Abstract
Mg2+ serves as an essential cofactor for numerous enzymes and its levels are tightly regulated by various Mg2+ transporters. Here, we analyzed Caenorhabditis elegans strains carrying mutations in genes encoding cyclin M (CNNM) Mg2+ transporters. We isolated inactivating mutants for each of the five Caenorhabditis elegans cnnm family genes, cnnm-1 through cnnm-5. cnnm-1; cnnm-3 double mutant worms showed various phenotypes, among which the sterile phenotype was rescued by supplementing the media with Mg2+. This sterility was caused by a gonadogenesis defect with severely attenuated proliferation of germ cells. Using this gonadogenesis defect as an indicator, we performed genome-wide RNAi screening, to search for genes associated with this phenotype. The results revealed that RNAi-mediated inactivation of several genes restores gonad elongation, including aak-2, which encodes the catalytic subunit of AMP-activated protein kinase (AMPK). We then generated triple mutant worms for cnnm-1; cnnm-3; aak-2 and confirmed that the aak-2 mutation also suppressed the defective gonadal elongation in cnnm-1; cnnm-3 mutant worms. AMPK is activated under low-energy conditions and plays a central role in regulating cellular metabolism to adapt to the energy status of cells. Thus, we provide genetic evidence linking Mg2+ homeostasis to energy metabolism via AMPK.
Author Summary
Mg2+ is the second most abundant cation in cells and serves as an essential cofactor for numerous enzymes. To avoid its shortage, cellular and organismal levels of Mg2+ are tightly regulated by the concerted actions of various Mg2+ transporters and channels. In this study, we analyzed Caenorhabditis elegans strains carrying mutations in genes encoding Mg2+ transporters and found that the mutations abrogated Mg2+ homeostasis. Additionally, these worms were sterile because of a developmental defect in the gonads with severely attenuated proliferation of germ cells. These abnormalities were rescued by additional Mg2+ supplementation to the medium, and thus were considered to be due to Mg2+ shortage. We investigated the mechanism of this Mg2+-associated attenuation of gonadal development, and found that disrupting of the function of AMP-activated protein kinase (AMPK) restored gonad elongation. It is well-known that AMPK is activated under low-energy conditions and plays a central role in regulating cellular metabolism to adapt to the energy status of cells. Thus, we demonstrated that Mg2+ homeostasis is intimately connected to energy metabolism via AMPK.
Introduction
Mg2+ is the second most abundant cation in cells and serves as an essential cofactor for numerous enzymes. In mammals, magnesium levels are primarily regulated by intestinal absorption and renal reabsorption, where the epithelial cell layer permits selective and regulated Mg2+ transport between apical and basolateral surfaces. There are two known pathways for Mg2+ transport through the epithelial cell layers: the paracellular and the transcellular pathways [1]. The transcellular pathway consists of apical entry and basolateral extrusion mediated by Mg2+-permeable cation channels and transporters. TRPM6, a member of the transient receptor potential channel (TRP) family, is a key molecule in the transcellular pathway [2]. TRPM6 localizes at the apical membrane of intestinal epithelial cells and distal convoluted tubule (DCT) cells in the kidney [3] and mediates Mg2+ absorption and reabsorption, respectively. Indeed, mutations in TRPM6 result in recessive familial hypomagnesemia with secondary hypocalcemia [4,5]. In addition, the related channel TRPM7 was also found to play an important role in magnesium homeostasis in mice [6]. These observations implicate TRPM6/7 in the apical entry of Mg2+ into epithelial cells.
Another key molecule in the transcellular pathway is the ancient conserved domain protein/cyclin M (CNNM) family. In mammals, the CNNM family consists of 4 integral membrane proteins (CNNM1–4) that possess an evolutionarily conserved domain from bacteria [7]. Recent genomic analyses have revealed that several single-nucleotide polymorphisms in CNNMs are linked to serum magnesium levels [8] and that mutations in CNNM2 are responsible for familial hypomagnesemia [9]. It was reported that CNNM4 extrudes Mg2+ from the basolateral membrane of intestinal epithelial cells and is involved in intestinal Mg2+ absorption [10]. Another family member, CNNM2, is strongly expressed at the basolateral membrane of DCT cells [9,11] and can extrude Mg2+ similarly to CNNM4 [12], suggesting that CNNM2 plays a similar role in basolateral Mg2+ extrusion in kidney DCT cells.
Two groups recently reported that CNNMs associate with phosphatase of regenerating liver (PRL), a cancer-associated tyrosine phosphatase [13,14]. One group found that PRL binds to CNNMs and inhibits the Mg2+-transporting function [13], whereas the other group reported that PRL stimulates this activity [14]. Therefore, how PRL affects the function of CNNMs remains unclear. In addition, it is unknown whether other molecules are involved in the regulation of CNNM function. To address these problems, comprehensive screening can be used to identify genes that functionally associate with CNNMs. Caenorhabditis elegans (C. elegans), which is a model organism commonly used for genetic analyses, also absorbs Mg2+ through a similar transcellular mechanism in the intestine. The apical entry step of the transcellular pathway is mediated by two TRPM family channels: GTL-1 and GON-2 [15]. C. elegans has an excretory canal that removes wastes from the body, wherein another C. elegans TRPM channel, GTL-2, plays an important role in magnesium homeostasis [16]. Thus, C. elegans possesses a system for regulating magnesium homeostasis that is similar to that in mammals. Taken together with the genetic tractability of C. elegans, this organism may serve as an ideal experimental model for investigating the regulatory mechanism and functional importance of magnesium homeostasis.
In this study, we performed functional analyses of the C. elegans CNNM family and found that cnnm-1; cnnm-3 double mutant worms displayed pleiotropic phenotypes. The sterile phenotype (due to defective gonadogenesis) was rescued by Mg2+ supplementation: oocyte development was restored and mutant worms became fertile. Detailed analyses of the gonadal phenotype revealed that the inactivating mutation of aak-2, which encodes the α subunit of AMP-activated protein kinase (AMPK), significantly rescued the gonadogenesis defect in cnnm-1; cnnm-3 mutants, thereby indicating a genetic interaction between CNNM and AMPK.
Results
cnnm family genes of C. elegans
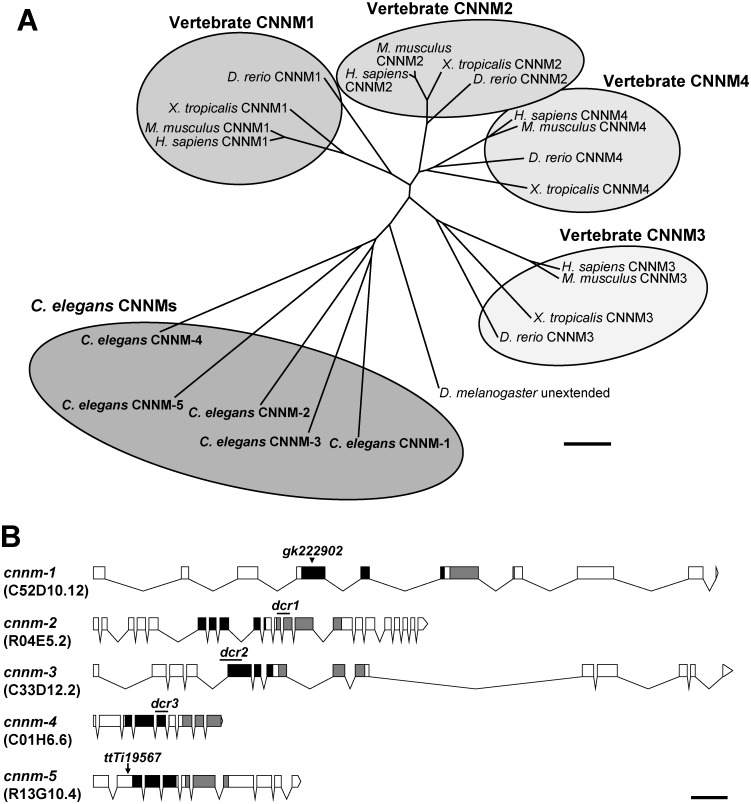
A homology search using BLAST with amino acid sequences of the human CNNM4 protein revealed that the C. elegans genome contains genes encoding 5 previously uncharacterized CNNM family proteins, which possess the functionally essential domains DUF21 and CBS [10,12] (S1 Fig). Each C. elegans CNNM protein showed significant identity with all human CNNM family members (24–47%). To determine the evolutionary relationship between C. elegans CNNM proteins and other CNNM homologs, we constructed a phylogenetic tree (Fig 1A). The vertebrates have 4 paralogs (CNNM1–4), and each of them is orthologous between different vertebrate species (human, mouse, frog, and zebrafish). In contrast, C. elegans CNNM-1−5 emerged independently of the vertebrate CNNMs. To investigate the in vivo functions of C. elegans CNNM family proteins, we obtained and generated mutant alleles for all cnnm family members (Fig 1B). These mutations abolish the function of each CNNM protein because the mutant proteins lack functionally essential part of either the DUF21 or CBS domains (S1 Fig, see its legend for details).
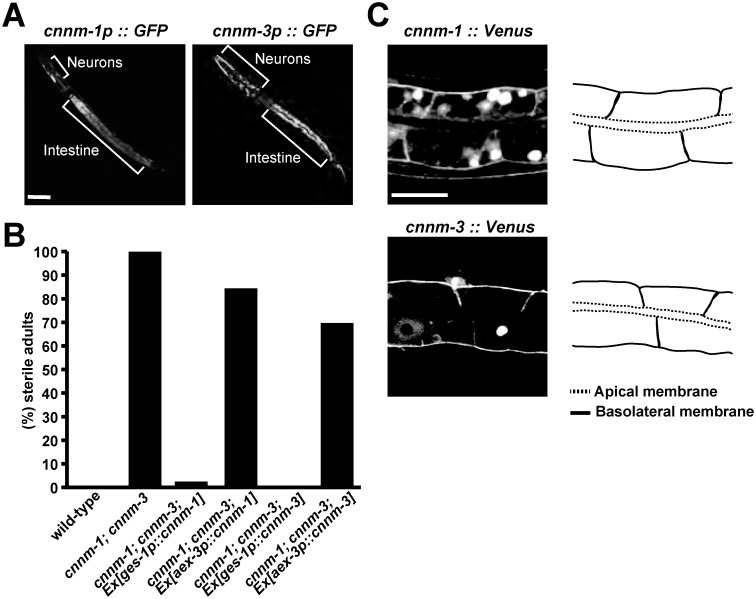
Fig 1. The C. elegans cnnm family.
