Abstract
Epilepsy is a prevalent chronic neurological disorder afflicting about 50 million people worldwide. There is evidence of a strong relationship between familial risk factors and epilepsy, as well as associations with substance use. The goal of this study was to explore the interactions between familial risk factors and substance use based on structured data from the family and social history modules of an electronic health record system for adult epilepsy patients. A total of 8,957patients with 38,802 family history entries and 8,822 substance use entries were gathered and mined for associations at different levels of granularity for three age groupings (>18, 18-64, and ≥65 years old). Our results demonstrate the value of an association rule mining approach to validate knowledge of familial risk factors. The preliminary findings also suggest that substance use does not demonstrate significant association between social and familial risk factors for epilepsy.
Introduction
Epilepsy is one of the most prevalent chronic neurological disorders with a high incidence worldwide1. Studies have focused on investigating associations between family history, social history, and the risk of developing epilepsy2 –9. Prior studies have been based on case control studies of small sets of patients3,5,8,9, literature review4, or regional epidemiologic studies10 to explore the effect of potential familial risk and social factors on epilepsy. Some studies have reported a strong association between positive history of epilepsy in first-, second-, or third-degree relatives and risk of epilepsy2,3,5,6,8,9. One study also reported a high epilepsy risk of certain types of smokers (e.g., acute secondhand smokers and chronic active smokers) based on literature review4.
The availability of information recorded in structured format for patients within the electronic health record (EHR) provides an opportunity for researchers to access a wide range of information about an individual’s family and social history. Moreover, some studies have utilized natural language processing (NLP) techniques to extract family and social history from unstructured documents11 –15. Another area of development has been work to improve the representation of family and social history with enhanced specifications and models16 –19. Together, these specifications and data mining approaches, such as association rule mining, may be used on both structured information and free-text EHR documents to discover interesting associations (e.g., disease-disease and disease-drug)20 –23.
In previous work, we developed an approach for association mining, visualizing, and evaluating rules representing pairwise interactions among familial risk factors stored in the electronic health record24. In this previous study, a pipeline was developed with Ruby (2.0.0) and R (3.1.2) that were interfaced using the RinRuby Ruby gem (2.0.3). Structured family history information from the EHR at the University of Vermont Medical Center was collected for pediatric asthma patients in 2014. A set of association rules was generated and the findings were validated using clinical textbooks, studies, and reports, as well as by clinical experts.
This study involved enhancing the previously developed association rule mining approach for generating, visualizing, and validating association rules between familial risk factors and substance use for adult epilepsy. We sought to gain more understanding on the potential of leveraging family and social history information in the EHR as a contribution to disease or outcome prediction.
Methods
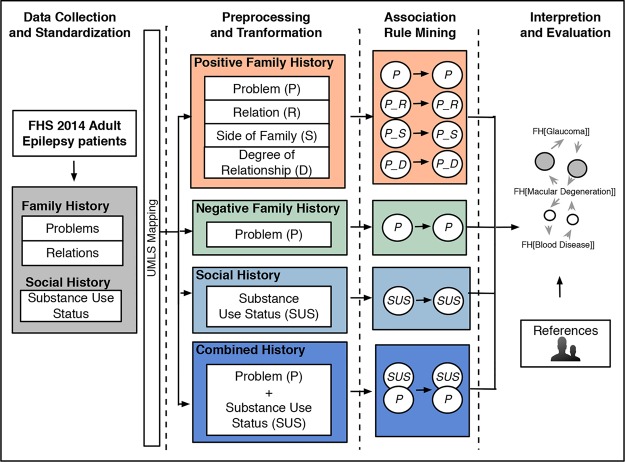
The datasets used in this study were collected from the University of Minnesota-affiliated Fairview Health Services (FHS). Figure 1 provides an overview of the approach used in this study. Similar steps from our previous study24 were followed for structured family history information extraction. Additionally, we included social history information about substance use status from the EHR. The four high level steps were as follows and are described in further detail in the following subsections:
Figure 1.
Overview of the association rule mining process.
(1) Data Collection and Standardization - identify adult epilepsy patients and extract associated family and social history entries from the EHR. The list of medical problems and relations used in the family history module of the EHR were standardized by mapping to the Unified Medical Language System (UMLS)25 Metathesaurus and HL7 Vocabulary for RoleCode respectively.
(2) Preprocessing and Transformation - the datasets were then prepared for mining and visualization with items classified by relationship, side of family, and degree of relationship or negation for family history. For substance use, items were classified for each substance (tobacco, alcohol, and drug) as positive or negative for current or past use.
(3) Association Rule Mining - basic statistics and association rules were generated.
