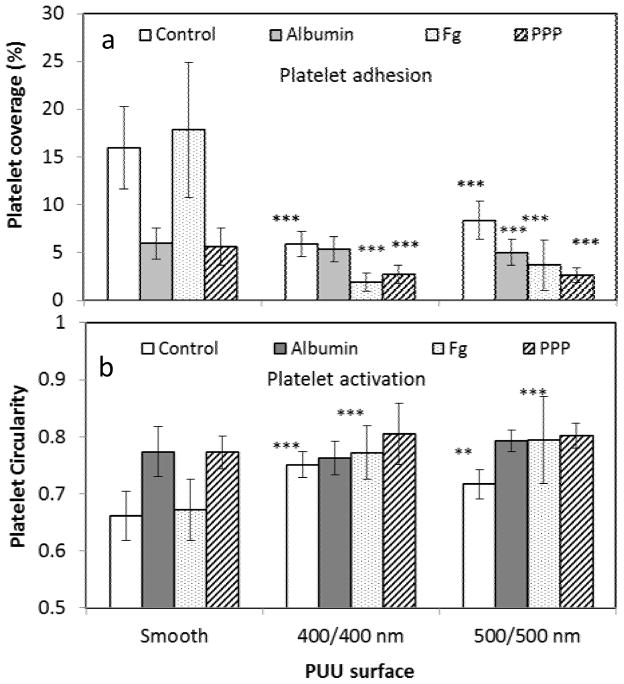
Protein adsorption affects platelet adhesion and activation on textured biomaterial surface. A series of PUU film surfaces were pre-adsorbed with proteins (human albumin, fibrinogen, and platelet poor plasma (PPP)) at 37°C for 1 hr. Samples were then incubated in platelet-containing solution (2.50×108/ml) at 37°C for 1 hr in a microwell plate195 under static condition. Samples were labeled for fluorescence microscopy to examine platelet adhesion and activation. Activation of platelets adherent on surfaces was inferred from platelet circularity, with higher circularity indicative of less active platelets. Results showed that the both 400/400 nm and 500/500 nm textured surfaces significantly reduced platelet adhesion and activation compared to the smooth surfaces (Fig. 14). Fibrinogen increases platelet adhesion on smooth surfaces while albumin and PPP suppress adhesion, as expected. It is interesting to see that platelet adhesion on fibrinogen-adsorbed textured surfaces is lower than that on smooth surfaces, and even lower than smooth surfaces pre-adsorbed with albumin. This suggests that fibrinogen adsorbed on textured surface may have less functional activity, resulting in lower platelet adhesion. This is also evidenced by the circularity of platelets, with those adhered on fibrinogen-textured surfaces having higher circularity than platelets on smooth or textured surfaces (without protein adsorption), indicating that textured surfaces reduce platelet activation.
Figure 14.
(a) Platelet adhesion and (b) activation on PUU surfaces with pre-adsorbed with plasma proteins. The statistical analysis is performed between textured and smooth PUU samples.
(**: p<0.01; ***:p<0.001, error bar: standard deviation)
Surface topography can potentially affect protein orientation and conformation structure upon adsorption. There is evidence to show that the curvature of features affects the unfolding of proteins (e.g., fibrinogen) at feature edges and make receptor ligands more (or less) accessible, resulting in differing cell adhesion196. As the dimension of top surface of submicron-sized pillars is larger than dimensions of proteins which lie in the nanometer range, the top surface is not as flat as smooth, but with a slight curve, due to the fabrication. Moreover, the edges of pillar features are generally on the length scale of a protein. Therefore, both top surface and edges may affect protein adsorption and its conformation, causing different platelet responses. Characterization of protein conformational structure and functional activity at local area on top surface of features will largely contribute to understand the cell behaviors on textured surface and will be more interesting to biomaterial scientists.
3. Blood coagulation at biomaterial interface
3.1 Blood coagulation cascade and contact activation of FXII
The process of biomaterial associated thrombosis consists of both platelet-mediated reactions (platelet adhesion, activation, and aggregation) and coagulation of blood plasma. Plasma protein interactions with surfaces trigger the coagulation cascade of blood, resulting in thrombus production and formation of a fibrin clot. Coagulation involves a series of self-amplifying, zymogen-enzyme conversions which are traditionally grouped as the intrinsic and extrinsic pathways197,198. Both pathways are initiated separately but merge into a common pathway leading to thrombin (FIIa), which hydrolyses fibrinogen into fibrin, which assembles and causes plasma to become clotted198–201. The extrinsic pathway is responsible for hemostatic control and response to vascular injury while the intrinsic coagulation cascade has little physiological significance under normal conditions. However, it is widely believed that the intrinsic pathway is trigged by blood contact with artificial materials and it is an important cause for poor hemocompatibility of biomaterials202–204.
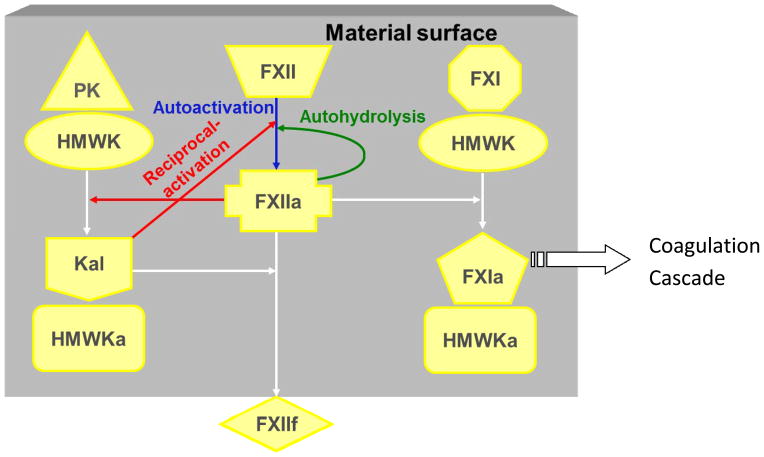
The initiation of the intrinsic pathway is generally referred as contact activation, and primarily mainly involves four proteins: coagulation factor XII (FXII, Hageman factor), prekallikrein (PK, Fletcher factor), high-molecular weight kininogen (HMWK, Fitzgerald factor), and coagulation factor XI (FXI, Plasma Thromboplastin Antecedent). The traditional biochemistry of contact activation shows that FXII is converted to the active enzyme form FXIIa. FXIIa can be produced by at least three different biochemical reactions (Fig. 15): (1) contact autoactivation process - FXII interacts with a procoagulant surface and converts into active enzyme form FXIIa through autoactivation due to conformational structural change upon binding of FXII to the surface (FXII → FXIIa)205; (2) reciprocal activation - the generated FXIIa can in turn cleave PK bound to the surface as a complex with HMWK to produce Kal which enzymatically acts on FXII upon binding on surface206; (3) autohydrolysis or self-amplification - FXIIa hydrolyzes FXII to produce FXIIa (FXIIa+FXII → 2FXIIa)207. Ultimately, FXIIa activates FXI bound at the surface as complex with HMWK to generate FXIa leading to propagation of subsequent coagulation cascade reactions198,208,209.
Figure 15.
Surface-mediated interactions in contact activation of plasma coagulation. PK: prekallikrein; HMWK: high molecular weight kinnogen; Kal: kallikrein; FXII: factor XII; FXI: Factor XI. Suffixes “a” and “f” represent activated and fragmented form. (Reprint from Vogler and Siedlecki 198 with permission from Elsevier)
Each of these three biochemical reactions can be observed in defined solutions, however, all three reactions do not occur in plasma to the same extent. For example, autohydrolysis is thought to be a contributing reaction in buffer solutions of FXIIa, but does appear to be a significant reaction in plasma, because the preferred FXIIa substrates PK and FXI are not available in neat buffer solutions of FXII207. Mathematical modeling of the intrinsic cascade predicts that the primary mechanism for activation of coagulation involves autoactivation of FXII by the procoagulant surface or kallikrein-mediated reciprocal activation of FXII, but FXIIa-induced self-amplification of FXII is insignificant210. In plasma, FXIIa is principally produced by autoactivation which is then substantially increased by reciprocal-activation pathways. Analysis of the amounts of FXIIa produced showed that kal-mediated reciprocal-activation pathway principally contributes to FXIIa generation at 75%211.
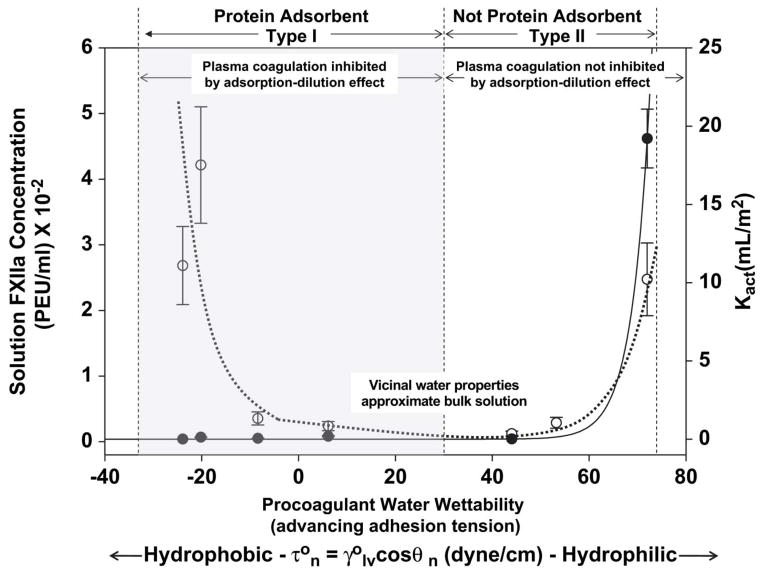
FXII contact activation is surface-dependent. A variety of parameters including surface energy or wettability, surface chemistry, and surface area have been well investigated on contact activation212–214. Since common observations clearly show that plasma coagulation is more efficient in activation by contact with anionic215,216 or hydrophilic surfaces13,217, it was concluded that contact autoactivation of FXII was more specific for hydrophilic surfaces than hydrophobic surfaces based on traditional biochemistry theory. However, experimental evidence demonstrated that hydrophobic and hydrophilic surfaces have nearly equal autoactivation properties in neat – buffer solution of FXII, i.e. contact activation of FXII is not specific to anionic hydrophilic surfaces in neat buffer214,218. In fact, contact activation of FXII in neat-buffer solution exhibits a parabolic profile when scaled as a function of surface energy. Nearly equal activation is observed at both extremes of activator water wettability, and falls through a broad minimum with water contact angle in the range of 20 < τ < 40 dyn/cm (55°< θ <75°), suggesting that materials with surface energy in this range exhibit minimal activation of blood-plasma coagulation (Fig. 16) 213. A similar result was obtained by preparing mixtures of various thiol-based self-assembled monolayers at different concentrations and the mixtures of thiols appear to prolong coagulation time beyond that seen with the background container alone (unpublished results).