(A) Phylogenetic tree of CNNM family. Amino acid sequences of C. elegans CNNMs, along with human, mouse, frog, zebrafish, and fruit fly CNNM orthologues, were aligned using Clustal W (version 2.1, http://clustalw.ddbj.nig.ac.jp/), and the phylogenetic tree was generated with neighbor-joining method by using Clustal W2—phylogeny (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/). Vertebrate CNNM orthologues are grouped, and C. elegans CNNMs are also grouped. Bar, 0.1 substitutions per amino acid. (B) Schematic illustrations of the cnnm family genes. Exons and introns are indicated by boxes and lines, respectively. The regions encoding the evolutionarily conserved DUF21 and CBS domains are indicated with black and gray boxes, respectively. Sequence names are shown in parentheses. The illustrations were generated using the Exon-Intron Graphic Maker by Nikhil Bhatla (http://www.wormweb.org/exonintron). Lines, deletion; arrowhead, point mutation; arrow, Mos1 insertion. Bar, 0.5 kb.
Pleiotropic phenotypes of cnnm-1; cnnm-3 mutant worms
Observation of these mutant worms revealed no obvious abnormalities except for in the cnnm-3 mutant worms, a few (5.3%) of which were sterile (Fig 2A). We speculated that functional redundancy among the cnnm family members may have masked the mutant phenotype. Thus, we generated double mutants for all possible combinations by crossing all single mutants. We found that cnnm-1; cnnm-3 and cnnm-2; cnnm-3 mutant worms were severely (100%) and moderately (22%) sterile, respectively (Fig 2A). Because of the completely sterile phenotype, we analyzed cnnm-1; cnnm-3 mutants in subsequent experiments.
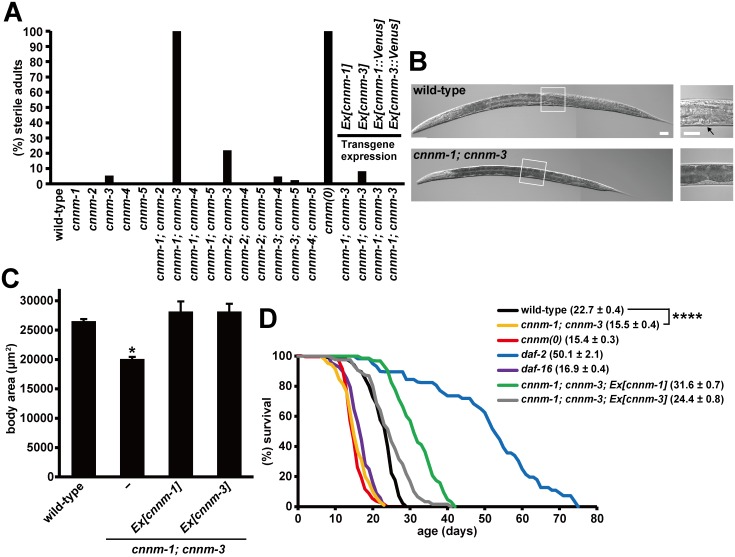
Fig 2. Pleiotropic phenotypes of cnnm-1; cnnm-3 mutant worms.
(A) Quantification of the sterile phenotype for each genotype. Eggs collected by synchronous laying were grown for 4 days and then examined for the presence of embryos in their uteri by microscopic observation. Worms lacking embryos were scored as sterile. More than 100 worms were analyzed for each genotype. cnnm(0): cnnm-1; cnnm-2; cnnm-3; cnnm-4; cnnm-5 mutant. (B) Nomarski images of wild-type and cnnm-1; cnnm-3 mutant worms grown for 3 days. High-magnification views of the boxed areas are also shown (right). Arrow indicates the vulva. Bar, 30 μm. (C) Mixed stage worms from L2 to adult (L4 to adult worms were most abundant) were examined for alae formation and subjected to body size measurement (n = 30 in each experiment). The body size at the transition from L4 to the adult molt was estimated by calculating the mean body size of the smallest three worms with alae and the largest three worms without alae. The data are shown as the means of three experiments. Error bars indicate SEM. p values were determined by ANOVA, followed by two-tailed multiple Student’s t-test with Tukey’s correction. *p < 0.05 versus wild-type. (D) Lifespan of worms with the indicated genotype. For each genotype, 45 synchronized L4/young adult worms were transferred to fresh plates (15 worms per plate) and were then scored daily for survival. The graph represents data combined from at least two experiments. Mean lifespan (± SEM) of worms is also indicated in parentheses. p values were determined by log rank (Mantel-Cox) test, and the Bonferroni method was then used to correct for multiple comparisons. ****p < 0.0001.
The cnnm-1; cnnm-3 mutant worms were significantly smaller than the wild-type N2 worms grown for the same amount of time (Fig 2B). Therefore, we compared the body sizes of stage-matched worms. cnnm-1; cnnm-3 mutant worms did not form the vulva (Fig 2B), the eversion of which determines the adult stage [17]. Therefore, we focused on the presence of alae, the longitudinal ridges present in adult worms but not in earlier L2−4 larvae as the marker to confirm whether the worms had reached the adult stage [18,19]. We examined alae formation and body sizes of mixed stage worms from L2 to adult, and then estimated the body size at the transition from L4 to adult molt, which was determined as the mean value of body size of the three smallest worms with alae and three largest worms without alae. The results showed that the body size of cnnm-1; cnnm-3 mutant worms was smaller than that of wild-type worms (Fig 2C). At 64 h when all wild-type worms had just reached the adult stage, approximately half of the cnnm-1; cnnm-3 mutant worms were alae-positive, indicating developmental delay. These phenotypes of cnnm-1; cnnm-3 mutant worms were rescued by the introduction of either cnnm-1 or cnnm-3 genomic DNA (Fig 2A and 2C), confirming that these abnormalities were caused by mutations in cnnm-1 and cnnm-3.
We also found that the color of the intestine in cnnm-1; cnnm-3 mutant worms was dark (Fig 2B). This was also observed in mutant worms of daf-2, which encodes the insulin-like receptor [20]. Because daf-2 mutations are well-known to extend lifespan, we next examined the lifespan of cnnm-1; cnnm-3 mutant worms (Fig 2D). Consistent with previous studies, daf-2 mutant worms showed much longer lifespans than wild-type worms. In contrast, cnnm-1; cnnm-3 mutant worms had shorter lifespans than wild-type worms, which were similar to those of daf-16 mutant worms; these worms were used here as an example of short-lived mutants [20]. Introduction of either cnnm-1 or cnnm-3 genomic DNA to cnnm-1; cnnm-3 mutant worms not only rescued the short-life phenotype, but also showed moderately longer lifespans than wild-type worms, particularly in the case of worms introduced with cnnm-1 (Fig 2D). Overexpression of either cnnm-1 or cnnm-3 in wild-type worms also prolonged the lifespan (S2 Fig), indicating the important role of these genes in determining lifespan.
We also analyzed whether the short-life phenotype of cnnm-1; cnnm-3 mutants could be exacerbated by further mutations in other CNNM family genes. For this, we generated cnnm-1; cnnm-2; cnnm-3; cnnm-4; cnnm-5 quintuple mutant worms (hereafter, cnnm(0)), which possessed mutations in all five C. elegans cnnm family genes, by mating. As expected, cnnm(0) worms showed complete sterility (Fig 2A) with a lifespan similar to that of cnnm-1; cnnm-3 mutants (Fig 2D).
Effects of Mg2+ supplementation on cnnm-1; cnnm-3 mutant worms
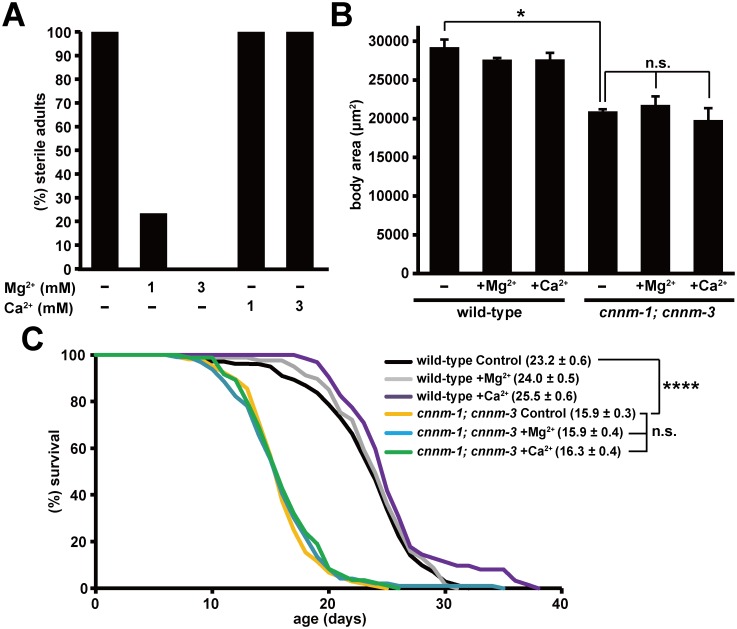
Because mammalian CNNM family proteins are involved in Mg2+ transport [10,12–14], the phenotypes of cnnm-1; cnnm-3 mutant worms may be related to abnormalities in magnesium homeostasis. Therefore, we tested the effects of Mg2+ supplementation in the media, and found that 76.6% and 100% of cnnm-1; cnnm-3 mutant worms became fertile following supplementation with 1 mM and 3 mM of Mg2+, respectively (Fig 3A). In contrast, the small body size and short lifespan were not affected by Mg2+ supplementation (Fig 3B and 3C). Supplementation of culture plates with Ca2+ did not affect any of the phenotypes. Collectively, these results suggest that the sterile phenotype of cnnm-1; cnnm-3 mutant worms is related to altered magnesium homeostasis. As the cnnm-1; cnnm-3 mutant worms did not produce oocytes or form a vulva (Fig 2B), which are formed during gonadal development, we next examined gonadal development in cnnm-1; cnnm-3 mutant worms.