(4) Interpretation and Evaluation - associations were visualized and validated with medical knowledge sources.
Data selection and standardization
Three datasets for adult epilepsy patients were created: (1) ≥ 18 years old (all adult), (2) 18 - 64 years old, and (3) ≥ 65 years old (senior adult). Each patient had at least one encounter in 2014 and either ICD-9-CM code 345.XX or 780.39 as an encounter diagnosis in 2014 or on the problem list. For each dataset, the latest family history information (problem and relation) and substance use status (for alcohol, tobacco, and drug) of patients in the dataset were collected from the EHR. For each family history relation, side of family and degree of relationship were inferred. For each substance type, a status of “Yes” or “Quit” indicating current or past use was included in the mining process while a status of “Never”, “Not Asked”, or “Passive” (for tobacco) was excluded. Table 1 shows examples of family history and substance use status entries.
Table 1.
Examples of family history and substance use status entries.
| Examples of family history entries | |||||
|---|---|---|---|---|---|
| Problem | Relation | Side of Family | Degree | ||
| Hypertension | Mother | Maternal | First | ||
| Diabetes | Neg Hx | - | - | ||
| Heart Disease | Brother | - | First | ||
| Examples of substance use status entries | |||||
| Tobacco Use | Alcohol Use | Drug Use | |||
| Quit | Yes | - | |||
| Passive | - | - | |||
“-” indicates not applicable or cannot be inferred from structured relation field
For standardization, the list of family history problems and relations were mapped to the UMLS Metathesaurus and HL7 Vocabulary for RoleCode respectively. Of the 24 relations (e.g., “Mother” or “Father”), 16 (66.7%) could be mapped to the HL7 Vocabulary for RoleCode. A total of 141 medical problems were collected from FHS EHR system. The medical problems were mapped to UMLS terms by using the MetaMap26 (2014) mapping tool, as well as manual search with the UMLS Metathesaurus (2014AB) browser for those terms that did not map using the following steps:
All problems were reviewed. Duplicates and synonyms (e.g., Chemical Addiction and Chemical Dependency) were merged.
- Local medical problem terms were mapped to UMLS terms using the following mapping approaches with MetaMap or the UMLS Metathesaurus (2014AB) browser:
- Mapping Approach 1: Run MetaMap for all local names.
-
Mapping Approach 2: For terms not mapped with approach 1, MetaMap was run with potential variant names.(e.g., “Cancer - Colorectal” ^ “Colorectal cancer” and “Pulm HTN” ^ “Pulmonary hypertension”)
- Mapping Approach 3: The UMLS Metathesaurus (2014AB) browser was used to manually search for names not mapped using Approaches 1 or 2, or those with questionable mappings using prior approaches.
Check if mapped UMLS terms can map to SNOMED CT (Systematized Nomenclature of Medicine--Clinical Terms).
For the 141 medical problems, 122 (86.5%) directly mapped using default MetaMap settings, another six (4.2%) terms were mapped by using variant names for MetaMap mapping, and a final four (2.8%) terms were mapped by using the UMLS browser. Three problems could not be mapped (e.g., “Unknown/Adopted” that was subsequently excluded from further analysis) and another five terms mapped with MetaMap, but needed verification with the UMLS browser.
Preprocessing and transformation
In the preprocessing step, family history entries in each dataset were first split into two subsets. One subset (positive family history) consists of all entries with a positive family history of a medical problem. The other subset (negative family history) consists of all entries with a relation “Neg Hx”. For example, as shown in Table 1, the second example entry of family history is split into the negative family history subset and the other two example entries were grouped into positive family history.
Four subsets were created from the positive family history subset of each patient group:
-
(1)
Subset with only problem. [pos_problem subset]
-
(2)
Subset with problem + relation, e.g. “Diabetes_Mother”. [pos_problem_relation subset]
-
(3)
Subset with problem + side of family, e.g., “Diabetes_Maternal”. [pos_problem_side subset]
-
(4)
Subset with problem + degree, e.g., “Diabetes_First”. [pos _problem_degree subset]
One subset was created from the negative family history subset of each patient group [neg_problem subset] and another created for the substance use status entries [substance_status subset]. An additional subset was created to combine the positive family history subset with only problem and the substance use status entries [pos_problem_substance_status subset]. In the transformation step, the seven created subsets for each of the three patients groups were transformed into an input format for the R association rule-mining package.