Figure 16.
Comparison of FXII activation in neat-buffer solution (left-hand ordinate, dashed line) to catalytic potential in plasma Kact (right-hand ordinate, solid line) for activator (procoagulant) particles exhibiting different surface energy (abscissa) expressed as water adhesion tension in dyne/cm (where is water interfacial tension in dyne/cm and θ is the advancing contact angle) (reprint from 213 with permission from Elsevier).
Contribution of surface area to contact activation of FXII is inconclusive. The plausible explanation consistent with current understanding of coagulation-cascade biochemistry is that procoagulant stimulus arising from the activation complex of the intrinsic pathway is dependent on activator surface area. In plasma, FXII autoactivation can be studied in vitro using activator surface-area titration of plasma coagulation and the titration plot of clotting time against activator surface areas is generally characterized to be asymptotic and leading to a lower clotting time plateau which depends on activator surface energy213,219. The total amount of FXIIa yield is also characterized by the FXIIa titration curve and results show that total contact activation of FXII scales with increasing surface area of procoagulant and the normalized amount of enzyme produced per-unit-area procoagulant is constant211. All these results suggested that contact activation of FXII increased with surface area in plasma depending on surface energy. However, recent results showed that activation of FXII dissolved in buffer, protein cocktail, heat-denatured serum, and FXI deficient plasma did not exhibit activator surface-area dependence, suggesting activator surface-area dependence observed in contact activation of plasma coagulation does not solely arise at the FXII activation step of the intrinsic pathway220. Other data show that contact activation of FXII either in plasma or in buffer is surface-area dependence for hydrophilic activator but independence for hydrophobic activator. This is most likely a result of synergistic effects of protein interaction (adsorption/desorption) and contact activation on surfaces.
Contact activation of blood FXII is moderated by the protein composition of the fluid phase and protein adsorption competition on surfaces depending on surface energy. Zhuo et al.221 found that FXIIa yield arising from FXII contact activation on hydrophilic surfaces in a protein cocktail was much greater than that obtained in buffer solution containing only FXII, but it was contrary on the hydrophobic surface. Results suggest that contact activation in the presence of proteins unrelated to the plasma coagulation cascade leads to an apparent specificity for hydrophilic surfaces due to a relative diminution of activation at hydrophobic surfaces and an enhancement at hydrophilic surfaces. It was also found that the rate of FXIIa accumulation decreased with time leading to a steady-state FXIIa yield for both hydrophilic and hydrophobic surfaces in protein and buffer solutions, suggesting that activation competed with a yet unknown autoinhibition reaction to inhibit the conversion of FXII to FXIIa. Thus plasma proteins play a dual role in moderating contact activation of the plasma coagulation cascade. The principle role is inhibiting FXII contact with activating surfaces, but these proteins also displace FXIIa from an activating surface into solution where FXIIa is involved in the subsequent steps of the plasma coagulation cascade222. The importance of protein adsorption competition in contact activation was also observed in moderating interactions between FXII and PK, which are components of the reciprocal-activations218. All these observations implicate protein adsorption competition is an important mediator of contact activation and must be included in any comprehensive mechanisms of surface-induced blood coagulation.
3.2 Blood coagulation in response to surface with chemical heterogeneity
The effects of surface chemistry or surface energy on blood coagulation is often investigated using surfaces with well-defined homogeneous chemistry such as silane or thiol SAMs with various terminating functional groups212,213,223. However, nanometer-scale patterning or domain formation with different chemistry is of interest as the block copolymers such as polyurethane have been widely used in biomedical applications. The microphase separation structures with nanoscale chemical domains contribute to the hemocompatibility of block biomaterial. Polyurethanes have shown to reduce contact activation with lower amidolytic activities by FXII and kallikrein compared to glass in plasma224. Our studies indicated that the microphase separation of polyurethanes produces a local surface microenvironment and influences protein and cellular responses to the surfaces164,171, suggesting the nanoscale chemistry heterogeneity may influence the blood coagulation. Miller er al.225 investigated silane-based mixed films on glass substrates having nanoscale chemical heterogeneity formed by the stepwise adsorption of an amine-terminated silane, 3-aminopropyltriethoxysilane (APS), and a methyl-terminated silane, n-butyltrichlorosilane (BTS), and found that the mixed films consists of 400 nm islands of an amine-terminated silane dispersed in a butyltrichlorosilane background on glass substrates and such nanometer-scale chemical heterogeneity were inefficient at activating the intrinsic coagulation of human blood plasma when compared to their one-component control surfaces. Results suggested that the nanoscale engineering of surface may provide a route to biomaterials with improved hemocompatibility.
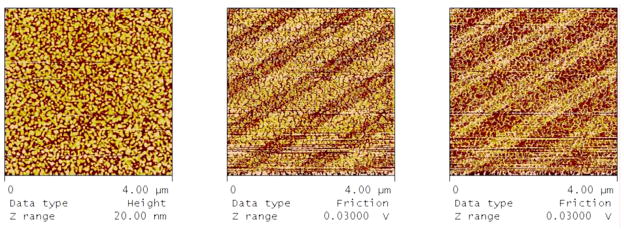
With the development of nanotechnology, it is possible to create surfaces having well-defined surface chemistry combined with nanometer scale patterning. Figure 17 illustrates an example of linear patterned surface chemistry consisting of carboxyl-terminated and methyl-terminated thiols on gold-coated glass coverslips prepared by microcontact printing. The wider bands in the friction trace and retrace microscopy images correspond to the octadecanethiol stamped areas, while the thinner bands correspond to the 11-mercaptoundecanoic acid backfilled areas. The cumulative area of 11-mercaptoundecanoic acid backfilled areas was estimated to be 40% by comparing band widths. The measured water contact angle of patterned surface is 73±4°, while the contact angles for octadecanethiol and mercaptoundecanoic acid controlled surfaces are 96±1° and 24±1°, as expected. The plasma coagulation characteristics of nanoscale chemical patterned surface were assayed as coagulation time of platelet poor plasma incubated with gold coverslips. Results show that nanometer-scale patterning of surfaces leads to statistically-significant increases in coagulation times when compared to carboxyl surface area (Fig 17a) or simply to the measured fraction of carboxyl in mixed film, relative to surfaces with equivalent, macro-scale areas of carboxyl and methyl-terminated thiol chemisorption. The increased coagulation times by patterning suggest that the patterning of the surface may affect activation of the intrinsic coagulation cascade and that it is not simply a matter of the cumulative carboxyl-terminated thiol backfilled area.
Figure 17.
From left to right – height, friction mode retrace, and friction mode trace AFM images of patterned thiol surfaces. Bare gold surface was stamped with a linear pattern PDMS stamp inked with octadecanethiol and backfilled with 11-mercaptoundecanoic acid. The light colored bands in the friction retrace image (middle) correspond to the stamped octadecanethiol.
4. Summary and prospective
Biocompatibility is the central theme for the design and application of polymeric biomaterials. Blood coagulation and thrombosis resulting from blood-material interactions remain a challenge in the use of blood-contacting materials. Protein adsorption is a critical early event during the interaction of blood with implanted biomaterials and it is the adsorbed proteins, rather than the surface itself, that mediates the subsequent biological responses, including platelet adhesion and thrombus formation on surfaces. However, surface properties including surface chemistry, surface energy, and surface topography determines the protein composition, structure, function of the adsorbed protein layer. Thus the biological events occurring on biomaterial surface are the results of interactions of material surface, proteins, and cells at interface. The future of research in this field will rely on a better analysis and understanding of the influence of surface properties from microscale to nanoscale on the quality, quantity, and conformation of proteins adsorbed on surfaces. The development of new techniques or the adaption of existing techniques as well as the development of new biomaterials with specific features will largely help to obtain these information.
Protein undergoes the conformational structural change upon adsorption, e.g., fibrinogen, and influences the subsequent biological responses. It would be more important to reveal the conformation state or functional activity of protein adsorbed than the amount of protein in mediating the platelet adhesion and activation127. The current understanding of plasma protein adsorption on biomaterial surfaces has primarily focused on an overall measurement of protein adsorption, primarily due to limitations in the analysis techniques, especially on topographical surfaces, while the precise effects of surface properties on protein structure at molecular scale is largely unknown. It is of fundamental interest to the biomaterials community to understand how molecular protein structure and activity are correlated to specific surface properties. This may call for new measurement techniques and analysis methods to measure the individual protein adsorption and its functional activity on surfaces, and to understand the role of surface property in protein adsorption and subsequent biological responses.