Fig 3. Effects of Mg2+ supplementation on cnnm-1; cnnm-3 mutant worms.
(A) cnnm-1; cnnm-3 mutant worms were grown on plates supplemented with the indicated concentrations of Mg2+ or Ca2+ for 4 days and then examined for sterility. More than 50 worms were analyzed for each experimental condition. (B) Wild-type and cnnm-1; cnnm-3 mutant worms were grown on plates supplemented with 1 mM Mg2+ or Ca2+ for at least two generations. Mixed stage worms from L2 to adult (L4 to adult worms comprise the most) were examined for alae formation and subjected to body size measurement (n = 30 per each experiment). Body size at the transition from L4 to adult molt was estimated as in Fig 2C. The data are shown as the means of three experiments. Error bars indicate SEM. p values were determined by ANOVA, followed by two-tailed multiple Student’s t-tests with Tukey’s correction. *p < 0.05. (C) 45 synchronized L4/young adult worms of the indicated genotype were transferred to fresh plates supplemented with 1 mM Mg2+ or Ca2+ (15 worms per plate), and then scored for survival daily. The graph represents data combined from at least two experiments. Mean lifespan (± SEM) of worms is also indicated in parentheses. p values were determined by log rank (Mantel-Cox) test, and the Bonferroni method was then used to correct for multiple comparisons. ****p < 0.0001.
cnnm-1 and cnnm-3 are required for postembryonic gonadal development
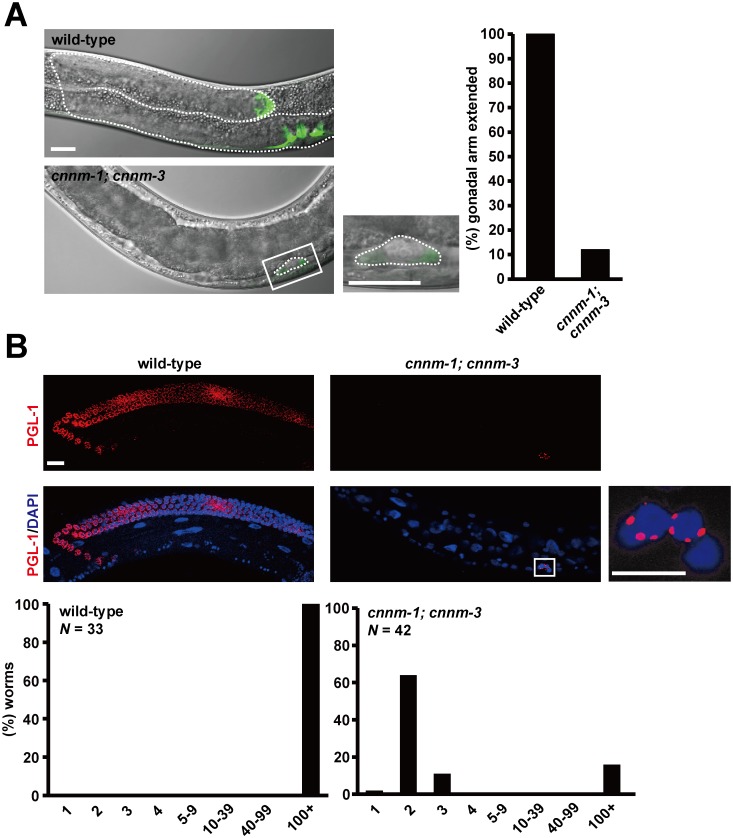
At hatching, the primordial gonad in C. elegans is composed of four cells: Z1–Z4 [21]. During larval development, Z1 and Z4 cells give rise to the somatic gonad including the distal tip cells (DTCs), uterus, sheath cells, and spermathecae, whereas the Z2 and Z3 cells give rise to the germ line. We examined the extent of gonadal development by expressing GFP under control of the lag-2 promoter, which drives gene expression in Z1/Z4 cells and DTCs [22,23] located at the distal end of two gonadal arms and lead the elongation of the arms to form the U-shaped hermaphrodite gonad. When the mutant worms hatched from eggs, two GFP-positive cells were observed at the appropriate positions, suggesting that primordial gonad development proceeded normally in mutants. However, the primordial gonad of mutants did not elongate, even in the L4/young adult stages (Fig 4A). Immunofluorescence analysis using an anti-PGL-1 antibody, which stains P-granules in germ cells, demonstrated that most of the cnnm-1; cnnm-3 mutant L4/young adult worms had only two germ cells corresponding to the Z2/Z3 cells (Fig 4B). Considering that Mg2+ supplementation restored the fertility of cnnm-1; cnnm-3 mutant worms, these results suggest that CNNM-1 and CNNM-3 promote postembryonic gonadal development by regulating Mg2+ levels. Previous studies demonstrated that mutants of gon-2, which encodes a TRPM channel protein, showed a severe gonadogenesis defect, which was partially restored by Mg2+ supplementation [15,24].
Fig 4. cnnm-1 and cnnm-3 are required for postembryonic development of the gonad.
(A) L4/young adult worms of the indicated genotype, carrying lag-2p::GFP, were examined for gonadal arm extension. Representative images and the high-magnification view of the boxed area are shown (left). In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. Bar, 20 μm. Worms with gonads of ≥ 60 μm in length of the long axis (3-fold as long as the primordial gonad) were determined as extended and the results (%) are shown on the graph (right). More than 100 worms were analyzed. (B) The worms were stained with DAPI (blue) and anti-PGL-1 antibody (red), and then the number of germ cells (PGL-1-positive cells) per worm was counted. Representative images and the high-magnification view of the boxed area are shown (top). Bar, 15 μm. The graph indicates the distribution of worms (%) with the indicated numbers of germ cells (bottom).
In addition to somatic gonad and germ cells, several types of cells, such as the ray cells of the male tail, are known to proliferate during postembryonic stages [25]. Our findings revealed no apparent abnormalities in the tail ray structures of cnnm-1; cnnm-3 mutant males (S3 Fig), suggesting that not all of the postembryonic cell divisions were affected in cnnm-1; cnnm-3 mutant worms.
Localization of CNNM-1 and CNNM-3 at the basolateral membrane of intestinal cells
To characterize the roles of CNNM-1 and CNNM-3, we first examined their expression patterns by generating transgenic worms expressing GFP under control of the cnnm-1 or cnnm-3 promoters. Unique GFP expression was observed in various tissues, such as the pharynx, hypodermis, rectum, and muscles, but strong expression was commonly observed in the intestine and neurons (Fig 5A). Given this expression pattern, we forced the expression of CNNM-1 or CNNM-3 in the intestine or neurons of cnnm-1; cnnm-3 mutant worms using the intestine-specific ges-1 promoter [26] or neuron-specific aex-3 promoter [27], respectively. The intestinal expression of either CNNM-1 or CNNM-3 nearly completely rescued the sterile phenotype of cnnm-1; cnnm-3 mutants, whereas their expression in neurons showed only a subtle effect (Fig 5B), suggesting that their expression in the intestine is important for gonadal development. The intestinal cells of C. elegans are attached to each other at the borders of the apical membrane via cell-cell junctions known as apical junctions, which have mixed traits of both the adherens junction and the tight junction in mammalian epithelial cells, and thus exhibit apico-basal polarity [28]. We subsequently examined the subcellular localization of CNNM-1 and CNNM-3 in intestinal cells using transgenic worms expressing their respective Venus-fusion proteins, which also rescued the sterile phenotype (Fig 2A). Excluding some large clumps in the cytoplasm, which are often observed following ectopic expression of Venus-fusion proteins, the fluorescent signal of Venus-fusion CNNM1 and CNNM3 was predominantly observed in the basolateral membrane of intestinal cells (Fig 5C). Therefore, both CNNM-1 and CNNM-3 are considered to extrude Mg2+ from intestinal cells to the pseudocoelom. Based on this hypothesis, cnnm-1; cnnm-3 mutant worms were expected to have higher levels of Mg2+ in intestinal cells and lower levels of Mg2+ in the pseudocoelom, which contains other tissues such as the gonad, possibly explaining why Mg2+ supplementation restored gonadal development (Fig 3A).
Fig 5. Localization of CNNM-1 and CNNM-3 at the basolateral membrane of intestinal cells.
(A) Fluorescent images of wild-type L1 larvae with cnnm-1p::GFP or cnnm-3p::GFP. Areas of neurons and intestines showing strong GFP signals are marked. Bar, 30 μm. (B) Quantification of sterile phenotype in wild-type, cnnm-1; cnnm-3 mutants, and cnnm-1; cnnm-3 mutants expressing either CNNM-1 or CNNM-3 under the control of the promoters for intestinal (ges-1p) or pan-neuronal (aex-3p) markers. More than 50 worms were analyzed for each genotype. (C) Fluorescent images of intestinal cells from wild-type L1 larvae expressing the Venus-fusion proteins of CNNM-1 or CNNM-3 (left). Schematic representation of the intestinal cell plasma membranes are also shown (right). Apical and basolateral membranes are indicated by dotted and solid lines, respectively. Bar, 10 μm.