Association rule mining & interpretation and evaluation
The arules R package (1.1-9) was used to generate rules for the seven subsets for each patient group. For each dataset, the mining process was performed for combinations of minimum support values (0.0 to 0.1 in 0.01 increments) and minimum confidence values (0.0 to 1.0 in 0.1 increments). In this study, we set the maximum rule length to two for focusing on only pairwise associations of familial and social risk factors. Existing knowledge sources including chapters in epilepsy textbooks 1,27–31 and published studies on epilepsy 4,5 were reviewed for known familial risk factors, social risk factors, and comorbidities.
Results
Overall, a total of 8,957 adult epilepsy patients were identified from FHS EHR system. Of all patients in the dataset, 986 (11%) patients were identified by encounter diagnosis code, 3196 (35.7%) patients were identified by problem list only, and 4,775 (53.3%) patients were identified by both fields. Within the FHS EHR system, not all patients had a family history entry or substance use entry. In our dataset, 6,082 (67.9%) of patients that had at least one family history entry in the EHR since 2011 and 5868 (65.5%) patients have at least one family history entry in the EHR in 2014. A total of 8,648 (96.5%) of the patients had at least one substance use entry in the EHR since 2014 and 8,822 (98.5%) patients had at least one substance use entry in the EHR since 2011. In total, 38,802 of the most recent family history entries of those patients were collected from the EHR, with 32,997 (85%) for positive family history and 5,805 (15%) for negative family history. A total of 8,822 substance use status entries were gathered for the patients. The distribution of family history and substance use status entries are summarized in Tables 2 and 3.
Table 2.
Distribution of entries, age, and sex for the full dataset, as well as positive and negative family history subsets for major age groupings.
| Epilepsy patients (≥18 years) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Total | # Entries/Patient | Age | Sex | ||||
| # Entries | # Patients | Range | Mean±SD | Range | Mean±SD | Female | Male | |
| All | 38,802 | 5868 | 1 - 67 | 6.1 ± 6.1 | 18 - 100 | 53 ± 9.1 | 3217 | 2651 |
| Positive | 32,997 | 5868 | 0 - 62 | 4.8 ± 4.1 | 18 - 100 | 51 ± 9.0 | 2970 | 2898 |
| Negative | 5805 | 1406 | 0 - 37 | 3.2 ± 3.0 | 18 - 100 | 56 ± 8.6 | 812 | 594 |
| Epilepsy patients (18 - 64 years) | ||||||||
| Dataset | Total | # Entries/Patient | Age | Sex | ||||
| # Entries | # Patients | Range | Mean±SD | Range | Mean±SD | Female | Male | |
| All | 30,453 | 4550 | 0 - 61 | 5.8 ± 4.3 | 18 - 64 | 56 ± 7.6 | 2373 | 2177 |
| Positive | 25,946 | 4337 | 0 - 53 | 5.1 ± 4.1 | 18 - 64 | 51 ± 5.2 | 1326 | 3011 |
| Negative | 4507 | 1120 | 0 - 37 | 3.1 ± 2.8 | 18 - 64 | 52 ± 6.1 | 472 | 648 |
| Epilepsy patients (≥65 years) | ||||||||
| Dataset | Total | # Entries/Patient | Age | Sex | ||||
| # Entries | # Patients | Range | Mean±SD | Range | Mean±SD | Female | Male | |
| All | 8349 | 1366 | 0 - 66 | 5.1 ± 3.7 | 65 - 100 | 76 ± 7.6 | 671 | 695 |
| Positive | 7051 | 1304 | 0 - 66 | 4.4 ± 4.1 | 65 - 100 | 69 ± 5.2 | 596 | 804 |
| Negative | 1298 | 293 | 0 - 37 | 3.3 ± 2.9 | 65 - 94 | 66 ± 6.1 | 135 | 158 |
Table 3.
Distribution of entries, age, and sex for substance use status subsets for each age grouping.
| Dataset | Total | Age | Sex | |||
|---|---|---|---|---|---|---|
| # Entries | # Patients | Range | Mean±SD | Female | Male | |
| ≥ 18 years | 8822 | 8957 | 18 - 100 | 50.2 ± 18.2 | 4915 | 4042 |
| 18 - 64 years | 6936 | 7408 | 18 - 64 | 41.4 ± 5.3 | 3857 | 4191 |
| ≥ 65 years | 1886 | 1909 | 65 - 100 | 87.7 ± 6.7 | 1058 | 851 |
The top ten family history problems, family history problems with relations, and negative family history problems were ranked by prevalence for all patients in the study (Table 4). Prevalence numbers were computed with a formula similar to term frequency-inverse document frequency (TF-IDF), where TF is term frequency and IDF is inverse document frequency, to reflect the importance of a term described in previous work24. The formula used for the prevalence calculation was:
Where pd represents patients with a family history of problem p in a cohort for a particular disease d (e.g., epilepsy), Nd is the total number of patients in the disease cohort, Ny is the number of patients with family history entries in 2014 and py is the number of patients with a family history of problem p in the year.