The formation of a thrombus on the surface of a blood-contacting material in vivo involves both the activation of the intrinsic coagulation cascade and the adhesion/activation of platelets. Platelet adhesion and contact activation are generally studied separately. However, platelet activation can trigger FXII-mediated contact activation on the surface, leading to generation of FXIIa-antithrombin and FXIa-antithrombin complex, and contributing to clot formation226. It is also reported that contact activation and platelet adhesion have a strong synergistic effect on coagulation on blood-contacting materials even though these events in isolation are not sufficient to induce substantial thrombus formation227. These results may motivate the study of contact activation and blood coagulation on biomaterial surfaces with incorporation of platelet, proteins, and surfaces. The systematic investigation of biological responses to surfaces will be the great beneficial for biomaterial designs with improved biocompatibility.
Highlights.
We highlight the current knowledge in the field of blood-materials interactions
We highlight the role of surface chemistry, energy and topography in mediating blood-material interactions
We highlight novel methods for measuring biological responses
Acknowledgments
These studies were supported in part by NIH RO1 HL69965, and the National Science Foundation (DMR-0804873). The authors would like to thank Dr. Keith Milner, Dr. Erwin Vogler and Dr. Avantika Golas for numerous discussions and technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 2.Peppas NA, Langer R. New challanges in biomaterials. Science. 1994;263:1715–1720. doi: 10.1126/science.8134835. [DOI] [PubMed] [Google Scholar]
- 3.Hu WJ, Eaton JW, Ugarova TP, Tang L. Molecular basis of biomaterial-mediated foreign body reactions. Blood. 2001;98:1231–1238. doi: 10.1182/blood.v98.4.1231. [DOI] [PubMed] [Google Scholar]
- 4.Anderson JM. Biological responses to materials. Annual Review of Materials Research. 2001;31:81–110. [Google Scholar]
- 5.Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Seminars in Immunology. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. doi: http://dx.doi.org/10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29:2941–2953. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Horbett TA. Principles underlying the role of adsorbed plasma-proteins in blood Interactions with foreign materials. Cardiovascular Pathology. 1993;2:S137–S148. [Google Scholar]
- 8.Thevenot P, Hu WJ, Tang LP. Surface chemistry influences implant biocompatibility. Current Topics in Medicinal Chemistry. 2008;8:270–280. doi: 10.2174/156802608783790901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Engineering. 2005;11:1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez-Gonzalez R, Boto GR, Perez-Zamarron A. Cerebrospinal fluid diversion devices and infection. A comprehensive review. European Journal of Clinical Microbiology & Infectious Diseases. 2012;31:889–897. doi: 10.1007/s10096-011-1420-x. [DOI] [PubMed] [Google Scholar]
- 11.Johnson B, Starks I, Bancroft G, Roberts PJ. The effect of care bundle development on surgical site infection after hemiarthroplasty: An 8-year review. Journal of Trauma and Acute Care Surgery. 2012;72:1375–1379. doi: 10.1097/TA.0b013e318245267c. [DOI] [PubMed] [Google Scholar]
- 12.Ruggeri ZM. Mechanisms initiating platelet thrombus formation. Thrombosis and haemostasis. 1997;78:611–616. [PubMed] [Google Scholar]
- 13.Vogler EA, et al. Contact activation of the plasma coagulation cascade. I. Procoagulant surface chemistry and energy. J Biomed Mater Res. 1995;29:1005–1016. doi: 10.1002/jbm.820290813. [DOI] [PubMed] [Google Scholar]
- 14.Rodrigues SN, Goncalves IC, Martins MCL, Barbosa MA, Ratner BD. Fibrinogen adsorption, platelet adhesion and activation on mixed hydroxyl-/methyl-terminated self-assembled monolayers. Biomaterials. 2006;27:5357–5367. doi: 10.1016/j.biomaterials.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Chuang WH, Lin JC. Surface characterization and platelet adhesion studies for the mixed self-assembled monolayers with amine and carboxylic acid terminated functionalities. Journal of Biomedical Materials Research Part A. 2007;82A:820–830. doi: 10.1002/jbm.a.31193. [DOI] [PubMed] [Google Scholar]
- 16.Wang LF, Wei YH. Effect of soft segment length on properties of fluorinated polyurethanes. Colloids and Surfaces B-Biointerfaces. 2005;41:249–255. doi: 10.1016/j.colsurfb.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Tsai WB, Grunkemeier JM, Horbett TA. Human plasma fibrinogen adsorption and platelet adhesion to polystyrene. Journal of Biomedical Materials Research. 1999;44:130–139. doi: 10.1002/(sici)1097-4636(199902)44:2<130::aid-jbm2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Chiumiento A, Lamponi S, Barbucci R. Role of fibrinogen conformation in platelet activation. Biomacromolecules. 2007;8:523–531. doi: 10.1021/bm060664m. [DOI] [PubMed] [Google Scholar]
- 19.Amiji MM, Kamath KR, Park K. In: Encyclopedic Handbook of Biomaterials and Bioebgineering. Wise DL, editor. CRC; 1995. [Google Scholar]
- 20.Hantgan RR, Francis CW, Mardner VJ. In: Hemostasis and Thrombosis: Basic Priciples and Clinical Practice. Colman Robert W, Hirsh Jack, Marder Victor J, Salzman Edwin W., editors. J.B. Lippincott Company; 1994. pp. 277–300. [Google Scholar]
- 21.Grunkemeier JM. Platelet adhesion and procoagulant activity induced by contact with radiofrequency glow discharge polymers: Roles of adsorbed fibrinogen and vWF. Journal of Biomedical Materials Research. 2000;51:669–679. doi: 10.1002/1097-4636(20000915)51:4<669::aid-jbm15>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Farrell DH, Thiagarajan P. Binding of recombinant fibrinogen mutants to platelets. Journal of Biological Chemistry. 1994;269:226–231. [PubMed] [Google Scholar]
- 23.Bastida E, Escolar G, Ordinas A, Sixma J. Fibronectin is required for platelet adhesion and for thrombus formation on subendothelium and collagen surfaces. Blood. 1987;70:1437–1442. [PubMed] [Google Scholar]
- 24.Bültmann A, et al. Impact of glycoprotein VI and platelet adhesion on atherosclerosis—A possible role of fibronectin. Journal of Molecular and Cellular Cardiology. 2010;49:532–542. doi: 10.1016/j.yjmcc.2010.04.009. doi: http://dx.doi.org/10.1016/j.yjmcc.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Gawaz M, et al. Vitronectin Receptor (αvβ3) Mediates Platelet Adhesion to the Luminal Aspect of Endothelial Cells : Implications for Reperfusion in Acute Myocardial Infarction. Circulation. 1997;96:1809–1818. doi: 10.1161/01.cir.96.6.1809. [DOI] [PubMed] [Google Scholar]
- 26.Asch E, Podack E. Vitronectin binds to activated human platelets and plays a role in platelet aggregation. Journal of Clinical Investigation. 1990;85:1372–1378. doi: 10.1172/jci114581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reininger AJ, et al. Mechanism of platelet adhesion to von Willebrand factor and microparticle formation under high shear stress. Blood. 2006;107:3537–3545. doi: 10.1182/blood-2005-02-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomokiyo K, et al. Von Willebrand factor accelerates platelet adhesion and thrombus formation on a collagen surface in platelet-reduced blood under flow conditions. Blood. 2005;105:1078–1084. doi: 10.1182/blood-2004-05-1827. [DOI] [PubMed] [Google Scholar]
- 29.Tsai W-B, Grunkemeier JM, McFarland CD, Horbett TA. Platelet adhesion to polystyrene-based surfaces preadsorbed with plasmas selectively depleted in fibrinogen, fibronectin, vitronectin, or von Willebrand’s factor. Journal of Biomedical Materials Research. 2002;60:348–359. doi: 10.1002/jbm.10048. [DOI] [PubMed] [Google Scholar]
- 30.Geer CB, Rus IA, Lord ST, Schoenfisch MH. Surface-dependent fibrinopeptide A accessibility to thrombin. Acta Biomaterialia. 2007;3:663–668. doi: 10.1016/j.actbio.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Evans-Nguyen KM, et al. Changes in adsorbed fibrinogen upon conversion to fibrin. Langmuir. 2006;22:5115–5121. doi: 10.1021/la053070y. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs B, et al. Flow-based measurements of von Willebrand factor (VWF) function: Binding to collagen and platelet adhesion under physiological shear rate. Thrombosis Research. 2010;125:239–245. doi: 10.1016/j.thromres.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Yago T, et al. Platelet glycoprotein lb alpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. Journal of Clinical Investigation. 2008;118:3195–3207. doi: 10.1172/jci35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai WB, Horbett TA. The role of fibronectin in platelet adhesion to plasma preadsorbed polystyrene. Journal of Biomaterials Science-Polymer Edition. 1999;10:163–181. doi: 10.1163/156856299x00117. [DOI] [PubMed] [Google Scholar]