Assessment of magnesium levels by ICP-MS
To assess the predicted regulatory model, we quantified the magnesium levels in wild-type worms and in cnnm-1; cnnm-3 mutant worms using inductively coupled plasma mass spectrometry (ICP-MS). As shown in Table 1, we found that cnnm-1; cnnm-3 mutant worms had higher magnesium levels (143% of wild-type worms). We then physically dissected the intestines from the worms using a scalpel and subjected the intestines to magnesium quantitation. The results indicated much higher levels of magnesium in cnnm-1; cnnm-3 mutant worms (195% of wild-type worms). It should be noted that there was no overlap in magnesium levels in wild-type and mutant worms. It was technically difficult to physically dissect other remaining tissues, which are much smaller than the intestine. Therefore, we estimated magnesium levels in other tissues by calculating the volumes of the total body and intestine, and found that the magnesium level was reduced in other tissues of cnnm-1; cnnm-3 mutant worms (67% of wild-type levels). Taken together with the result that the infertile phenotype was rescued by artificially increasing Mg2+ levels (Fig 3), the abnormalities in cnnm-1; cnnm-3 mutant worms may have been caused by Mg2+ deficiency.
Table 1. Magnesium levels in wild-type and cnnm-1; cnnm-3 mutant worms.
| wild-type | cnnm-1; cnnm-3 | p-value | |
|---|---|---|---|
| Total (ppb/μg) | 3.9 ± 0.2 | 5.6 ± 0.4 | 0.014 |
| Intestine (ppb/μg) | 4.0 ± 0.4 | 7.8 ± 0.8 | 0.015 |
| Other tissues (ppb/μg) (estimated value) | 3.9 | 2.6 | ND |
The total and intestinal levels of magnesium were directly determined by ICP-MS using L4/young adult worms. The data are shown as the means (± SEM) of three experiments. Magnesium levels in the other remaining tissues were estimated by calculating the total body volume (wild-type: 1,170 ± 61 nl, n = 10; cnnm-1; cnnm-3: 638 ± 38 nl, n = 10) and intestine (wild-type: 458 ± 22 nl, n = 10; cnnm-1; cnnm-3: 357 ± 13 nl, n = 10). p values were determined by Student’s two tailed t-test.
RNAi screening for genes that functionally associated with cnnm-1 and cnnm-3
Next, we searched for genes that are functionally associated with cnnm-1 and cnnm-3. For this purpose, we performed RNAi-based screening, because (i) an RNAi feeding library targeting 86% of the open reading frames of C. elegans is available and widely used in genome-wide screening [29] and (ii) unlike conventional forward genetics approaches, it is not necessary to collect the worms and their progenies after screening, making this method applicable to sterile cnnm-1; cnnm-3 strains. To increase RNAi efficacy, screening was performed using worms carrying the rrf-3 mutation, which renders the worms hypersensitive to RNAi treatment [30]. Two rounds of screening identified 31 genes, of which RNAi treatment reproducibly resulted in elongation of the gonadal arm in more than 50% of cnnm-1; cnnm-3; rrf-3 mutant worms (Table 2 and S4 Fig). These 31 genes are involved in a variety of biological processes, including protein transport, metabolism, mitochondrial function, signal transduction, gene expression, ion transport, immune response, and the cell cycle. Among these, we performed detailed analyses of aak-2, which encodes the α-subunit of AMPK. AMPK is the key energy sensor in most eukaryotic cells and is activated under low-energy conditions such as decreased ATP levels [31]. Most intracellular ATP is known to form complexes with Mg2+, which is required for numerous enzymatic reactions involving ATP [32,33]. We hypothesized that dysregulation of cellular Mg2+ levels in cnnm-1; cnnm-3 mutant worms could affect AMPK activity.
Table 2. Genes of which RNAi suppressed the gonadogenesis defect in cnnm-1; cnnm-3 mutant worms.
| (%) gonadal arm extended | ||||
|---|---|---|---|---|
| Sequence name | Gene name | Description | 1st round | 2nd round |
| C10E2.6 | mct-6 | Monocarboxylate transporter | 100 | 100 |
| R160.1 | dpy-23 | AP-2 complex subunit mu2 | 100 | 100 |
| C06B8.7 | Scavenger receptor cysteine-rich domain | 100 | 100 | |
| F55D10.3 | glit-1 | Thyroglobulin | 80 | 93 |
| T05H4.5 | hpo-19 | NADH-cytochrome b5 reductase | 90 | 90 |
| C55B7.8 | dbr-1 | RNA-lariat debranching enzyme | 90 | 87 |
| F02E8.3 | aps-2 | AP-2 complex subunit sigma2 | 90 | 87 |
| T01C8.1 | aak-2 | AMPK alpha2 | 80 | 87 |
| C34C6.6 | prx-5 | Peroxisomal targeting signal 1 receptor | 80 | 87 |
| R03E1.1 | sym-4 | WD repeat-containing protein | 80 | 83 |
| F29A7.6 | M-phase phosphoprotein 6 | 100 | 83 | |
| T14G10.7 | hpo-5 | GPI transamidase component PIG-S | 70 | 83 |
| C07A9.11 | ncx-7 | Sodium/potassium/calcium exchanger | 90 | 80 |
| R12C12.2 | ran-5 | RanBP1 domain | 80 | 77 |
| F11E6.5 | elo-2 | palmitic acid elongase | 50 | 73 |
| F36H12.5 | Transcription initiation factor TFIID subunit 3 | 80 | 73 | |
| F44A6.2 | sex-1 | Nuclear hormone receptor | 60 | 73 |
| F58B3.4 | Nucleolar pre-rRNA processing protein | 80 | 70 | |
| C27F2.8 | Transmembrane protein 131 | 80 | 67 | |
| T26A8.4 | Zinc finger CCCH domain-containing protein 4 | 80 | 67 | |
| K04E7.2 | pept-1 | Oligopeptide transporter | 80 | 67 |
| Y65B4BR.4 | wwp-1 | NEDD4-like E3 ubiquitin-protein ligase WWP1 | 50 | 63 |
| C09H6.3 | mau-2 | MAU2 chromatid cohesion factor homolog | 70 | 60 |
| C27B7.8 | rap-1 | Ras-related protein Rap-1b | 50 | 60 |
| F23C8.6 | did-2 | Charged multivesicular body protein 1b | 50 | 57 |
| F08B12.2 | prx-12 | Peroxisomal biogenesis factor 12 | 60 | 57 |
| ZK418.4 | lin-37 | LIN37 family protein | 70 | 53 |
| R07H5.8 | Adenosine kinase | 60 | 53 | |
| Y71H10B.1 | Cytosolic purine 5'-nucleotidase | 50 | 53 | |
| Y40B1B.7 | Coiled-coil domain-containing protein 86 | 50 | 53 | |
| F46F11.4 | ubl-5 | Ubiquitin-like protein 5 | 50 | 50 |
Genome wide RNAi screening identified 31 genes that functionally associate with cnnm-1 and cnnm-3. The information is derived from either Wormbase or InterProScan.
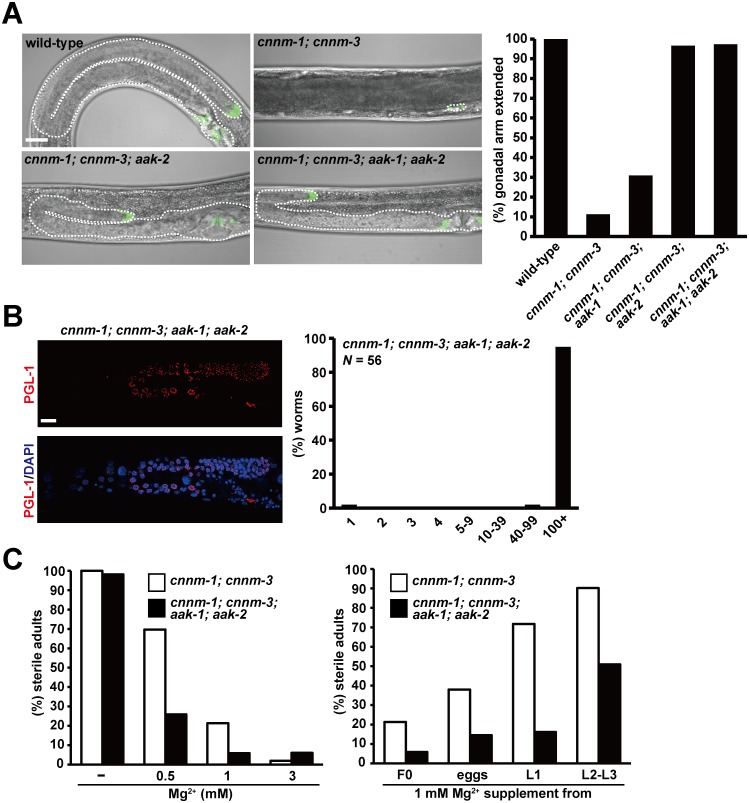
AMPK mediates the gonadogenesis defect caused by cnnm-1; cnnm-3 mutation
AMPK is a heterotrimeric kinase consisting of a catalytic subunit (α) and two regulatory subunits (β and γ). In C. elegans, there are two α subunits, AAK-1 and AAK-2, which are encoded by different genes [34]. Therefore, we examined whether the predicted null mutations for aak-1 and/or aak-2 could suppress the gonadogenesis defect in cnnm-1; cnnm-3 mutant worms (Fig 6A). The additional mutation in aak-2 or in both aak-1 and aak-2 nearly completely restored the gonadal arm extension, while the aak-1 mutation showed only a marginal effect. We also analyzed germ cell proliferation in cnnm-1; cnnm-3; aak-1; aak-2 quadruple mutant worms and found that most contained many germ cells (> 100 cells, Fig 6B). Moreover, when we examined the fertility of quadruple mutant worms by supplementation with various concentrations of Mg2+, fertility was restored at lower concentrations of Mg2+ (Fig 6C, left). We also performed rescue experiments by changing the initiation timing of Mg2+ supplementation. The results showed that fertility was restored even when supplementation was started at later stages (Fig 6C, right). Therefore, the additional mutations in aak-1 and aak-2 significantly augmented the effect of Mg2+ on fertility, implicating that AMPK mediates the effect of Mg2+ on gonadogenesis.
Fig 6. AMPK mediates the gonadogenesis defect of cnnm-1; cnnm-3 mutant worms.