Table 4.
Ranking of family history problems and relations by prevalence ([n] indicates ranking by frequency).
| Family History of Problem P | Family History of Problem P in Relation R | Negative Family History of Problem P |
|---|---|---|
Pairwise association rules were generated for the seven subsets using different thresholds for support and confidence. Table 5 shows the number of association rules generated with different thresholds for the pos _problem subset for all adults (≥18 years old). Table 6 summarizes numbers of association rules generated with different thresholds for the pos_problem subset of senior adults (≥65 years old). Figure 2 shows the number of association rules generated with different support values for confidence thresholds of 0.2 and 0.4 for each age grouping. As shown in Figure 3, the number of rules generated for the pos_problem subset of all age groups had similar trends. For each subset of each age grouping, the rules generated with “low” and “intermediate” thresholds were selected for evaluation. For the ≥18 years old and the 18-64 years old groups, we observed a similar matrix (Table 5), and the same “low” (minimum support of 0.01 and confidence of 0.2) and “intermediate” (minimum support of 0.04 and confidence of 0.4) thresholds were selected. For the senior adult (65+) patient group, we chose different thresholds for “low” (minimum support of 0.02 and confidence of 0.1) and “intermediate” (minimum support of 0.04 and confidence of 0.3).
Table 5.
Number of rules generated with combinations of minimum support and confidence for pos_problem subset of all adult patients. Highlighted cells are rules generated with “low” (minimum support of 0.01 and confidence of 0.2) and “intermediate” (minimum support of 0.04 and confidence of 0.4).
| Support | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Confidence | 0 | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.08 | 0.09 | 0.1 | |
| 0 | 9506 | 590 | 308 | 170 | 108 | 76 | 50 | 32 | 28 | 24 | 22 | |
| 0.1 | 1684 | 490 | 271 | 155 | 108 | 76 | 50 | 32 | 28 | 24 | 22 | |
| 0.2 | 980 | 283 | 177 | 106 | 72 | 53 | 40 | 30 | 28 | 24 | 22 | |
| 0.3 | 622 | 168 | 123 | 87 | 59 | 45 | 33 | 23 | 21 | 19 | 18 | |
| 0.4 | 440 | 109 | 86 | 63 | 44 | 37 | 25 | 17 | 15 | 14 | 13 | |
| 0.5 | 327 | 43 | 32 | 24 | 16 | 14 | 8 | 4 | 4 | 3 | 3 | |
| 0.6 | 134 | 6 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| 0.7 | 86 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| 0.8 | 78 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0.9 | 77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Table 6.
Number of rules generated with combinations of minimum support and confidence for pos_problem subset of senior adult patients. Highlighted cells are rules generated with “low” (minimum support of 0.01 and confidence of 0.2) and “intermediate” (minimum support of 0.04 and confidence of 0.4).
| Support | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Confidence | 0 | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.08 | 0.09 | 0.1 | |
| 0 | 6480 | 476 | 236 | 152 | 76 | 58 | 36 | 28 | 28 | 26 | 22 | |
| 0.1 | 1178 | 377 | 191 | 136 | 75 | 58 | 36 | 28 | 28 | 26 | 22 | |
| 0.2 | 747 | 233 | 135 | 92 | 54 | 45 | 33 | 28 | 28 | 26 | 22 | |
| 0.3 | 550 | 167 | 114 | 82 | 47 | 38 | 27 | 24 | 24 | 23 | 20 | |
| 0.4 | 374 | 107 | 75 | 50 | 28 | 22 | 15 | 12 | 12 | 12 | 12 | |
| 0.5 | 290 | 43 | 32 | 17 | 7 | 3 | 2 | 0 | 0 | 0 | 0 | |
| 0.6 | 158 | 14 | 12 | 5 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0.7 | 116 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0.8 | 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0.9 | 106 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 106 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Figure 2.
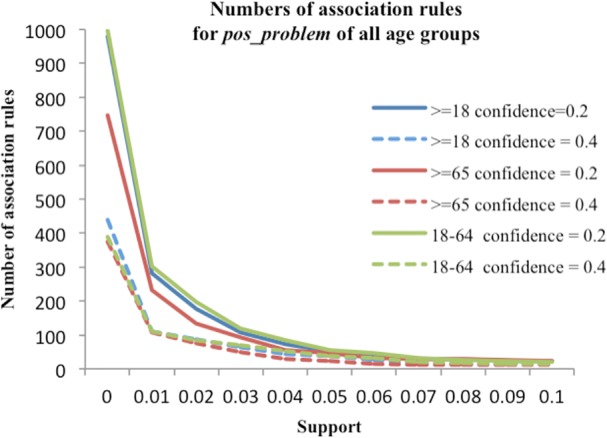
Numbers of association rules with different support value at confidence of 0.2 and 0.4 for all age groupings.