- 35.Asch E, Podack E. Vitronectin binds to activated human platelets and plays a role in platelet aggregation. The Journal of Clinical Investigation. 1990;85:1372–1378. doi: 10.1172/jci114581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YP, et al. Fibrin-incorporated vitronectin is involved in platelet adhesion and thrombus formation through homotypic interactions with platelet-associated vitronectin. Blood. 2004;104:1034–1041. doi: 10.1182/blood-2003-12-4293. [DOI] [PubMed] [Google Scholar]
- 37.Rabe M, Verdes D, Seeger S. Understanding protein adsorption phenomena at solid surfaces. Advances in Colloid and Interface Science. 2011;162:87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Cornelius RM, Archambault JG, Berry L, Chan AKC, Brash JL. Adsorption of proteins from infant and adult plasma to biomaterial surfaces. Journal of Biomedical Materials Research. 2002;60:622–632. doi: 10.1002/jbm.10117. [DOI] [PubMed] [Google Scholar]
- 39.Ziats NP, Pankowsky DA, Tierney BP, Ratnoff OD, Anderson JM. Adsorption of Hageman factor (factor XII) and other human plasma proteins to biomedical polymers. The Journal of laboratory and clinical medicine. 1990;116:687–696. [PubMed] [Google Scholar]
- 40.Brash JL, ten Hove P. Effect of plasma dilution on adsorption of fibrinogen to solid surfaces. Thromb Haemost. 1984;51:326–330. [PubMed] [Google Scholar]
- 41.Balasubramanian V, Grusin NK, Bucher RW, Turitto VT, Slack SM. Residence-time dependent changes in fibrinogen adsorbed to polymeric biomaterials. Journal of Biomedical Materials Research. 1999;44:253–260. doi: 10.1002/(sici)1097-4636(19990305)44:3<253::aid-jbm3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 42.Chinn JA, Posso SE, Horbett TA, Ratner BD. Postadsorptive transition in fibrinogen adsorbed to polyurethanes: Changes in antibody binding and sodium dodecy1 sulfate elutability. Journal of Biomedical Materials Research. 1992;26:757–778. doi: 10.1002/jbm.820260606. [DOI] [PubMed] [Google Scholar]
- 43.Horbett TA, Lew KR. Residence time effects on monoclonal antibody binding to adsorbed fibrinogen. Journal of Biomaterials Science, Polymer Edition. 1995;6:15–33. doi: 10.1163/156856295x00724. [DOI] [PubMed] [Google Scholar]
- 44.Balasubramanian V, Slack SM. Effects of fibrinogen residence time and shear rate on the morphology and procoagulant activity of human platelets adherent to polymeric biomaterials. Asaio Journal. 2001;47:354–360. doi: 10.1097/00002480-200107000-00012. [DOI] [PubMed] [Google Scholar]
- 45.Horbett TA. Principles underlying the role of adsorbed plasma proteins in blood interactions with foreign materials. Cardiovascular Pathology. 1993;2:137S–148S. [Google Scholar]
- 46.Brash JL. Exploiting the current paradigm of blood-material interactions for the rational design of blood-compatible materials. Journal of Biomaterials Science, Polymer Edition. 2000;11:1135–1146. doi: 10.1163/156856200744237. [DOI] [PubMed] [Google Scholar]
- 47.Roach P, Farrar D, Perry CC. Surface Tailoring for Controlled Protein Adsorption: Effect of Topography at the Nanometer Scale and Chemistry. Journal of the American Chemical Society. 2006;128:3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- 48.Denis FA, et al. Protein adsorption on model surfaces with controlled nanotopography and chemistry. Langmuir. 2002;18:819–828. doi: 10.1021/la011011o. [DOI] [Google Scholar]
- 49.Wang YX, Robertson JL, Spillman WB, Claus RO. Effects of the chemical structure and the surface properties of polymeric biomaterials on their biocompatibility. Pharm Res. 2004;21:1362–1373. doi: 10.1023/b:pham.0000036909.41843.18. [DOI] [PubMed] [Google Scholar]
- 50.Ruckenstein E, Gourisankar SV. Preparation and characterization of thin film surface coatings for biological environments. Biomaterials. 1986;7:403–422. doi: 10.1016/0142-9612(86)90028-1. doi: http://dx.doi.org/10.1016/0142-9612(86)90028-1. [DOI] [PubMed] [Google Scholar]
- 51.Krishnan A, Cha P, Liu Y-H, Allara D, Vogler EA. Interfacial energetics of blood plasma and serum adsorption to a hydrophobic self-assembled monolayer surface. Biomaterials. 2006;27:3187–3194. doi: 10.1016/j.biomaterials.2005.12.032. doi: http://dx.doi.org/10.1016/j.biomaterials.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 52.Krishnan A, Liu Y-H, Cha P, Allara D, Vogler EA. Interfacial energetics of globular–blood protein adsorption to a hydrophobic interface from aqueous-buffer solution. Journal of The Royal Society Interface. 2006;3:283–301. doi: 10.1098/rsif.2005.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cha P, Krishnan A, Fiore VF, Vogler EA. Interfacial energetics of protein adsorption from aqueous buffer to surfaces with varying hydrophilicity. Langmuir. 2008;24:2553–2563. doi: 10.1021/la703310k. [DOI] [PubMed] [Google Scholar]
- 54.Comelles J, Estévez M, Martínez E, Samitier J. The role of surface energy of technical polymers in serum protein adsorption and MG-63 cells adhesion. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6:44–51. doi: 10.1016/j.nano.2009.05.006. doi: http://dx.doi.org/10.1016/j.nano.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Quinn A, Mantz H, Jacobs K, Bellion M, Santen L. Protein adsorption kinetics in different surface potentials. EPL. 2008;81 doi: 10.1209/0295-5075/81/56003. [DOI] [Google Scholar]
- 56.dos Santos EA, Farina M, Soares GA, Anselme K. Surface energy of hydroxyapatite and ss-tricalcium phosphate ceramics driving serum protein adsorption and osteoblast adhesion. Journal of Materials Science-Materials in Medicine. 2008;19:2307–2316. doi: 10.1007/s10856-007-3347-4. [DOI] [PubMed] [Google Scholar]
- 57.Baszkin A, Lyman DJ. The interaction of plasma proteins with polymers. I. Relationship between polymer surface energy and protein adsorption/desorption. Journal of Biomedical Materials Research. 1980;14:393–403. doi: 10.1002/jbm.820140406. [DOI] [PubMed] [Google Scholar]
- 58.Vogler EA. Protein adsorption in three dimensions. Biomaterials. 2012;33:1201–1237. doi: 10.1016/j.biomaterials.2011.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogler EA. In: Water in Biomaterials Surface Science. Morra M, editor. John Wiley and Sons; 2001. pp. 269–290. [Google Scholar]
- 60.Vogler EA. Water and the acute biological response to surfaces. Journal of Biomaterials Science-Polymer Edition. 1999;10:1015–1045. doi: 10.1163/156856299x00667. [DOI] [PubMed] [Google Scholar]
- 61.Vogler EA. The Goldilocks surface. Biomaterials. 2011;32:6670–6675. doi: 10.1016/j.biomaterials.2011.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elbert DL, Hubbell JA. Surface Treatments of Polymers for Biocompatibility. Annual Review of Materials Science. 1996;26:365–294. doi: 10.1146/annurev.ms.26.080196.002053. [DOI] [Google Scholar]
- 63.Vogler EA, Martin DA, Montgomery DB, Graper J, Sugg HW. A Graphical-Method for Predicting Surfactant and Protein Adsorption Properties. Langmuir. 1993;9:497–507. [Google Scholar]
- 64.Parhi P, Golas A, Barnthip N, Noh H, Vogler EA. Volumetric interpretation of protein adsorption: Capacity scaling with adsorbate molecular weight and adsorbent surface energy. Biomaterials. 2009;30:6814–6824. doi: 10.1016/j.biomaterials.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noh H, Vogler EA. Volumetric interpretation of protein adsorption: Mass and energy balance for albumin adsorption to particulate adsorbents with incrementally increasing hydrophilicity. Biomaterials. 2006;27:5801–5812. doi: 10.1016/j.biomaterials.2006.08.005. doi: http://dx.doi.org/10.1016/j.biomaterials.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 66.Hayashi T, Tanaka Y, Koide Y, Tanakac M, Hara M. Mechanism underlying bioinertness of self-assembled monolayers of oligo(ethyleneglycol)-terminated alkanethiols on gold: protein adsorption, platelet adhesion, and surface forces. Physical Chemistry Chemical Physics. 2012;14:10196–10206. doi: 10.1039/c2cp41236e. [DOI] [PubMed] [Google Scholar]
- 67.Kidoaki S, Matsuda T. Mechanistic aspects of protein/material interactions probed by atomic force microscopy. Colloids and Surfaces B-Biointerfaces. 2002;23:153–163. [Google Scholar]
- 68.Xu LC, Siedlecki CA. Effects of surface wettability and contact time on protein adhesion to biomaterial surfaces. Biomaterials. 2007;28:3273–3283. doi: 10.1016/j.biomaterials.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sethuraman A, Han M, Kane RS, Belfort G. Effect of surface wettability on the adhesion of proteins. Langmuir. 2004;20:7779–7788. doi: 10.1021/la049454q. [DOI] [PubMed] [Google Scholar]
- 70.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. A survey of structure-property relationships of surfaces that resist the adsorption of protein. Langmuir. 2001;17:5605–5620. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 71.Evans-Nguyen KM, Tolles LR, Gorkun OV, Lord ST, Schoenfisch MH. Interactions of Thrombin with Fibrinogen Adsorbed on Methyl-, Hydroxyl-, Amine-, and Carboxyl-Terminated Self-Assembled Monolayers. Biochemistry. 2005;44:15561–15568. doi: 10.1021/bi0514358. [DOI] [PubMed] [Google Scholar]
- 72.Benesch J, et al. Protein adsorption to oligo(ethylene glycol) self-assembled monolayers: Experiments with fibrinogen, heparinized plasma, and serum. Journal of Biomaterials Science, Polymer Edition. 2001;12:581–597. doi: 10.1163/156856201316883421. [DOI] [PubMed] [Google Scholar]