(A) L4/young adult worms of the indicated genotype, carrying lag-2p::GFP, were examined for gonadal arm extension. Representative images are shown (left). In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. Bar, 20 μm. Worms with gonads of ≥ 60 μm in length of the long axis (3-fold as long as the primordial gonad) were determined as extended and the results (%) are shown in the graph (right). More than 100 worms were analyzed. (B) cnnm-1; cnnm-3; aak-1; aak-2 mutant worms were stained with DAPI (blue) and anti-PGL-1 antibody (red), and then the number of germ cells (PGL-1-positive cells) per worm was counted. Representative images are shown (left). Bar, 15 μm. The graph indicates the distribution of worms (%) with the indicated numbers of germ cells (right). (C) Worms of the indicated genotype were grown on plates supplemented with the indicated concentrations of Mg2+ from the F0 generation (left) or grown on plates supplemented with 1 mM Mg2+ starting from the indicated stages (right). More than 50 worms were analyzed for each experimental condition.
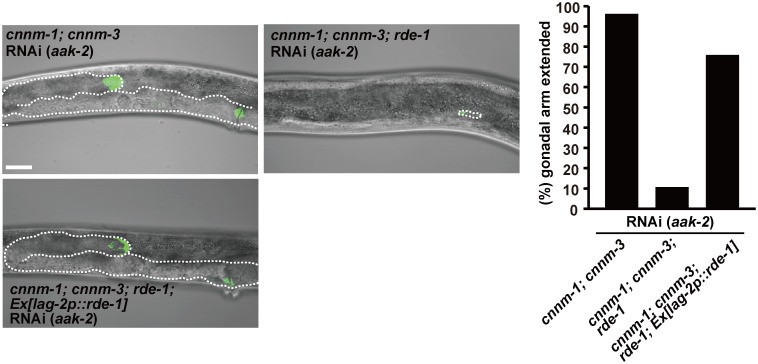
To identify the cell type that is primarily affected by the cnnm-1; cnnm-3 mutations and responsible for the gonadogenesis defect, we performed tissue-specific RNAi experiments. It has been reported that somatic gonad-specific RNAi can be achieved using a strain that carries both a mutation in the rde-1 gene, which encodes an Argonaute protein required for siRNA maturation [35], and a transgene qIs140[lag-2p::rde-1] that drives the expression of wild-type rde-1 under the control of the lag-2 promoter [36]. We generated cnnm-1; cnnm-3; rde-1; qIs140 strains and performed feeding RNAi experiments of aak-2. As shown in Fig 7, somatic gonad-specific RNAi of aak-2 restored the gonadal arm extension in most (75.9% extended) cnnm-1; cnnm-3; rde-1; qIs140 worms, while worms without the transgene (cnnm-1; cnnm-3; rde-1) failed to show restored extension. These results clearly show the importance of aak-2 function in the somatic gonad.
Fig 7. Primary defect occurs in somatic gonad.
L4/young adult worms of the indicated genotype, carrying lag-2p::GFP, were fed with bacteria carrying the dsRNA corresponding to aak-2. In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. Bar, 20 μm. Worms with gonads of ≥ 60 μm in length of the long axis (3-fold as long as the primordial gonad) were determined as extended and the results (%) are shown in the graph (right). More than 50 worms were analyzed for each condition.
AMPK can regulate multiple downstream molecules, including the target of rapamycin complex (TORC)1, which is well-known to be important in cell growth [37]. Therefore, we performed RNAi experiments to investigate the importance of TORC1 in gonadogenesis. Because RNAi of either let-363 or daf-15 is known to cause L3 larval arrest [38], we observed worms in the L2–L3 stages. We found that inhibition of daf-15 (encoding Raptor ortholog) or let-363 (encoding ortholog of mammalian TOR kinase) efficiently suppressed gonadal arm extension in cnnm-1; cnnm-3; aak-1; aak-2 quadruple mutant worms (S5 Fig). In contrast, inhibition of rheb-1 (encoding Rheb ortholog) showed minimal suppression of gonadal arm extension. Biochemical analyses using mammalian cultured cells showed that AMPK inhibited TOR kinase via phosphorylation of TSC (the upstream regulator of Rheb [39]) and Raptor [40], but there is no TSC homolog in C. elegans [41]. Taken together, the results of our RNAi experiments suggest the involvement of dysregulated AMPK-TORC1 signaling in the gonadogenesis defect of cnnm-1; cnnm-3 mutants.
Discussion
In this study, we showed that cnnm-1; cnnm-3 mutant worms displayed pleiotropic phenotypes, such as infertility due to a gonadogenesis defect, shortened lifespan, and small body size (Fig 2). Among these, the gonadogenesis defect was completely restored by adding Mg2+ to the culture media (Fig 3). These results suggest that abnormal Mg2+ regulation in cnnm-1; cnnm-3 mutant worms affected gonadal development. In contrast, Mg2+ supplementation affected neither the lifespan nor the body size of cnnm-1; cnnm-3 mutants (Fig 3). Whether the lifespan and body size phenotypes are related to altered magnesium homeostasis remains unknown, and further analyses are required to clarify the relationship with CNNM functions.
Our elemental analyses suggested that the cnnm-1; cnnm-3 mutant worms had higher levels of Mg2+ in intestinal cells and lower levels of Mg2+ in the pseudocoelom, which contains other tissues such as the gonad (Table 1). This is consistent with the presumed molecular function of CNNM proteins at the basolateral membrane of the intestinal epithelia. In addition, we found that cnnm-1; cnnm-3 mutants exhibited a severe gonadogenesis defect (Fig 4), which was completely restored by additional Mg2+ supplementation to the media. Collectively, these results strongly suggest that the gonadogenesis defect is due to Mg2+ deficiency in the gonad. Because of experimental limitations, we could only estimate magnesium levels in non-intestinal tissues, which showed moderate reduction in cnnm-1; cnnm-3 mutant worms (~67% of wild-type worms). Whether this level of reduction alone can explain the pleiotropic phenotypes of mutant worms is unclear. However, a decrease of total magnesium levels by ~20% can cause proliferation arrest of HEK 293 human cultured cells [10]. More detailed studies of the Mg2+ distribution are required to precisely characterize the mechanism of how Mg2+ deficiency affects worm development.
The importance of Mg2+ in the regulation of various cell functions, such as proliferation, was predicted previously [42,43]. A study of chicken DT40 cells lacking TRPM7, a Mg2+-permeable cation channel, revealed the significance of Mg2+ influx in maintaining cell proliferation [44]. However, the mechanism of Mg2+ action is poorly understood. Our previous study showed that intracellular Mg2+ levels significantly affect ATP levels in cultured mammalian cells [13]. Because AMPK is an energy sensor kinase that is directly regulated by cellular ATP levels [39], AMPK was thought to play an important role in mediating the effect of Mg2+. In this study, we performed a genome-wide RNAi screen to identify genes involved in Mg2+-associated regulation of cell proliferation, which yielded 31 candidate genes including aak-2, which encodes a catalytic subunit of AMPK (Table 2). We confirmed the importance of AMPK by showing that additional mutations in aak-1 and aak-2 restored gonadal arm extension in cnnm-1; cnnm-3 mutants (Fig 6). Moreover, in cnnm-1; cnnm-3; aak-1; aak-2 quadruple mutant worms, fertility was restored at lower levels of Mg2+ supplementation (Fig 6). Therefore, we provide genetic evidence linking Mg2+ homeostasis to the AMPK function. In addition, tissue-specific RNAi experiments clearly located the rescue function of AMPK in somatic gonad (Fig 7). Therefore, Mg2+ decrease presumably causes gonadogenesis defect by affecting the AMPK function in somatic gonad, which then affects proliferation of germ cells.
One of the important downstream targets of AMPK is TORC1: activated AMPK suppresses the function of TORC1 [37]. Our RNAi experiments implicated daf-15 (encoding Raptor ortholog) and let-363 (encoding ortholog of mammalian TOR kinase) in the restored elongation of gonads by aak-1/aak-2 mutation (S5 Fig), suggesting the importance of AMPK-TORC1 signaling in mediating gonadogenesis. It should be noted that Mg2+ influx through TRPM7 in chicken culture cells is essential for sustained activation of TORC1 and cell proliferation [44]. This Mg2+-dependent TORC1 signaling is presumed to be mediated by phosphoinositide 3-kinase and Akt. Whether it is functionally related to the Mg2+-dependent AMPK-TORC1 signaling in C. elegans remains unknown, but it is intriguing that Mg2+ perturbation convergently affects the same target molecule in distant species.
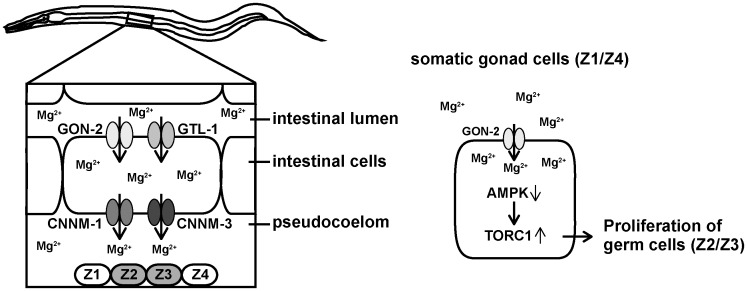
Gonadal development in C. elegans is severely affected by the nutrition status, and starvation forces primordial gonad to stop cell proliferation due to the checkpoint activation. Reportedly, the starvation-induced proliferation arrest of germ cells can be partly rescued by aak-1/2 mutation [45], as in the case of cnnm-1; cnnm-3 mutant worms. Therefore, the arrest of germ cell proliferation in cnnm-1; cnnm-3 mutants are presumed to occur by similar AMPK-dependent mechanism. In more detail, gonad cells are arrested at different stages of the cell cycle by starvation, depending on the cell types: Z1/Z4 cells (somatic gonad cells) are arrested at the G1 phase, while Z2/Z3 cells (germ cells) are arrested at the G2 phase [46–48]. It is not determined at which stage cells are arrested in cnnm-1; cnnm-3 mutants, and thus, it is impossible to further evaluate the similarities between the starved worms and cnnm-1; cnnm-3 mutant worms. However, it is reported that inactivation of GON-2 Mg2+ channel causes G1 arrest in Z1/Z4 cells by upregulating the G1/S checkpoint molecule CKI-1 [46]. Also, in chicken cell culture experiments, TRPM7-deficiency caused downregulation of TORC1 signaling and G1 arrest [44]. Taken together, it is plausible that Mg2+ shortage in the pseudocoelom of cnnm-1; cnnm-3 mutant worms causes G1 arrest in Z1/Z4 cells by the AMPK-TORC1-dependent checkpoint control mechanism, and subsequently causes G2 arrest in Z2/Z3 cells (Fig 8).