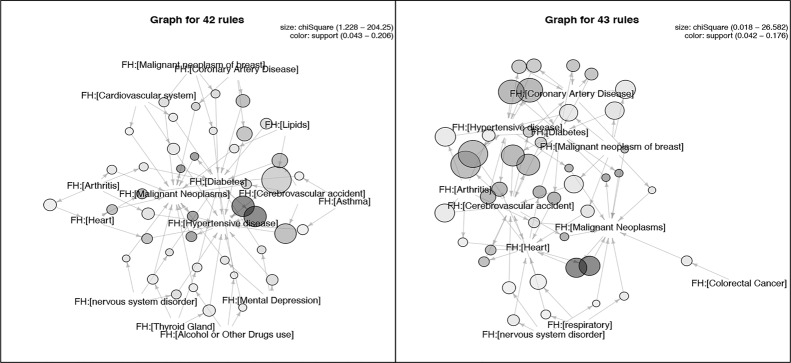
Figure 3.
Graph-based visualization of rules for pos_problem with minimum support of 0.04 and confidence of 0.4 [42 rules] for 18-64 year old adult patients (a) and rules for pos_problem with minimum support of 0.04 and confidence of 0.3 [43 rules] for senior patients (b).
Figure 3 shows the graph-based visualizations (generated using the “arulesViz” R package [10.2]) of the top 50 and 47 rules generated with the intermediate thresholds for the pos_problem subset of the 18-64 years old patient group and the senior adult patient group, respectively. In this representation, family history problems and rules are represented as vertices and edges indicate the relationship between problems in the rules. The size of the vertex specifies the value where a larger circle indicates a higher value. The color corresponds with support value where a darker shade indicates a higher value. For example, the top rule based on in Figure 3a is {FH:[Lipids]} => {FH:[Hypertension]} with support=0.1 and . The top rule in Figure 3b is also {FH:[Lipids]} => {FH:[Hypertension]} with support=0.05 and . This suggests that similar rules apply to both age groups when the number of rules generated for the two groups are close.
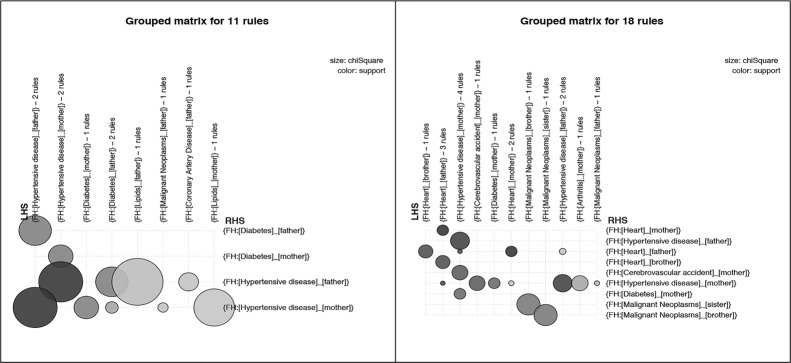
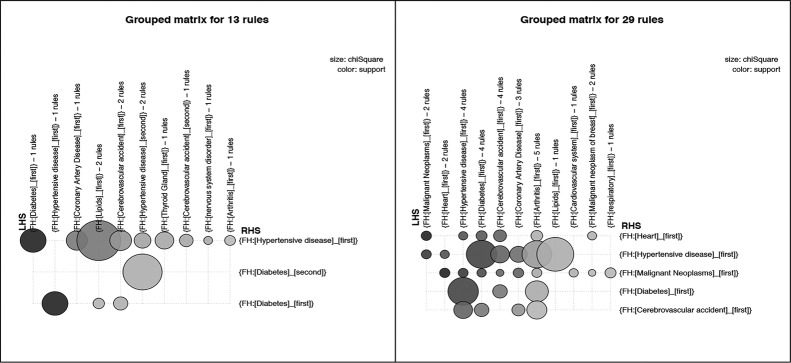
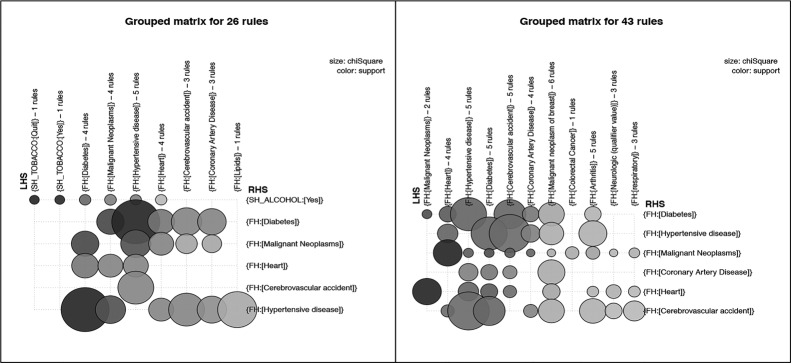
Similar to Figure 3, Figure 4 presents grouped matrix-based visualizations for the 18 and 13 rules generated using the intermediate thresholds for the pos_problem_relation subsets of two patient groups, and Figure 5 shows grouped matrix-based visualizations of rules generated for the pos_problem_degree subsets of the two patient groups. Figure 6 shows the that association rules generated for the two age groups (senior patients and 18-64 year old patients) for the combined subset pos_problem_substance_status
Figure 4.