- 73.Martins MCL, Ratner BD, Barbosa MA. Protein adsorption on mixtures of hydroxyl- and methylterminated alkanethiols self-assembled monolavers. Journal of Biomedical Materials Research Part A. 2003;67A:158–171. doi: 10.1002/jbm.a.10096. [DOI] [PubMed] [Google Scholar]
- 74.Agashe M, Raut V, Stuart SJ, Latour RA. Molecular simulation to characterize the adsorption behavior of a fibrinogen gamma-chain fragment. Langmuir. 2005;21:1103–1117. doi: 10.1021/la0478346. [DOI] [PubMed] [Google Scholar]
- 75.Tidwell CD, et al. Endothelial Cell Growth and Protein Adsorption on Terminally Functionalized, Self-Assembled Monolayers of Alkanethiolates on Gold. Langmuir. 1997;13:3404–3413. doi: 10.1021/la9604341. [DOI] [Google Scholar]
- 76.Gonçalves IC, Martins MCL, Barbosa MA, Ratner BD. Protein adsorption on 18-alkyl chains immobilized on hydroxyl-terminated self-assembled monolayers. Biomaterials. 2005;26:3891–3899. doi: 10.1016/j.biomaterials.2004.10.006. doi: http://dx.doi.org/10.1016/j.biomaterials.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 77.Tegoulia VA, Rao W, Kalambur AT, Rabolt JF, Cooper SL. Surface Properties, Fibrinogen Adsorption, and Cellular Interactions of a Novel Phosphorylcholine-Containing Self-Assembled Monolayer on Gold. Langmuir. 2001;17:4396–4404. doi: 10.1021/la001790t. [DOI] [Google Scholar]
- 78.Kidoaki S, Matsuda T. Adhesion Forces of the Blood Plasma Proteins on Self-Assembled Monolayer Surfaces of Alkanethiolates with Different Functional Groups Measured by an Atomic Force Microscope. Langmuir. 1999;15:7639–7646. doi: 10.1021/la990357k. [DOI] [Google Scholar]
- 79.Arima Y, Iwata H. Effect of wettability and surface functional groups on protein adsorption and cell adhesion using well-defined mixed self-assembled monolayers. Biomaterials. 2007;28:3074–3082. doi: 10.1016/j.biomaterials.2007.03.013. doi: http://dx.doi.org/10.1016/j.biomaterials.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Barrias CC, Martins MCL, Almeida-Porada G, Barbosa MA, Granja PL. The correlation between the adsorption of adhesive proteins and cell behaviour on hydroxyl-methyl mixed self-assembled monolayers. Biomaterials. 2009;30:307–316. doi: 10.1016/j.biomaterials.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 81.Afara N, Omanovic S, Asghari-Khiavi M. Functionalization of a gold surface with fibronectin (FN) covalently bound to mixed alkanethiol self-assembled monolayers (SAMs): The influence of SAM composition on its physicochemical properties and FN surface secondary structure. Thin Solid Films. 2012;522:381–389. doi: 10.1016/j.tsf.2012.08.025. [DOI] [Google Scholar]
- 82.Brittain WJ, Minko S. A structural definition of polymer brushes. Journal of Polymer Science Part a-Polymer Chemistry. 2007;45:3505–3512. doi: 10.1002/pola.22180. [DOI] [Google Scholar]
- 83.Ayres N. Polymer brushes: Applications in biomaterials and nanotechnology. Polymer Chemistry. 2010;1:769–777. doi: 10.1039/b9py00246d. [DOI] [Google Scholar]
- 84.Ikada Y. Surface modification of polymers for medical applications. Biomaterials. 1994;15:725–736. doi: 10.1016/0142-9612(94)90025-6. doi: http://dx.doi.org/10.1016/0142-9612(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 85.Alibeik S, Zhu S, Brash JL. Surface modification with PEG and hirudin for protein resistance and thrombin neutralization in blood contact. Colloids Surf B Biointerfaces. 2010;81:389–396. doi: 10.1016/j.colsurfb.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 86.Nagaoka S, Nakao A. Clinical application of antithrombogenic hydrogel with long poly (ethylene oxide) chains. Biomaterials. 1990;11:119–121. doi: 10.1016/0142-9612(90)90126-b. doi: http://dx.doi.org/10.1016/0142-9612(90)90126-B. [DOI] [PubMed] [Google Scholar]
- 87.Sofia SJ, Premnath V, Merrill EW. Poly(ethylene oxide) Grafted to Silicon Surfaces: Grafting Density and Protein Adsorption. Macromolecules. 1998;31:5059–5070. doi: 10.1021/ma971016l. [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Zhang Z, Chen Y, Brook MA, Sheardown H. Protein repellant silicone surfaces by covalent immobilization of poly(ethylene oxide) Biomaterials. 2005;26:2391–2399. doi: 10.1016/j.biomaterials.2004.07.068. doi: http://dx.doi.org/10.1016/j.biomaterials.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 89.Chen H, et al. Effect of chain density and conformation on protein adsorption at PEG-grafted polyurethane surfaces. Colloids and Surfaces B-Biointerfaces. 2008;61:237–243. doi: 10.1016/j.colsurfb.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 90.Archambault JG, Brash JL. Protein repellent polyurethane-urea surfaces by chemical grafting of hydroxyl-terminated poly(ethylene oxide): effects of protein size and charge. Colloids and Surfaces B: Biointerfaces. 2004;33:111–120. doi: 10.1016/j.colsurfb.2004.08.009. doi: http://dx.doi.org/10.1016/j.colsurfb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 91.Archambault JG, Brash JL. Protein resistant polyurethane surfaces by chemical grafting of PEO: amino-terminated PEO as grafting reagent. Colloids and Surfaces B: Biointerfaces. 2004;39:9–16. doi: 10.1016/j.colsurfb.2004.08.009. doi: http://dx.doi.org/10.1016/j.colsurfb.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 92.Huang N-P, et al. Poly(l-lysine)-g-poly(ethylene glycol) Layers on Metal Oxide Surfaces: Surface-Analytical Characterization and Resistance to Serum and Fibrinogen Adsorption. Langmuir. 2001;17:489–498. doi: 10.1021/la000736+. [DOI] [Google Scholar]
- 93.Jeon SI, Andrade JD. Protein—surface interactions in the presence of polyethylene oxide: II. Effect of protein size. Journal of Colloid and Interface Science. 1991;142:159–166. doi: http://dx.doi.org/10.1016/0021-9797(91)90044-9. [Google Scholar]
- 94.Jeon SI, Lee JH, Andrade JD, De Gennes PG. Protein—surface interactions in the presence of polyethylene oxide: I. Simplified theory. Journal of Colloid and Interface Science. 1991;142:149–158. doi: http://dx.doi.org/10.1016/0021-9797(91)90043-8. [Google Scholar]
- 95.Gombotz WR, Guanghui W, Horbett TA, Hoffman AS. Protein adsorption to poly(ethylene oxide) surfaces. Journal of Biomedical Materials Research. 1991;25:1547–1562. doi: 10.1002/jbm.820251211. [DOI] [PubMed] [Google Scholar]
- 96.Jin J, Jiang W, Yin J, Ji X, Stagnaro P. Plasma Proteins Adsorption Mechanism on Polyethylene-Grafted Poly(ethylene glycol) Surface by Quartz Crystal Microbalance with Dissipation. Langmuir. 2013;29:6624–6633. doi: 10.1021/la4017239. [DOI] [PubMed] [Google Scholar]
- 97.Szott LM, Stein MJ, Ratner BD, Horbett TA. Complement activation on poly(ethylene oxide)-like radiofrequency glow discharge–deposited surfaces. Journal of Biomedical Materials Research Part A. 2011;96A:150–161. doi: 10.1002/jbm.a.32954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arima Y, Toda M, Iwata H. Complement activation on surfaces modified with ethylene glycol units. Biomaterials. 2008;29:551–560. doi: 10.1016/j.biomaterials.2007.10.015. doi: http://dx.doi.org/10.1016/j.biomaterials.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 99.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 100.Alves NM, Pashkuleva I, Reis RL, Mano JF. Controlling Cell Behavior Through the Design of Polymer Surfaces. Small. 2010;6:2208–2220. doi: 10.1002/smll.201000233. [DOI] [PubMed] [Google Scholar]
- 101.Lord MS, Foss M, Besenbacher F. Influence of nanoscale surface topography on protein adsorption and cellular response. Nano Today. 2010;5:66–78. doi: 10.1016/j.nantod.2010.01.001. [DOI] [Google Scholar]
- 102.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. doi: http://dx.doi.org/10.1016/S0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 103.Su W-T, Chu IM, Yang J-Y, Lin C-D. The geometric pattern of a pillared substrate influences the cell-process distribution and shapes of fibroblasts. Micron. 2006;37:699–706. doi: 10.1016/j.micron.2006.03.007. doi: http://dx.doi.org/10.1016/j.micron.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Lim JY, et al. Osteoblast adhesion on poly(L-lactic acid)/polystyrene demixed thin film blends: Effect of nanotopography, surface chemistry, and wettability. Biomacromolecules. 2005;6:3319–3327. doi: 10.1021/bm0503423. [DOI] [PubMed] [Google Scholar]
- 105.Milner KR, Siedlecki CA. Submicron poly(L-lactic acid) pillars affect fibroblast adhesion and proliferation. Journal of Biomedical Materials Research Part A. 2007;82A:80–91. doi: 10.1002/jbm.a.31049. [DOI] [PubMed] [Google Scholar]
- 106.Lim JY, Hansen JC, Siedlecki CA, Runt J, Donahue HJ. Human foetal osteoblastic cell response to polymer-demixed nanotopographic interfaces. Journal of the Royal Society Interface. 2005;2:97–108. doi: 10.1098/rsif.2004.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dalby MJ, et al. Increasing Fibroblast Response to Materials Using Nanotopography: Morphological and Genetic Measurements of Cell Response to 13-nm-High Polymer Demixed Islands. Experimental Cell Research. 2002;276:1–9. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 108.Magin CM, Cooper SP, Brennan AB. Non-toxic antifouling strategies. Materials Today. 2010;13:36–44. [Google Scholar]