Fig 8. Proposed model for the role of C. elegans CNNM proteins.
(Left) Mg2+ in the intestinal lumen enters into the intestinal cells through GON-2 and GTL-1, the apically-localized Mg2+-permeable channels. Mg2+ is then extruded from the intestinal cells to the pseudocoelom by CNNM-1 and CNNM-3, the Mg2+ transporters localized at the basolateral membrane. Pseudocoelom is in contact with many tissues/cells including primordial gonad cells (Z1−Z4). (Right) Through GON-2, somatic gonad cells (Z1/Z4) take up Mg2+, which suppresses the AMPK function and in turn augments the TORC1 function, ultimately stimulating the proliferation of adjacent germ cells (Z2/Z3).
Finally, besides aak-2, our RNAi screen yielded several other candidate genes, which are involved in various biochemical/biological processes. Further characterizations of the relationship between Mg2+ and these genes may reveal new roles for Mg2+ in the regulation of biological functions.
Materials and Methods
C. elegans strains and general methods for handling worms
All C. elegans strains used in this study were derived from wild-type C. elegans var. Bristol (N2). Unless otherwise indicated, the worms were grown at 20°C on NGM-lite plates seeded with OP-50 E. coli, as described previously [24]. The following mutations, rearrangement, and transgenes were used in this study: LGI cnnm-4(dcr3) and daf-16(mgDf50); LGII rrf-3(pk1426); LGIII daf-2(e1370), cnnm-5(ttTi19567), and aak-1(tm1944); LGIV cnnm-1(gk222902) and nT1[qIs51] (IV; V); LGV rde-1(ne219); LGX cnnm-3(dcr2), cnnm-2(dcr1), and aak-2(ok524); qIs56[lag-2p::GFP]; qIs140[lag-2p::rde-1]; sEx rCesC33D12.2::GFP 14584[rCesC33D12.2rCesC33D12.2::GFP:: GFP]. Strains carrying each mutation and rearrangement were either isolated from the Trimethylpsoralen/UV-mutagenized library (see the “isolation of cnnm mutant strains” section for details), provided by the Caenorhabditis Genetics Center which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), or obtained from NEMAGENETAG Project funded by the European Community [49]. All isolated/obtained strains were backcrossed at least 4 times with N2 before use. Double homozygous worms for cnnm-1(gk222902) and cnnm-3(dcr2) are completely sterile, and therefore, cnnm-1(gk222902)/+; cnnm-3(dcr2) worms were maintained using the nT1[qIs51] (IV; V) balancer. Synchronization of worm development was achieved by egg laying of gravid adults for 6 h unless otherwise indicated.
Generation of plasmids and transgenic C. elegans lines
To generate cnnm-1p::GFP, the 4,333-bp fragment of the 5′ region of cnnm-1 was amplified by PCR and then inserted into the GFP expression vector pPD95.77 (kindly provided by A. Fire). To generate the cnnm-1 genomic construct, the cnnm-1 genomic fragment (−4,933 to +7,492 relative to the ATG start codon) was obtained by restriction enzyme digestion from the fosmid WRM0636cE07 (Dnaform). The remaining portion of the cnnm-1 genomic fragment (+7,493 to +8,924) was generated by PCR amplification. For the cnnm-3 genomic construct, the cnnm-3 genomic fragment (−2,935 to +4,090) was obtained by PCR amplification. Thereafter, each genomic fragment was inserted into pBluescript KS (Stratagene). For the cnnm-1::Venus translational fusion construct, the same fosmid fragment as that used for the cnnm-1 genomic construct was linked to the cnnm-1 genomic fragment (+7,493 to +8,610) generated by PCR amplification. The fragments were then inserted into pPD95.79-venus (kindly provided by T. Ishihara). To express the venus-fusion protein of CNNM-3, the cnnm-3 genomic fragment (−2,935 to −1) and the cnnm-3 cDNA were both prepared by PCR, and these fragments were then inserted into pPD95.79-venus. To express cnnm-1 or cnnm-3 under the control of the ges-1 promoter or aex-3 promoter, cnnm-1 or cnnm-3 cDNA was prepared by RT-PCR and then inserted into pDEST-ges-1p and pDEST-aex-3p, kindly provided by H. Kuroyanagi [50,51]. To generate plasmids for feeding RNAi experiments (to target genes that are not included in the Ahringer library), cDNA fragments for let-363 and daf-15 were prepared by RT-PCR with the following primer sets: let-363; 5′-ACTAGTGCCGATAGACAGAACAAAGCAGCC-3′ and 5′-GTGGTACCGGACAAGCCATTCAACACCTTC-3′; daf-15; 5′-GTGCTAGCCCTCGTTTGCAGAACGTTTGAC-3′ and 5′-AGGTACCCCAGTTGAGCTCTCCGAGCACAG-3′. Amplified fragments were then inserted into L4440 (kindly provided by A. Fire). DNA fragments were inserted by conventional methods utilizing the restriction enzymes and ligases, with the exception of the expression construct for venus-fused CNNM-3, which was generated using the Gibson assembly method [52]. The DNA sequences of all PCR products were confirmed by sequencing. To generate transgenic lines, plasmids were injected into N2 or cnnm-1(gk222902)/nT1[qIs51]; cnnm-3(dcr2) along with rol-6(su1006) [53] or rab-3p::mCherry (Addgene) as an injection marker.
Isolation of cnnm mutant strains
The mutant strains of cnnm-2(dcr1), cnnm-3(dcr2), and cnnm-4(dcr3) in this study were isolated from the Trimethylpsoralen/UV-mutagenized library by performing nested PCR as described previously [54]. The primer sets used for screening were as follows: cnnm-2 first round; 5′-TGTCCCGTTTGATGGAAAAT-3′ and 5′-TTTGGAACTATCGTGCCTCC-3′; cnnm-2 second round; 5′-CGAGGATGGTAGAAATGCTCA-3′ and 5′-TACCTGTGGCATCATGGTTG-3′. cnnm-3 first round; 5′-TTGATTAGCGGCAATAAGGG-3′ and 5′-ATATGCCAAAATGGCTTTCG-3′; cnnm-3 second round; 5′-GCTCACCATTCAACGATTCA-3′ and 5′-ATGAACTCACGAGGTGTCGG-3′. cnnm-4 first round; 5′-CATTTTTCAGCGAGCCTTTC-3′ and 5′-CCCATCTTCTTCCGAATCAA-3′; cnnm-4 second round; 5′-CTTTGCCTCGGTTTATCTGC-3′ and 5′-AGACGTGAATGGCCTTGTTC-3′. The cnnm-1(gk222902) and the cnnm-5(ttTi19567) alleles were generated by the C. elegans Reverse Genetics Core Facility at the University of British Columbia and the NEMAGENETAG Project, respectively.
Germ cell counts
Germ cells were stained as previously described [55] with slight modifications. L4/young adults were permeabilized using the freeze–crack method and sequentially fixed in cold methanol for 10 min and in cold acetone for 10 min. The samples were blocked with 2% bovine serum albumin in PBS-T (PBS containing 0.05% Tween 20) for 30 min at room temperature, and then incubated overnight at 4°C with mouse anti-PGL-1 antibody K76 (1:20 dilution), developed by S. Strome [56] and provided by the Developmental Studies Hybridoma Bank. This was followed by incubation with Alexa Fluor 568 goat anti-mouse IgG (1:2000 dilution, Invitrogen) for 2 h at room temperature. Coverslips were mounted on a microscopic glass slide. Next, the samples were observed using a microscope, and PGL-1-positive cells were counted as germ cells.
Gonadal arm extension
To analyze gonadal arm extension, lag-2p::GFP was used to visualize the Z1/Z4 cells and DTCs. The worms were analyzed at the L4/young adult stage, unless otherwise noted, using a microscope. Based on visual observation of the lag-2p::GFP signals and the phase contrast view, we determined the outline of the gonad, and worms with gonads of ≥ 60 μm in length of the long axis (3-fold length of the primordial gonad) were defined as extended.
Lifespan assay
A lifespan assay was performed as described previously [20] with slight modifications, starting with L4/young adults. To remove contamination with progeny, worms were transferred to fresh NGM-lite plates seeded with OP-50 every 2 days until day 8, after which only those worms on plates where progeny was observed were transferred. Survival was monitored daily. Worms that did not move, respond to nose touch with a platinum picker, or exhibit pumping were determined as dead and were removed. Worms that crawled off the plate, had a protruded vulva, or died by internal hatching were excluded.
Body size measurement
Mixed stage worms from L2 to adult (L4 to adult worms were most abundant) were anesthetized with M9 buffer containing 50 mM NaN3 and were mounted on a 3% agarose pad on a microscopic glass slide. Thereafter, the worms were examined for alae formation using a microscope and then photographed for body size measurement. The area of the worms was directly measured from the images using Image J (NIH software). The body size at the transition from L4 to adult molt was determined by calculating the mean body size of the smallest three worms with alae and the largest three worms without alae.
Male tail observation
Observation of the male tail was performed as previously described [57]. Adult worms were anesthetized with 10 mM sodium azide and transferred to a 5% agar pad. The worms were turned over with a pick to the ventral side up and immediately covered with a coverslip.