Grouped matrix-based visualization of rules for pos_problem_relation with minimum support of 0.03 and confidence of 0.2 [11 rules] for 18-64 years adult patients (a) and rules for pos_problem_relation with minimum support of 0.03 and confidence of 0.2 [18 rules] for senior patients (b).
Figure 5.
Grouped matrix-based visualization of rules for pos_problem_degree with minimum support of 0.04 and confidence of 0.4 [13 rules] for 18-64 year old adult patients (a) and rules for pos_problem_degree with minimum support of 0.04 and confidence of 0.3 [29 rules] for senior patients (b).
Figure 6.
Graph-based visualization of rules for pos_problem_substance_status with minimum support of 0.04 and confidence of 0.4 [29 rules] for 18-64 years adult patients (a) and rules for pos_problem_substance_status with minimum support of 0.04 and confidence of 0.3 [50 rules] for senior patients (b).
The generated association rules for pos_problem_relation, pos_problem_side, and pos_problem_degree present more granular associations in comparison with those from pos_problem. The three sets of rules convey the presence of family history problems in particular relatives, side of family (maternal or paternal relative), and degree of relationship (first or second degree relative).
We observed that association rules generated for the two age groups (senior patients and 18-64 year old patients) for the same subsets (e.g., pos_problem or pos_problem_side) are similar when the numbers of rules generated for each group are close as shown in Figure 3a and 3b.
A body of published work has described familial risk factors of epilepsy related to family history of epilepsy in first-, second- or third-degree relatives3,5,8,9. While not directly family history, comorbidities of epilepsy include conditions such as cancer, cardiovascular disease, and hypertension, in several reviewed references27 –30. The top family history problems like the ones shown in Table 4 and association rules in Figures 3–5 are consistent with these known comorbidities of epilepsy. As presented in Figure 6, the interactions among social and behavioral factors such as alcohol, tobacco, and drug use and family history problems overall were not strong compared with rules among familial factors.
Discussion
In this study, we enhanced an existing approach for association rule mining to analyze interactions between familial risk factors and substance use for epilepsy using different age grouping. For family history, medical problems and relations associated with local codes were mapped to the UMLS and HL7 Vocabulary to promote generalizability of the approach as well as comparison of findings across EHR systems and institutions. Substance use status (for tobacco, alcohol, and drug use) was incorporated into the association rule mining process to enable exploration of interactions between substance use and family history. The preliminary results demonstrated a potential of the approach to validate known knowledge. While not known family history associations, Table 4 and Figure 3-5 show a good coverage of epilepsy comorbidities (e.g., Cancer, Hypertension, Stroke, Diabetes, and Arthritis). In addition, a number of association rules were generated for “FH:[Alcohol or Other Drugs use]” as shown in Figure 3 for pos_problem subset, suggesting that our approach can discover rules for family history of substance use and other family history problems.
We observed that unlike in our previous study on pediatric asthma, there is no specific entry for epilepsy within the list of medical problems used for family history stored in FHS EHR system. Consequently, the family history entries gathered from the EHR did not contain specific family history reference pertaining to the presence of epilepsy in first-, second-, or other degree family member. As a result, the datasets used in this study could not be used to validate familial risk factors such as relation degree (e.g., First or Second) for epilepsy. However, we observed that “nervous system disorder” was in the list of available medical problems, which included a range of neurologic diseases such as epilepsy. This was found to be a prevalent family history problem in Table 4 and some rules shown in Figure 5 and 6 were associated with this problem. To some degree, this type of finding is supported by other published studies3,5,8,9 Such issues underscore the need for enhanced tools that enable clinicians to document family history with the right level of granularity. To address this limitation, future work is planned that includes development of additional approaches for finding more specific family history problems from other structured fields or from free text notes.