- 109.Hsu LC, Fang J, Borca-Tasciuc DA, Worobo RW, Moraru CI. Effect of Micro- and Nanoscale Topography on the Adhesion of Bacterial Cells to Solid Surfaces. Applied and Environmental Microbiology. 2013;79:2703–2712. doi: 10.1128/aem.03436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Milner KR, Siedlecki CA, Snyder AJ. Development of novel submicron textured polyether(urethane urea) for decreasing platelet adhesion. Asaio Journal. 2005;51:578–584. doi: 10.1097/01.mat.0000171594.44974.89. [DOI] [PubMed] [Google Scholar]
- 111.Milner KR, Snyder AJ, Siedlecki CA. Sub-micron texturing for reducing platelet adhesion to polyurethane biomaterials. Journal of Biomedical Materials Research Part A. 2006;76A:561–570. doi: 10.1002/jbm.a.30554. [DOI] [PubMed] [Google Scholar]
- 112.Koc Y, et al. Nano-scale superhydrophobicity: suppression of protein adsorption and promotion of flow-induced detachment. Lab on a Chip. 2008;8:582–586. doi: 10.1039/b716509a. [DOI] [PubMed] [Google Scholar]
- 113.Mitragotri S, Lahann J. Physical approaches to biomaterial design. Nature Materials. 2009;8:15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luong-Van E, et al. Review: Micro- and nanostructured surface engineering for biomedical applications. Journal of Materials Research. 2013;28:165–174. doi: 10.1557/jmr.2012.398. [DOI] [Google Scholar]
- 115.Vlachopoulou ME, Petrou PS, Kakabakos SE, Tserepi A, Gogolides E. High-aspect-ratio plasma-induced nanotextured poly(dimethylsiloxane) surfaces with enhanced protein adsorption capacity. Journal of Vacuum Science & Technology B. 2008;26:2543–2548. doi: 10.1116/1.3010723. [DOI] [Google Scholar]
- 116.Song WL, Mano JF. Interactions between cells or proteins and surfaces exhibiting extreme wettabilities. Soft Matter. 2013;9:2985–2999. doi: 10.1039/c3sm27739a. [DOI] [Google Scholar]
- 117.Koh LB, Rodriguez I, Venkatraman SS. The effect of topography of polymer surfaces on platelet adhesion. Biomaterials. 2010;31:1533–1545. doi: 10.1016/j.biomaterials.2009.11.022. doi: http://dx.doi.org/10.1016/j.biomaterials.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 118.Leibner ES, et al. Superhydrophobic effect on the adsorption of human serum albumin. Acta Biomaterialia. 2009;5:1389–1398. doi: 10.1016/j.actbio.2008.11.003. doi: http://dx.doi.org/10.1016/j.actbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang Q, et al. Role of trapped air in the formation of cell-and-protein micropatterns on superhydrophobic/superhydrophilic microtemplated surfaces. Biomaterials. 2012;33:8213–8220. doi: 10.1016/j.biomaterials.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 120.Liu C. Rapid fabrication of microfluidic chip with three-dimensional structures using natural lotus leaf template. Microfluid Nanofluid. 2010;9:923–931. doi: 10.1007/s10404-010-0615-2. [DOI] [Google Scholar]
- 121.Chen H, et al. The effect of surface microtopography of poly(dimethylsiloxane) on protein adsorption, platelet and cell adhesion. Colloids and Surfaces B: Biointerfaces. 2009;71:275–281. doi: 10.1016/j.colsurfb.2009.02.018. doi: http://dx.doi.org/10.1016/j.colsurfb.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 122.Rechendorff K, Hovgaard MB, Foss M, Zhdanov VP, Besenbacher F. Enhancement of Protein Adsorption Induced by Surface Roughness. Langmuir. 2006;22:10885–10888. doi: 10.1021/la0621923. [DOI] [PubMed] [Google Scholar]
- 123.Scopelliti PE, et al. The Effect of Surface Nanometre-Scale Morphology on Protein Adsorption. Plos One. 2010;5 doi: 10.1371/journal.pone.0011862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goodman SL, Cooper SL, Albrecht RM. Integrin Receptors and Platelet-Adhesion to Synthetic Surfaces. Journal of Biomedical Materials Research. 1993;27:683–695. doi: 10.1002/jbm.820270516. [DOI] [PubMed] [Google Scholar]
- 125.Farrell DH, Thiagarajan P, Chung DW, Davie EW. Role of fibrinogen alpha-chain and gamma-chain sites in platelet aggregation. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10729–10732. doi: 10.1073/pnas.89.22.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu YG, Simonovsky FI, Ratner BD, Horbett TA. The role of adsorbed fibrinogen in platelet adhesion to polyurethane surfaces: A comparison of surface hydrophobicity, protein adsorption, monoclonal antibody binding, and platelet adhesion. Journal of Biomedical Materials Research Part A. 2005;74A:722–738. doi: 10.1002/jbm.a.30381. [DOI] [PubMed] [Google Scholar]
- 127.Sivaraman B, Latour RA. The relationship between platelet adhesion on surfaces and the structure versus the amount of adsorbed fibrinogen. Biomaterials. 2010;31:832–839. doi: 10.1016/j.biomaterials.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wagner MS, Castner DG. Characterization of Adsorbed Protein Films by Time-of-Flight Secondary Ion Mass Spectrometry with Principal Component Analysis. Langmuir. 2001;17:4649–4660. doi: 10.1021/la001209t. [DOI] [PubMed] [Google Scholar]
- 129.Sivaraman B, Fears KP, Latour RA. Investigation of the Effects of Surface Chemistry and Solution Concentration on the Conformation of Adsorbed Proteins Using an Improved Circular Dichroism Method. Langmuir. 2009;25:3050–3056. doi: 10.1021/la8036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Slack SM, Horbett TA. Changes in fibrinogen adsorbed to segmented polyurethanes and hydroxyethylmethacrylate-ethylmethacrylate copolymers. Journal of Biomedical Materials Research. 1992;26:1633–1649. doi: 10.1002/jbm.820261208. [DOI] [PubMed] [Google Scholar]
- 131.Wertz CF, Santore MM. Fibrinogen adsorption on hydrophilic and hydrophobic surfaces: Geometrical and energetic aspects of interfacial relaxations. Langmuir. 2002;18:706–715. [Google Scholar]
- 132.Sit PS, Marchant RE. Surface-dependent conformations of human fibrinogen observed by atomic force microscopy under aqueous conditions. Thrombosis and Haemostasis. 1999;82:1053–1060. [PubMed] [Google Scholar]
- 133.Agnihotri A, Siedlecki CA. Time-dependent conformational changes in fibrinogen measured by atomic force microscopy. Langmuir. 2004;20:8846–8852. doi: 10.1021/la049239+. [DOI] [PubMed] [Google Scholar]
- 134.Santore MM, Wertz CF. Protein spreading kinetics at liquid-solid interfaces via an adsorption probe method. Langmuir. 2005;21:10172–10178. doi: 10.1021/la051059s. [DOI] [PubMed] [Google Scholar]
- 135.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 136.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophysical Journal. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lo YS, Zhu YJ, Beebe TP. Loading-rate dependence of individual ligand-receptor bond-rupture forces studied by atomic force microscopy. Langmuir. 2001;17:3741–3748. [Google Scholar]
- 138.Bell GI. Models for Specific Adhesion of Cells to Cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 139.Xu LC, Siedlecki CA. Atomic Force Microscopy Studies of the Initial Interactions between Fibrinogen and Surfaces. Langmuir. 2009;25:3675–3681. doi: 10.1021/la803258h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hemmersam AG, Foss M, Chevallier J, Besenbacher F. Adsorption of fibrinogen on tantalum oxide, titanium oxide and gold studied by the QCM-D technique. Colloids and Surfaces B-Biointerfaces. 2005;43:208–215. doi: 10.1016/j.colsurfb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 141.Barattin R, Voyer N. Chemical modifications of AFM tips for the study of molecular recognition events. Chemical Communications. 2008:1513–1532. doi: 10.1039/b614328h. [DOI] [PubMed] [Google Scholar]
- 142.Dufrene YF, Hinterdorfer P. Recent progress in AFM molecular recognition studies. Pflugers Archiv-European Journal of Physiology. 2008;456:237–245. doi: 10.1007/s00424-007-0413-1. [DOI] [PubMed] [Google Scholar]
- 143.Dupres V, Verbelen C, Dufrene YF. Probing molecular recognition sites on biosurfaces using AFM. Biomaterials. 2007;28:2393–2402. doi: 10.1016/j.biomaterials.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 144.Soman P, Rice Z, Siedlecki CA. Immunological identification of fibrinogen in dual-component protein films by AFM imaging. Micron. 2008;39:832–842. doi: 10.1016/j.micron.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Agnihotri A, Siedlecki CA. Adhesion mode atomic force microscopy study of dual component protein films. Ultramicroscopy. 2005;102:257–268. doi: 10.1016/j.ultramic.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 146.Fotiadis D, Scheuring S, Muller SA, Engel A, Muller DJ. Imaging and manipulation of biological structures with the AFM. Micron. 2002;33:385–397. doi: 10.1016/s0968-4328(01)00026-9. [DOI] [PubMed] [Google Scholar]
- 147.Michael KE, et al. Adsorption-induced conformational changes in fibronectin due to interactions with well-defined surface chemistries. Langmuir. 2003;19:8033–8040. doi: 10.1021/la034810a. [DOI] [Google Scholar]
- 148.Soman P, Rice Z, Siedlecki CA. Measuring the Time-Dependent Functional Activity of Adsorbed Fibrinogen by Atomic Force Microscopy. Langmuir. 2008;24:8801–8806. doi: 10.1021/la801227e. [DOI] [PubMed] [Google Scholar]