Genome wide RNAi screening
Feeding RNAi was performed as described previously [58]. In total, 15,357 bacterial RNAi feeding strains from the Ahringer library [29] were tested as follows in the first round screening (n = 5–10 worms per strain), using bacteria carrying the empty vector L4440 as the negative control. cnnm-1(gk222902)/nT1[qIs51]; cnnm-3(dcr2); rrf-3(pk1426); qIs56[lag-2p::GFP] gravid adults were bleached, and synchronized P0 worms at the L1 stage were transferred to RNAi plates. F1 progeny lacking the balancer nT1[qIs51] (without pharyngeal GFP expression) were phenotypically scored at the L4/young adult stage as described in the “Gonadal arm extension” section. The wells were scored as positive if gonadal arms extended in more than 50% of worms in the F1 generation. Some wells showed larval arrest or sterile phenotypes in the P0 generation, and therefore, these wells were scored in the P0 generation. The first round screening led to the identification of 119 positive wells, which was followed by sequence analyses to identify the RNAi clone in each well. Because some wells contained multiple clones, we re-transformed each of the 135 sequence-verified clones into the HT115 E. coli strain and performed the second round screening as described above (n = 30 worms per clone).
ICP-MS
To measure magnesium levels in whole worms, 300 synchronized L4/young adult worms were incubated for 30 min with washing buffer containing 110 mM HNO3 (semiconductor grade, Wako) and 187 mM NH3 (ultrapure grade, Kanto Chemical), which corresponds to approximately 300 mOsm/l and pH 7.0–8.0 at room temperature, and were then washed 5 times with washing buffer. Subsequently, worms were boiled at 95°C for 5 min and sonicated using Bioruptor (UCD-250HSA; Cosmo Bio). The homogenates were completely dried by incubation at 98°C, and then subjected to treatment with 100 μl of 40% HNO3 at 95°C for 2 h. The solution was diluted to 1 ml with ddH2O and magnesium levels were determined using ICP-MS (7700x; Agilent), according to the manufacturer’s instructions. The magnesium levels were normalized to total protein levels, which were determined using the BCA assay kit (Thermo Scientific). A blank sample was prepared using the same procedure without worms. To measure magnesium levels in the intestine, approximately 300 synchronized L4/young adult worms were cut with a scalpel just behind the pharynx in a drop of washing buffer. The extruded intestine was cut away from the remnants of the body, and the isolated intestines were then washed twice with washing buffer. Magnesium levels were analyzed as described above.
Volume calculation
Body volume was measured as described previously [59,60] with slight modifications. L4/young adults were transferred to M9 buffer containing 50 mM NaN3 on coverslips and then photographed. The total body volume of the worms was calculated by assuming that the body shape was composed of two cones (from the tip of the nose to the anterior end of the intestine, and from the posterior end of the intestine to the tip of the tail) and a cylinder (remaining body part) and by measuring each length and radius. Intestine volume was calculated by assuming that the shape was cylindrical.
Microscopy
Fluorescence images were acquired using an inverted microscope (IX81; Olympus) equipped with a laser scanning confocal imaging system (FluoView FV1000; Olympus). Nomarski images were collected concurrently or alone using the same microscope using Nomarski optics. A multiline argon laser and image analysis system (FV10-ASW; Olympus) were also used for image acquisition. To analyze gonadal arm extension, lag-2p::GFP fluorescence was observed using a stereo microscope (SZX7; Olympus) equipped with a U-RFL-T 100W mercury lamp (U-RFL-T; Olympus).
Statistics
All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software) and are presented as the mean ± SEM. p values were obtained by Student’s two-tailed t-test for Table 1 and by ANOVA, followed by two-tailed multiple Student’s t-test with Tukey’s correction for Figs 2C and 3B. For lifespan assays (Figs 2D, 3C and S2 Fig), we used the log rank (Mantel-Cox) test. Bonferroni correction was applied to multiple comparisons of lifespans.
Supporting Information
Asterisk (*) and colon (:) denote identical residues and conserved substitutions, respectively. The sequence of H. sapiens CNNM4 is also shown. The regions for functionally essential DUF21 and CBS domains, and the amino acids for which coding nucleotide sequences were directly lost/changed by each genetic alteration, are highlighted. cnnm-1(gk222902) contains a point mutation that led to a premature stop codon in place of arginine residue 255 in the DUF21 domain. cnnm-2(dcr1) contains a deletion of 162 nucleotides from the inside of exon 11 to the inside of exon 12, leading to the loss of 40 amino acids in the CBS domain. cnnm-3(dcr2) contains a deletion of 289 nucleotides that include the splice acceptor site of intron 4 and 183 nucleotides in the following exon 5. This deletes 61 amino acids in the DUF21 domain and causes additional deletion/alterations because of incorrect splicing. cnnm-4(dcr3) contains a deletion of 173 nucleotides that include the entire exon 5 and the splice acceptor and donor sites in adjacent introns. This deletes exon 5-encoded 41 amino acids in the DUF21 domain, and causes frameshifts if RNA splicing occurs by directly linking exon 4 and exon 6. cnnm-5(ttTi19567) contains the Mos 1 sequence inserted in exon 2, resulting in a truncated product that lacks both the DUF21 and CBS domains.
(TIF)
For each genotype, 45 synchronized L4/young adult worms were transferred to fresh plates (15 worms per plate) and then scored daily for survival. The graph represents data combined from at least two experiments. Mean lifespan (± SEM) of worms is also indicated in parentheses. p values were determined by log rank (Mantel-Cox) test, and the Bonferroni method was then used to correct for multiple comparisons. ****p < 0.0001.
(TIF)
Nomarski images of the ventral view of adult wild-type and cnnm-1; cnnm-3 mutant mail tails. The anterior side of the worm is positioned to the left. Bar, 20 μm.
(TIF)
Representative images of L4/young adult worms of cnnm-1; cnnm-3; rrf-3 mutants with Ex[lag-2p::GFP], which were fed with bacteria carrying the dsRNA corresponding to the indicated genes (the results of the top 5 genes in second round screening are shown). In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. As a negative control, the worms were fed with bacteria carrying the empty vector L4440. Bar, 20 μm.
(TIF)
cnnm-1; cnnm-3; aak-1; aak-2 mutant worms, carrying lag-2p::GFP, were fed with bacteria carrying the dsRNA corresponding to the indicated genes. Because RNAi of either let-363 or daf-15 is known to cause L3 larval arrest [38], we observed the worms at the L2–L3 stages. In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. Bar, 20 μm. Worms with gonads of ≥ 60 μm in length of the long axis (3-fold as long as the primordial gonad) were determined as extended and the results (%) are shown in the right graph. More than 50 worms were analyzed for each condition.
(TIF)
The underlying numerical data for each figure or table are shown in separate sheets.
(XLSX)
Acknowledgments
We thank T. Hayashi, H. Yamamoto, and Y. Tsumori (Osaka University, Japan) for their technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files. C. elegans cnnm mutants isolated and used in this study are available on Wormbase (http://www.wormbase.org) under the following ID numbers: dcr1 (cnnm-2): WBVar02146596; dcr2 (cnnm-3): WBVar02146597; dcr3 (cnnm-4): WBVar02146598.