Since existing family history modules available in many EHR systems may not accurately reflect family history problems and give little details, NLP tools or modules are needed to obtain this information from clinical notes. In our previous studies32,33, NLP modules were developed to identify family history and social history related information from clinical notes and extract detailed information (e.g., side of family and disease for family history and status, type, and frequency for tobacco, alcohol, and drug use). As part of next steps, these modules could be used to help collect specific family and social history information from the EHR.
A control population could be used to analyze rules generated from different populations to filter common rules and highlight those unique to the epilepsy cohort. In this study, we compared rules generated for epilepsy patients in different age groupings. Our results show similar rules generated for these groups. Additional evaluations utilizing more explicitly defined case and control populations are needed to further enhance the approach and validate the findings. As a preliminary study, this study showed the potential of using association rule mining to explore familial, social, and behavioral risk factors for epilepsy as well as some challenges (e.g., semantic granularity).
This feasibility study focused on pairwise associations, with the initial results suggesting that these associations have potential for discovering and validating knowledge of familial risk factors. Next steps include extending the maximum rule length in the “arules” R package to generate more than dichotomous associations. In addition, further work is needed to develop a strategy for determining optimal thresholds for confidence and support as well as to explore additional metrics for assessing and comparing the strength of rules.
Conclusion
In this study, we enhanced an association rule mining approach for generating interactions among potential familial risk factors and substance use for epilepsy. The promising results suggest that structured family and social history information from the EHR can indeed be used to validate known evidence as well as potentially support disease prediction and detection of novel associations.
Acknowledgements
The authors thank Diantha Howard, MS from the University of Vermont for code that was adapted for the data selection step. The National Institutes of Health through the National Library of Medicine (R01LM011364 and R01GM102282), Clinical and Translational Science Award (8UL1TR000114-02) supported this work. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
References
- 1.Thurman DJ, Beghi E, Begley CE, Berg AT, Buchhalter JR, Ding D, et al. Standards for epidemiologic studies and surveillance of epilepsy. Epilepsia. 2011;52:2–26. doi: 10.1111/j.1528-1167.2011.03121.x. [DOI] [PubMed] [Google Scholar]
- 2.Callenbach PMC, Geerts A, Arts WFM, Donselaar CAv, Peters ACB, Stroink H, et al. Familial occurrence of epilepsy in children with newly diagnosed multiple seizures: Dutch Study of Epilepsy in Childhood. Epilepsia. 1998;39(3):6. doi: 10.1111/j.1528-1157.1998.tb01382.x. [DOI] [PubMed] [Google Scholar]
- 3.Khan H, Mohamed A, Zina-Al-Sakini, Zulfiquar K, Sohail A, Shaikh RB, et al. Consanguinity, family history and risk of epilepsy: A case control study. Gulf Medical Journal. 2012;1(1):4. [Google Scholar]
- 4.Rong L, Jr. ATF, Benbadis SR. Tobacco smoking, epilepsy, and seizures. Epilepsy & Behavior. 2014;31:8. doi: 10.1016/j.yebeh.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Ogunrin OA, Obiabo OY, Obehigie E. Risk factors for epilepsy in Nigerians -a cross-sectional case- control study. Acta Neurologica Scandinavica. 2014;129(2):109–13. doi: 10.1111/ane.12192. [DOI] [PubMed] [Google Scholar]
- 6.Macintosh DS, Camfield PR, Camfield CS. Children With Familial Cryptogenic Epilepsy Have a Favorable Seizure Prognosis. Journal of Child Neurology. 1998;13(8):5. doi: 10.1177/088307389801300802. [DOI] [PubMed] [Google Scholar]