- 149.Pandey LM, Pattanayek SK. Properties of competitively adsorbed BSA and fibrinogen from their mixture on mixed and hybrid surfaces. Applied Surface Science. 2013;264:832–837. doi: http://dx.doi.org/10.1016/j.apsusc.2012.10.150. [Google Scholar]
- 150.Soman P, Siedlecki CA. Effects of Protein Solution Composition on the Time-Dependent Functional Activity of Fibrinogen on Surfaces. Langmuir. 2011;27:10814–10819. doi: 10.1021/la201111r. [DOI] [PubMed] [Google Scholar]
- 151.Berglin M, et al. Fibrinogen adsorption and conformational change on model polymers: novel aspects of mutual molecular rearrangement. Langmuir. 2009;25:5602–5608. doi: 10.1021/la803686m. [DOI] [PubMed] [Google Scholar]
- 152.Zdrahala RJ, Zdrahala IJ. Biomedical applications of polyurethanes: A review of past promises, present realities, and a vibrant future. Journal of Biomaterials Applications. 1999;14:67–90. doi: 10.1177/088532829901400104. [DOI] [PubMed] [Google Scholar]
- 153.Xue L, Greisler HP. Biomaterials in the development and future of vascular grafts. Journal of Vascular Surgery. 2003;37:472–480. doi: 10.1067/mva.2003.88. [DOI] [PubMed] [Google Scholar]
- 154.Kanyanta V, Ivankovic A. Mechanical characterisation of polyurethane elastomer for biomedical applications. Journal of the Mechanical Behavior of Biomedical Materials. 2010;3:51–62. doi: 10.1016/j.jmbbm.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 155.Lamba NMK, Woodhouse KA, Cooper SL. Polyurethanes in Biomedical Applications. CRC Press; 1998. [Google Scholar]
- 156.Huang SL, Chao MS, Ruaan RC, Lai JY. Microphase separated structure and protein adsorption of polyurethanes with butadiene soft segment. European Polymer Journal. 2000;36:285–294. [Google Scholar]
- 157.Hsu SH, Kao YC. Biocompatibility of poly(carbonate urethane)s with various degrees of nanophase separation. Macromolecular Bioscience. 2005;5:246–253. doi: 10.1002/mabi.200400163. [DOI] [PubMed] [Google Scholar]
- 158.Goodman SL, Simmons SR, Cooper SL, Albrecht RM. Preferential Adsorption of Plasma-Proteins onto Apolar Polyurethane Microdomains. Journal of Colloid and Interface Science. 1990;139:561–570. [Google Scholar]
- 159.Groth T, et al. Protein Adsorption, Lymphocyte Adhesion and Platelet-Adhesion Activation on Polyurethane Ureas Is Related to Hard Segment Content and Composition. Journal of Biomaterials Science-Polymer Edition. 1994;6:497–510. doi: 10.1163/156856294x00464. [DOI] [PubMed] [Google Scholar]
- 160.Grasel TG, Cooper SL. Surface-Properties and Blood Compatibility of Polyurethaneureas. Biomaterials. 1986;7:315–328. doi: 10.1016/0142-9612(86)90002-5. [DOI] [PubMed] [Google Scholar]
- 161.Goodman SL, Grasel TG, Cooper SL, Albrecht RM. Platelet Shape Change and Cytoskeletal Reorganization on Polyurethaneureas. Journal of Biomedical Materials Research. 1989;23:105–123. doi: 10.1002/jbm.820230109. [DOI] [PubMed] [Google Scholar]
- 162.Hsu SH, Kao YC. Cell attachment and proliferation on poly(carbonate urethanes) with various degrees of nanophase separation. Macromolecular Bioscience. 2004;4:891–900. doi: 10.1002/mabi.200400062. [DOI] [PubMed] [Google Scholar]
- 163.Goodman SL, Cooper SL, Albrecht RM. Polyurethane Support Films - Structure and Cellular Adhesion. Scanning Microscopy. 1989:285–295. [PubMed] [Google Scholar]
- 164.Xu L-C, Siedlecki CA. Microphase separation structure influences protein interactions with poly(urethane urea) surfaces. Journal of Biomedical Materials Research Part A. 2010;92A:126–136. doi: 10.1002/jbm.a.32340. [DOI] [PubMed] [Google Scholar]
- 165.Hernandez R, et al. A Comparison of Phase Organization of Model Segmented Polyurethanes with Different Intersegment Compatibilities. Macromolecules. 2008;41:9767–9776. doi: 10.1021/ma8014454. [DOI] [Google Scholar]
- 166.Simmons A, et al. Long-term in vivo biostability of poly(dimethylsiloxane)/poly(hexamethylene oxide) mixed macrodiol-based polyurethane elastomers. Biomaterials. 2004;25:4887–4900. doi: 10.1016/j.biomaterials.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 167.Xu LC, Soman P, Agnihotri A, Siedlecki CA. Biological Interactions on Materials Surfaces. In: Puleo DA, Bizios R, editors. Understandig and Controlling Protein, Cell, and Tissue Responses. Springer; 2009. pp. 44–69. [Google Scholar]
- 168.Tingey KG, Andrade JD. Probing Surface Microheterogeneity of Poly(Ether Urethanes) in an Aqueous Environment. Langmuir. 1991;7:2471–2478. [Google Scholar]
- 169.Agnihotri A, Garrett JT, Runt J, Siedlecki CA. Atomic force microscopy visualization of poly(urethane urea) microphase rearrangements under aqueous environment. Journal of Biomaterials Science-Polymer Edition. 2006;17:227–238. doi: 10.1163/156856206774879036. [DOI] [PubMed] [Google Scholar]
- 170.Xu LC, Soman P, Runt J, Siedlecki CA. Characterization of surface microphase structures of poly(urethane urea) biomaterials by nanoscale indentation with AFM. Journal of Biomaterials Science-Polymer Edition. 2007;18:353–368. doi: 10.1163/156856207780425013. [DOI] [PubMed] [Google Scholar]
- 171.Xu LC, Runt J, Siedlecki CA. Dynamics of hydrated polyurethane biomaterials: Surface microphase restructuring, protein activity and platelet adhesion. Acta Biomaterialia. 2010;6:1938–1947. doi: 10.1016/j.actbio.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 172.Magnani A, Barbucci R, Lewis KB, Leachscampavia D, Ratner BD. Surface-properties and restructuring of a cross-linked polyurethane-poly(amido-amine) network. Journal of Materials Chemistry. 1995;5:1321–1330. [Google Scholar]
- 173.Scardino AJ, Hudleston D, Peng Z, Paul NA, de Nys R. Biomimetic characterisation of key surface parameters for the development of fouling resistant materials. Biofouling. 2009;25:83–93. doi: 10.1080/08927010802538480. Pii 905067268. [DOI] [PubMed] [Google Scholar]
- 174.Scardino AJ, Zhang H, Cookson DJ, Lamb RN, de Nys R. The role of nano-roughness in antifouling. Biofouling. 2009;25:757–767. doi: 10.1080/08927010903165936. [DOI] [PubMed] [Google Scholar]
- 175.Barthlott W, Neinhuis C. Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta. 1997;202:1–8. doi: 10.1007/s004250050096. [DOI] [Google Scholar]
- 176.Bhushan B, Jung YC. Natural and biomimetic artificial surfaces for superhydrophobicity, self-cleaning, low adhesion, and drag reduction. Progress in Materials Science. 2011;56:1–108. doi: 10.1016/j.pmatsci.2010.04.003. [DOI] [Google Scholar]
- 177.Mao Y, et al. In vivo nanomechanical imaging of blood-vessel tissues directly in living mammals using atomic force microscopy. Applied Physics Letters. 2009;95:013704–013703. [Google Scholar]
- 178.Sun T, Qing G, Su B, Jiang L. Functional biointerface materials inspired from nature. Chemical Society Reviews. 2011;40:2909–2921. doi: 10.1039/c0cs00124d. [DOI] [PubMed] [Google Scholar]
- 179.Schumacher JF, et al. Engineered antifouling microtopographies - effect of feature size, geometry, and roughness on settlement of zoospores of the green alga Ulva. Biofouling. 2007;23:55–62. doi: 10.1080/08927010601136957. [DOI] [PubMed] [Google Scholar]
- 180.Scardino AJ, de Nys R. Mini review: Biomimetic models and bioinspired surfaces for fouling control. Biofouling. 2011;27:73–86. doi: 10.1080/08927014.2010.536837. [DOI] [PubMed] [Google Scholar]
- 181.Kirschner CM, Brennan AB. In: Annual Review of Materials Research, Vol 42 Vol. 42 Annual Review of Materials Research. Clarke DR, editor. 2012. pp. 211–229. [Google Scholar]
- 182.Chung KK, et al. Impact of engineered surface microtopography on biofilm formation of Staphylococcus aureus. Biointerphases. 2007;2:89–94. doi: 10.1116/1.2751405. [DOI] [PubMed] [Google Scholar]
- 183.Xu L-C, Siedlecki CA. Submicron-textured biomaterial surface reduces staphylococcal bacterial adhesion and biofilm formation. Acta Biomaterialia. 2012;8:72–81. doi: 10.1016/j.actbio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 184.Xu L-C, Siedlecki CA. Staphylococcus epidermidis adhesion on hydrophobic and hydrophilic textured biomaterial surfaces. Biomedical Materials. 2014 doi: 10.1088/1748-6041/9/3/035003. in press. [DOI] [PubMed] [Google Scholar]
- 185.Anselme K, et al. The interaction of cells and bacteria with surfaces structured at the nanometre scale. Acta Biomaterialia. 2010;6:3824–3846. doi: 10.1016/j.actbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 186.Campoccia D, et al. Study of Staphylococcus aureus adhesion on a novel nanostructured surface by chemiluminometry. International Journal of Artificial Organs. 2006;29:622–629. doi: 10.1177/039139880602900612. [DOI] [PubMed] [Google Scholar]
- 187.Rothstein JP. Annual Review of Fluid Mechanics Vol. 42 Annual Review of Fluid Mechanics. 2010. pp. 89–109. [Google Scholar]
- 188.Sapan CV, Lundblad RL, Price NC. Colorimetric protein assay techniques. Biotechnology and Applied Biochemistry. 1999;29:99–108. doi: 10.1111/j.1470-8744.1999.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 189.Deligianni DD, et al. Effect of surface roughness of the titanium alloy Ti–6Al–4V on human bone marrow cell response and on protein adsorption. Biomaterials. 2001;22:1241–1251. doi: 10.1016/s0142-9612(00)00274-x. doi: http://dx.doi.org/10.1016/S0142-9612(00)00274-X. [DOI] [PubMed] [Google Scholar]