Funding Statement
This study was supported by KAKENHI Grant Numbers 261704 (TI), 26460364 (YF), 26291042 and 26111007 (HM) from Japan Society for the Promotion of Science (https://www.jsps.go.jp). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schweigel M, Martens H. Magnesium transport in the gastrointestinal tract. Front Biosci. 2000;5: D666–D677. [DOI] [PubMed] [Google Scholar]
- 2.Dimke H, Hoenderop JG, Bindels RJ. Molecular basis of epithelial Ca2+ and Mg2+ transport: insights from the TRP channel family. J Physiol. 2011;589: 1535–1542. 10.1113/jphysiol.2010.199869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voets T, Nilius B, Hoefs S, van der Kemp AW, Droogmans G, Bindels RJ, et al. TRPM6 Forms the Mg2+ Influx Channel Involved in Intestinal and Renal Mg2+ Absorption. J Biol Chem. 2004;279: 19–25. [DOI] [PubMed] [Google Scholar]
- 4.Schlingmann KP, Weber S, Peters M, Niemann Nejsum L, Vitzthum H, Klingel K, et al. Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat Genet. 2002;31: 166–170. [DOI] [PubMed] [Google Scholar]
- 5.Walder RY, Landau D, Meyer P, Shalev H, Tsolia M, Borochowitz Z, et al. Mutation of TRPM6 causes familial hypomagnesemia with secondary hypocalcemia. Nat Genet. 2002;31: 171–174. [DOI] [PubMed] [Google Scholar]
- 6.Ryazanova LV, Rondon LJ, Zierler S, Hu Z, Galli J, Yamaguchi TP, et al. TRPM7 is essential for Mg(2+) homeostasis in mammals. Nat Commun. 2010;1: 109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang CY, Shi JD, Yang P, Kumar PG, Li QZ, Run QG, et al. Molecular cloning and characterization of a novel gene family of four ancient conserved domain proteins (ACDP). Gene. 2003;306: 37–44. [DOI] [PubMed] [Google Scholar]
- 8.Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet. 2010;6: e1001045 10.1371/journal.pgen.1001045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuiver M, Lainez S, Will C, Terryn S, Günzel D, Debaix H, et al. CNNM2, encoding a basolateral protein required for renal Mg2+ handling, is mutated in dominant hypomagnesemia. Am J Hum Genet. 2011;88: 333–343. 10.1016/j.ajhg.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazaki D, Funato Y, Miura J, Sato S, Toyosawa S, Furutani K, et al. Basolateral Mg2+ extrusion via CNNM4 mediates transcellular Mg2+ transport across epithelia: a mouse model. PLoS Genet. 2013;9: e1003983 10.1371/journal.pgen.1003983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Baaij JH, Stuiver M, Meij IC, Lainez S, Kopplin K, Venselaar H, et al. Membrane topology and intracellular processing of cyclin M2 (CNNM2). J Biol Chem. 2012;287: 13644–13655. 10.1074/jbc.M112.342204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata Y, Funato Y, Takano Y, Miki H. Mg2+-dependent interactions of ATP with the cystathionine- β-synthase (CBS) domains of a magnesium transporter. J Biol Chem. 2014;289: 14731–14739. 10.1074/jbc.M114.551176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funato Y, Yamazaki D, Mizukami S, Du L, Kikuchi K, Miki H. Membrane protein CNNM4-dependent Mg2+ efflux suppresses tumor progression. J Clin Invest. 2014;124: 5398–5410. 10.1172/JCI76614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy S, Uetani N, Wong N, Kostantin E, Labbé DP, Bégin LR, et al. The protein tyrosine phosphatase PRL-2 interacts with the magnesium transporter CNNM3 to promote oncogenesis. Oncogene. 2015;34: 986–995. 10.1038/onc.2014.33 [DOI] [PubMed] [Google Scholar]
- 15.Teramoto T, Lambie EJ, Iwasaki K. Differential regulation of TRPM channels governs electrolyte homeostasis in the C. elegans intestine. Cell Metab. 2005;1: 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teramoto T, Sternick LA, Kage-Nakadai E, Sajjadi S, Siembida J, Mitani S, et al. Magnesium Excretion in C. elegans Requires the Activity of the GTL-2 TRPM Channel. PLoS One. 2010;5: e9589 10.1371/journal.pone.0009589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma-Kishore R, White JG, Southgate E, Podbilewicz B. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development. 1999;126: 691–699. [DOI] [PubMed] [Google Scholar]
- 18.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56: 110–156. [DOI] [PubMed] [Google Scholar]
- 19.Singh RN, Sulston JE. Some Observations On Moulting in Caenorhabditis Elegans. Nematologica. 1978;24: 63–71. [Google Scholar]
- 20.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366: 461–464. [DOI] [PubMed] [Google Scholar]
- 21.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70: 396–417. [DOI] [PubMed] [Google Scholar]
- 22.Blelloch R, Anna-Arriola SS, Gao D, Li Y, Hodgkin J, Kimble J. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol. 1999;216: 382–393. [DOI] [PubMed] [Google Scholar]
- 23.Mathies LD, Schvarzstein M, Morphy KM, Blelloch R, Spence AM, Kimble J. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development. 2004;131: 4333–4343. [DOI] [PubMed] [Google Scholar]
- 24.Sun AY, Lambie EJ. gon-2, a gene required for gonadogenesis in Caenorhabditis elegans. Genetics. 1997;147: 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol. 1980;78: 542–576. [DOI] [PubMed] [Google Scholar]
- 26.Edgar LG, McGhee JD. Embryonic expression of a gut-specific esterase in Caenorhabditis elegans. Dev Biol. 1986;114: 109–118. [DOI] [PubMed] [Google Scholar]
- 27.Iwasaki K, Staunton J, Saifee O, Nonet M, Thomas JH. aex-3 encodes a novel regulator of presynaptic activity in C. elegans. Neuron. 1997;18: 613–622. [DOI] [PubMed] [Google Scholar]
- 28.Cox EA, Hardin J. Sticky worms: adhesion complexes in C. elegans. J Cell Sci. 2004;117: 1885–1897. [DOI] [PubMed] [Google Scholar]
- 29.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421: 231–237. [DOI] [PubMed] [Google Scholar]
- 30.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1: E12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13: 251–262. 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romani AM. Cellular magnesium homeostasis. Arch Biochem Biophys. 2011;512: 1–23. 10.1016/j.abb.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Günther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+. Magnes Res. 2006;19: 225–236. [PubMed] [Google Scholar]
- 34.Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18: 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127: 747–757. [DOI] [PubMed] [Google Scholar]
- 36.Martynovsky M, Wong MC, Byrd DT, Kimble J, Schwarzbauer JE. mig-38, a novel gene that regulates distal tip cell turning during gonadogenesis in C. elegans hermaphrodites. Dev Biol. 2012;368: 404–414. 10.1016/j.ydbio.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie DG. AMPK-sensing energy while talking to other signaling pathways. Cell Metab. 2014;20: 939–952. 10.1016/j.cmet.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110: 177–189. [DOI] [PubMed] [Google Scholar]
- 39.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115: 577–590. [DOI] [PubMed] [Google Scholar]
- 40.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30: 214–226. 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, et al. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell. 2014;53: 209–220. 10.1016/j.molcel.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin H. Central role for magnesium in coordinate control of metabolism and growth in animal cells. Proc Natl Acad Sci U S A. 1975;72: 3551–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubin H. The logic of the Membrane, Magnesium, Mitosis (MMM) model for the regulation of animal cell proliferation. Arch Biochem Biophys. 2007;458: 16–23. [DOI] [PubMed] [Google Scholar]
- 44.Sahni J, Scharenberg AM. TRPM7 ion channels are required for sustained phosphoinositide 3-kinase signaling in lymphocytes. Cell Metab. 2008;8: 84–93. 10.1016/j.cmet.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuyama M, Sakuma K, Park R, Kasuga H, Nagaya R, Atsumi Y, et al. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol Open. 2012;1: 929–936. 10.1242/bio.2012836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- 47.Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16: 773–779. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe S, Yamamoto TG, Kitagawa R. Spindle assembly checkpoint gene mdf-1 regulates germ cell proliferation in response to nutrition signals in C. elegans. EMBO J. 2008;27: 1085–1096. 10.1038/emboj.2008.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallin E, Gallagher J, Granger L, Martin E, Belougne J, Maurizio J, et al. A genome-wide collection of Mos1 transposon insertion mutants for the C. elegans research community. PLoS One. 2012;7: e30482 10.1371/journal.pone.0030482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods. 2006;3: 909–915. [DOI] [PubMed] [Google Scholar]
- 51.Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc. 2010;5: 1495–1517. 10.1038/nprot.2010.107 [DOI] [PubMed] [Google Scholar]
- 52.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 53.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kubota Y, Kuroki R, Nishiwaki K. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr Biol. 2004;14: 2011–2018. [DOI] [PubMed] [Google Scholar]
- 55.Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94: 635–645. [DOI] [PubMed] [Google Scholar]
- 56.Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35: 15–25. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, et al. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126: 241–250. [DOI] [PubMed] [Google Scholar]
- 58.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2: RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choe KP, Strange K. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am J Physiol Cell Physiol. 2007;293: C915–C927. [DOI] [PubMed] [Google Scholar]
- 60.Banerjee S, Versaw WK, Garcia LR. Imaging Cellular Inorganic Phosphate in Caenorhabditis elegans Using a Genetically Encoded FRET-Based Biosensor. PLoS One. 2015;10: e0141128 10.1371/journal.pone.0141128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Asterisk (*) and colon (:) denote identical residues and conserved substitutions, respectively. The sequence of H. sapiens CNNM4 is also shown. The regions for functionally essential DUF21 and CBS domains, and the amino acids for which coding nucleotide sequences were directly lost/changed by each genetic alteration, are highlighted. cnnm-1(gk222902) contains a point mutation that led to a premature stop codon in place of arginine residue 255 in the DUF21 domain. cnnm-2(dcr1) contains a deletion of 162 nucleotides from the inside of exon 11 to the inside of exon 12, leading to the loss of 40 amino acids in the CBS domain. cnnm-3(dcr2) contains a deletion of 289 nucleotides that include the splice acceptor site of intron 4 and 183 nucleotides in the following exon 5. This deletes 61 amino acids in the DUF21 domain and causes additional deletion/alterations because of incorrect splicing. cnnm-4(dcr3) contains a deletion of 173 nucleotides that include the entire exon 5 and the splice acceptor and donor sites in adjacent introns. This deletes exon 5-encoded 41 amino acids in the DUF21 domain, and causes frameshifts if RNA splicing occurs by directly linking exon 4 and exon 6. cnnm-5(ttTi19567) contains the Mos 1 sequence inserted in exon 2, resulting in a truncated product that lacks both the DUF21 and CBS domains.
(TIF)
For each genotype, 45 synchronized L4/young adult worms were transferred to fresh plates (15 worms per plate) and then scored daily for survival. The graph represents data combined from at least two experiments. Mean lifespan (± SEM) of worms is also indicated in parentheses. p values were determined by log rank (Mantel-Cox) test, and the Bonferroni method was then used to correct for multiple comparisons. ****p < 0.0001.
(TIF)
Nomarski images of the ventral view of adult wild-type and cnnm-1; cnnm-3 mutant mail tails. The anterior side of the worm is positioned to the left. Bar, 20 μm.
(TIF)
Representative images of L4/young adult worms of cnnm-1; cnnm-3; rrf-3 mutants with Ex[lag-2p::GFP], which were fed with bacteria carrying the dsRNA corresponding to the indicated genes (the results of the top 5 genes in second round screening are shown). In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. As a negative control, the worms were fed with bacteria carrying the empty vector L4440. Bar, 20 μm.
(TIF)
cnnm-1; cnnm-3; aak-1; aak-2 mutant worms, carrying lag-2p::GFP, were fed with bacteria carrying the dsRNA corresponding to the indicated genes. Because RNAi of either let-363 or daf-15 is known to cause L3 larval arrest [38], we observed the worms at the L2–L3 stages. In each image, the anterior side of the worm is positioned to the left. Dotted lines indicate the outlines of the gonad. Bar, 20 μm. Worms with gonads of ≥ 60 μm in length of the long axis (3-fold as long as the primordial gonad) were determined as extended and the results (%) are shown in the right graph. More than 50 worms were analyzed for each condition.
(TIF)
The underlying numerical data for each figure or table are shown in separate sheets.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. C. elegans cnnm mutants isolated and used in this study are available on Wormbase (http://www.wormbase.org) under the following ID numbers: dcr1 (cnnm-2): WBVar02146596; dcr2 (cnnm-3): WBVar02146597; dcr3 (cnnm-4): WBVar02146598.