- 7.Ferro MA. A population-based study of the prevalence and sociodemographic risk factors of self-reported epilepsy among adults in the United Kingdom. Seizure. 2011;20(10):784–8. doi: 10.1016/j.seizure.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Cansu A, Serdaroğlu A, Yüksel D, Doğan V, Özkan S, Hirfanoğlu T, et al. Prevalence of some risk factors in children with epilepsy compared to their controls. Seizure. 2007;16(4):338–44. doi: 10.1016/j.seizure.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Asadi-Pooya AA, Hojabri K. Risk factors for childhood epilepsy: a case-control study. Epilepsy & Behavior. 2004;16(1):6. doi: 10.1016/j.yebeh.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Peljto AL, Barker-Cummings C, Vasoli VM, Leibson CL, Hauser WA, Buchhalter JR, et al. Familial risk of epilepsy: a population-based study. Brain. 2014;137(3):795–805. doi: 10.1093/brain/awt368. 2014-03-01 00:00:00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C-Y, Chang C-K, Robson D, Jackson R, Chen S-J, Hayes RD, et al. Evaluation of smoking status identification using electronic health records and open-text information in a large mental health case register. PLoS One. 2013;8(9):e74262. doi: 10.1371/journal.pone.0074262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wicentowski R, Sydes MR. Using implicit information to identify smoking status in smoke-blind medical discharge summaries. J Am Med Inform Assoc. 2008;15(1):29–31. doi: 10.1197/jamia.M2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long W. Extracting diagnoses from discharge summaries. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2005:470–4. [PMC free article] [PubMed] [Google Scholar]
- 15.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med. 1998;37(1):1–7. [PubMed] [Google Scholar]
- 16.Melton GB, Manaktala S, Sarkar IN, Chen ES. Social and behavioral history information in public health datasets. AMIA Annu Symp Proc. 2012;2012:625–34. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ES, Manaktala S, Sarkar IN, Melton GB. A multi-site content analysis of social history information in clinical notes; AMIA Annual Symposium proceedings /AMIA Symposium AMIA Symposium; 2011. pp. 227–36. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen E, Garcia-Webb M. An analysis of free-text alcohol use documentation in the electronic health record: early findings and implications. Appl Clin Inform. 2014;5(2):402–15. doi: 10.4338/ACI-2013-12-RA-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melton GB, Raman N, Chen ES, Sarkar IN, Pakhomov S, Madoff RD. Evaluation of family history information within clinical documents and adequacy of HL7 clinical statement and clinical genomics family history models for its representation: a case report. J Am Med Inform Assoc. 2010 May-Jun;17(3):337–40. doi: 10.1136/jamia.2009.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform. 2010 Dec;43(6):891–901. doi: 10.1016/j.jbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Roque FS, Jensen PB, Schmock H, Dalgaard M, Andreatta M, Hansen T, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011 Aug;7(8):e1002141. doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanauer DA, Rhodes DR, Chinnaiyan AM. Exploring clinical associations using ‘-omics’ based enrichment analyses. PLoS One. 2009;4(4):e5203. doi: 10.1371/journal.pone.0005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen ES, Melton GB, Wasserman RC, Rosenau PT, Howard DB, Sarkar IN. Mining and Visualizing Family History Associations in the Electronic Health Record: A Case Study for Pediatric Asthma; Proceedings of the American Medical Informatics Association Symposium; 2015. p. 2015. [PMC free article] [PubMed] [Google Scholar]
- 25.Unified Medical Language System (UMLS) Available from: http://www.nlm.nih.gov/research/umls/
- 26.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welty TE, Eng ML. Epilepsy in the Older Adult. Pharmacotherapy Self Assessment Program. 2011 [Google Scholar]
- 28.(CDC) CfDCaP. Comorbidity in adults with epilepsy--United States. MMWR Morb Mortal Wkly Rep 2013 Nov 1. 2010;62(43):849–53. [PMC free article] [PubMed] [Google Scholar]
- 29.Boro A, Haut S. Medical comorbidities in the treatment of epilepsy. Epilepsy & Behavior. 2003;4:2–12. doi: 10.1016/j.yebeh.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 30.ANNEGERS JF, ROCCA WA, HAUSER WA. Causes of Epilepsy: Contributions of the Rochester Epidemiology Project. Mayo Clinic Proceedings. 1996;71(6):570–5. doi: 10.4065/71.6.570. [DOI] [PubMed] [Google Scholar]
- 31.Aicardi J, Bancaud J, Beaussart M, Cohadon F, Courjon J, Favel P, et al. General Conclusions concerning Familial Factors in Epilepsy. Epilepsia. 1969;10(1):65–8. doi: 10.1111/j.1528-1157.1969.tb03830.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Chen ES, Pakhomov S, Arsoniadis E, Carter EW, Lindemann E, et al. Automated Extraction of Substance Use Information from Clinical Texts. Proceedings of the American Medical Informatics Association Symposium 2015. 2015 [PMC free article] [PubMed] [Google Scholar]
- 33.Bill R, Serguei P, Chen ES, Winden TJ, Carter EW, Melton GB. Automated Extraction of Family History Information from Clinical Notes. Proceedings of the American Medical Informatics Association Symposium. 2014 [PMC free article] [PubMed] [Google Scholar]