- 190.Aubin-Tam ME, Hamad-Schifferli K. Structure and function of nanoparticle–protein conjugates. Biomedical Materials. 2008;3:034001. doi: 10.1088/1748-6041/3/3/034001. [DOI] [PubMed] [Google Scholar]
- 191.Panyala NR, Maria Pena-Mendez E, Havel J. Gold and nano-gold in medicine: overview, toxicology and perspectives. Journal of Applied Biomedicine. 2009;7:75–91. [Google Scholar]
- 192.Koh LB, Rodriguez I, Venkatraman SS. A novel nanostructured poly(lactic-co-glycolic-acid)–multi-walled carbon nanotube composite for blood-contacting applications: Thrombogenicity studies. Acta Biomaterialia. 2009;5:3411–3422. doi: 10.1016/j.actbio.2009.06.003. doi: http://dx.doi.org/10.1016/j.actbio.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 193.Minelli C, Kikuta A, Yamamoto A. International Conference on Nanoscience and Nanotechnology; 2006. ICONN ‘06. [Google Scholar]
- 194.Ye X, Shao Y-l, Zhou M, Li J, Cai L. Research on micro-structure and hemo-compatibility of the artificial heart valve surface. Applied Surface Science. 2009;255:6686–6690. doi: http://dx.doi.org/10.1016/j.apsusc.2009.02.068. [Google Scholar]
- 195.Linnes JC, Mikhova K, Bryers JD. Adhesion of Staphylococcus epidermidis to biomaterials is inhibited by fibronectin and albumin. Journal of Biomedical Materials Research Part A. 2012;100A:1990–1997. doi: 10.1002/jbm.a.34036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yamamoto S, et al. Relationship between adsorbed fibronectin and cell adhesion on a honeycomb-patterned film. Surface Science. 2006;600:3785–3791. doi: http://dx.doi.org/10.1016/j.susc.2006.01.085. [Google Scholar]
- 197.Gorbet MB, Sefton MVMV. Biomaterial-associated thrombosis: roles of coagulation factors, complement, platelets and leukocytes. Biomaterials. 2004;25:5681–5703. doi: 10.1016/j.biomaterials.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 198.Vogler EA, Siedlecki CA. Contact activation of blood-plasma coagulation. Biomaterials. 2009;30:1857–1869. doi: 10.1016/j.biomaterials.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 200.Dahlbäck B. Blood coagulation. The Lancet. 2000;355:1627–1632. doi: 10.1016/S0140-6736(00)02225-X. doi: http://dx.doi.org/10.1016/S0140-6736(00)02225-X. [DOI] [PubMed] [Google Scholar]
- 201.Jenny NS, Mann KG. In: Thrombosis and Hemorrhage. Loscalzo J, Schafer A, editors. Lippincott, Williams & Wilkins; 2002. pp. 1–21. [Google Scholar]
- 202.RWC, VJM, EWS, JH . In: Hemostasis and Thrombosis: Basic Principles and Clinical Practice. Colman RW, Marder VJ, Salzman EW, Hirsh J, editors. J.B. Lippincott Company; 1994. pp. 3–18. [Google Scholar]
- 203.Colman RW, et al. Initiation of Blood Coagulation at Artificial Surfaces. Annals of the New York Academy of Sciences. 1987;516:253–267. doi: 10.1111/j.1749-6632.1987.tb33046.x. [DOI] [PubMed] [Google Scholar]
- 204.Pokhilko AV, Ataullakhanov FI. Contact Activation of Blood Coagulation: Trigger Properties and Hysteresis Hypothesis: Kinetic Recognition of Foreign Surfaces upon Contact Activation of Blood Coagulation: A Hypothesis. Journal of Theoretical Biology. 1998;191:213–219. doi: 10.1006/jtbi.1997.0584. doi: http://dx.doi.org/10.1006/jtbi.1997.0584. [DOI] [PubMed] [Google Scholar]
- 205.Griffin JH. Role of surface in surface-dependent activation of Hageman factor (blood coagulation Factor XII) Proceedings of the National Academy of Sciences. 1978;75:1998–2002. doi: 10.1073/pnas.75.4.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meier HL, Pierce JV, Colman RW, Kaplan AP. Activation and function of human Hageman factor. The role of high molecular weight kininogen and prekallikrein. Journal of Clinical Investigation. 1977;60:18–31. doi: 10.1172/jci108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhuo R, Vogler EA. Practical application of a chromogenic FXIIa assay. Biomaterials. 2006;27:4840–4845. doi: 10.1016/j.biomaterials.2006.05.008. doi: http://dx.doi.org/10.1016/j.biomaterials.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 208.Wiggins RC, Cochrane CC. The autoactivation of rabbit Hageman factor. J Exp Med. 1979;150:1122–1133. doi: 10.1084/jem.150.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Miller G, Silverberg M, Kaplan AP. Autoactivatability of human Hageman factor (factor XII) Biochemical and Biophysical Research Communications. 1980;92:803–810. doi: 10.1016/0006-291x(80)90774-3. doi: http://dx.doi.org/10.1016/0006-291X(80)90774-3. [DOI] [PubMed] [Google Scholar]
- 210.Guo Z, et al. Mathematical modeling of material-induced blood plasma coagulation. Biomaterials. 2006;27:796–806. doi: 10.1016/j.biomaterials.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 211.Chatterjee K, Guo Z, Vogler EA, Siedlecki CA. Contributions of contact activation pathways of coagulation factor XII in plasma. J Biomed Mater Res A. 2009;90:27–34. doi: 10.1002/jbm.a.32076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vogler EA. Contact Activation of the Plasma Coagulation Cascade.1. Procoagulant Surface-Chemistry and Energy. Journal of Biomedical Materials Research. 1995;29:1005–1016. doi: 10.1002/jbm.820290813. [DOI] [PubMed] [Google Scholar]
- 213.Golas A, Parhi P, Dimachkie ZO, Siedlecki CA, Vogler EA. Surface-energy dependent contact activation of blood factor XII. Biomaterials. 2010;31:1068–1079. doi: 10.1016/j.biomaterials.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhuo R, Siedlecki CA, Vogler EA. Autoactivation of blood factor XII at hydrophilic and hydrophobic surfaces. Biomaterials. 2006;27:4325–4332. doi: 10.1016/j.biomaterials.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 215.Griep MA, Fujikawa K, Nelsestuen GL. Possible basis for the apparent surface selectivity of the contact activation of human blood coagulation factor XII. Biochemistry. 1986;25:6688–6694. doi: 10.1021/bi00369a054. [DOI] [PubMed] [Google Scholar]
- 216.Mitropoulos KA. High affinity binding of factor XIIa to an electronegative surface controls the rates of factor XII and prekallikrein activation in vitro. Thrombosis research. 1999;94:117–129. doi: 10.1016/s0049-3848(98)00207-2. [DOI] [PubMed] [Google Scholar]
- 217.Vogler EA, Graper JC, Sugg HW, Lander LM, Brittain WJ. Contact activation of the plasma coagulation cascade. II. Protein adsorption to procoagulant surfaces. J Biomed Mater Res. 1995;29:1017–1028. doi: 10.1002/jbm.820290814. [DOI] [PubMed] [Google Scholar]
- 218.Chatterjee K, Thornton JL, Bauer JW, Vogler EA, Siedlecki CA. Moderation of prekallkrein-factor XII interactions in surface activation of coagulation by protein-adsorption competition. Biomaterials. 2009;30:4915–4920. doi: 10.1016/j.biomaterials.2009.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhuo R, Miller R, Bussard KM, Siedlecki CA, Vogler EA. Procoagulant stimulus processing by the intrinsic pathway of blood plasma coagulation. Biomaterials. 2005;26:2965–2973. doi: 10.1016/j.biomaterials.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 220.Golas A, Yeh C-HJ, Pitakjakpipop H, Siedlecki CA, Vogler EA. A comparison of blood factor XII autoactivation in buffer, protein cocktail, serum, and plasma solutions. Biomaterials. 2013;34:607–620. doi: 10.1016/j.biomaterials.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhuo R, Siedlecki CA, Vogler EA. Competitive-protein adsorption in contact activation of blood factor XII. Biomaterials. 2007;28:4355–4369. doi: 10.1016/j.biomaterials.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yeh C-HJ, et al. Contact activation of blood plasma and factor XII by ion-exchange resins. Biomaterials. 2012;33:9–19. doi: 10.1016/j.biomaterials.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sperling C, Schweiss RB, Streller U, Werner C. In vitro hemocompatibility of self-assembled monolayers displaying various functional groups. Biomaterials. 2005;26:6547–6557. doi: 10.1016/j.biomaterials.2005.04.042. doi: http://dx.doi.org/10.1016/j.biomaterials.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 224.van der Kamp KWHJ, Hauch KD, Feijen J, Horbett TA. Contact activation during incubation of five different polyurethanes or glass in plasma. Journal of Biomedical Materials Research. 1995;29:1303–1306. doi: 10.1002/jbm.820291018. [DOI] [PubMed] [Google Scholar]
- 225.Miller R, Guo Z, Vogler EA, Siedlecki CA. Plasma coagulation response to surfaces with nanoscale chemical heterogeneity. Biomaterials. 2006;27:208–215. doi: 10.1016/j.biomaterials.2005.05.087. [DOI] [PubMed] [Google Scholar]
- 226.Bäck J, Sanchez J, Elgue G, Ekdahl KN, Nilsson B. Activated human platelets induce factor XIIa-mediated contact activation. Biochemical and Biophysical Research Communications. 2010;391:11–17. doi: 10.1016/j.bbrc.2009.10.123. doi: http://dx.doi.org/10.1016/j.bbrc.2009.10.123. [DOI] [PubMed] [Google Scholar]
- 227.Sperling C, Fischer M, Maitz MF, Werner C. Blood coagulation on biomaterials requires the combination of distinct activation processes. Biomaterials. 2009;30:4447–4456. doi: 10.1016/j.biomaterials.2009.05.044. doi: http://dx.doi.org/10.1016/j.biomaterials.2009.05.044. [DOI] [PubMed] [Google Scholar]