Abstract
Sentence boundary detection (SBD) is a critical preprocessing task for many natural language processing (NLP) applications. However, there has been little work on evaluating how well existing methods for SBD perform in the clinical domain. We evaluate five popular off-the-shelf NLP toolkits on the task of SBD in various kinds of text using a diverse set of corpora, including the GENIA corpus of biomedical abstracts, a corpus of clinical notes used in the 2010 i2b2 shared task, and two general-domain corpora (the British National Corpus and Switchboard). We find that, with the exception of the cTAKES system, the toolkits we evaluate perform noticeably worse on clinical text than on general-domain text. We identify and discuss major classes of errors, and suggest directions for future work to improve SBD methods in the clinical domain. We also make the code used for SBD evaluation in this paper available for download at http://github.com/drgriffis/SBD-Evaluation.
Introduction
With the growth in adoption of Electronic Health Records (EHRs) there is an increase in the number of studies that make use of clinical data for various purposes. While structured data such as lab values, medications and diagnosis codes are quick to document and look up, medical nuances are often found in free text clinical notes, which is a preferred means of documentation for physicians1. Thus, researchers are using natural language processing (NLP) techniques to process clinical notes and extract knowledge from them. The sentence is a fundamental unit in NLP research. A typical NLP pipeline in the biomedical domain consists of identifying medical concepts, parsing, part of speech tagging, negation identification, word sense disambiguation, etc. as pre-processing tasks. Most of these begin with the fundamental task of identifying sentences, or what is more formally referred to as sentence boundary detection (SBD). SBD is a non-trivial problem, since the end-of-sentence punctuation marks are ambiguous. For example, a period can refer to an abbreviation, the end of a sentence, or a numeric decimal point. Exclamation points, question marks, quotation marks, ellipses, parentheses or a combination of these make the problem even more challenging.
Although SBD is an important task, it has not recently received much attention from the NLP community, for a variety of reasons2,3. Previous work in SBD is mostly restricted to the news domain and limited datasets such as the Wall Street Journal (WSJ) corpus4. These studies discuss problems with SBD assuming properties of good quality text, such as proper formatting, grammatical sentence construction, and the absence of incorrect spellings. Clinical text, however, does not possess these desirable properties5. The challenges associated with SBD are thus elevated in the clinical domain, since basic assumptions of sentence construction and punctuation do not hold true; for example, only 50% of the sentences in our clinical notes corpus end in periods, compared to 99% in the GENIA corpus. It is likely that models developed for SBD on open domain data may not generalize well to clinical texts. Popular shared tasks in the clinical domain, such as the i2b2 challenges6 –8, provide participants with gold-standard sentence boundaries to evaluate isolated NLP tasks. Previous studies in the open domain have shown that the impact of error propagation originating from SBD into other NLP tasks is substantial9,10. Despite this, SBD has been a neglected task in the biomedical NLP community, with very few studies reviewing or addressing the problem. Therefore, the problem of SBD in the clinical domain deserves a comprehensive assessment with existing NLP tools and directions for development of appropriate solutions.
In this paper, we identify multiple popular NLP tools used by the biomedical community and evaluate their performance on the SBD task. Our primary focus is on comparing performance between general-domain text and clinical text. We use the toolkits off-the-shelf, i.e. without additional training or modification, and compare their performance over a variety of openly available datasets. More specifically, we evaluate cTAKES11, Stanford CoreNLP12, Lingpipe13, SPECIALIST14 and Splitta2 on the general-domain British National Corpus, transcripts from the Switchboard corpus17, the GENIA corpus15 of biomedical abstracts, and a set of clinical notes from the 2010 i2b2 challenge6. We have made the source code of our evaluation available for use by the research communitya.
The paper is organized as follows: we review related work in both the open domain and clinical domain for the task of SBD. This is followed by an overview of the different tools and datasets used in this study, and a description of our evaluation method. We then present the results of our quantitative evaluation, and demonstrate that SBD performance varies significantly across corpora and tools. Performance of most tools on our clinical corpus is noticeably lower than on our other corpora. We further include a qualitative evaluation of common sources of error across different tool-corpus combinations. Finally, we discuss directions for future work to address this problem in the clinical domain.
Related Work
The task of SBD has been addressed by relatively few studies in the NLP community. Interestingly, machine learning was the earliest approach for SBD, with rule-based systems being developed later. There have been some attempts exploring the use of unsupervised techniques as well. One of the first SBD systems, by Riley18, uses a decision tree classifier to address the “not-so-simple problem of deciding when a period in text corresponds to the end of a declarative sentence.” Thus, his work investigates only a single punctuation mark: namely, the period. The decision tree model uses features such as length and the case of the words before and after the period and probabilities of words beginning or ending a sentence. Evaluation using a model trained on 25 million words of newswire text achieved an accuracy of 99.8%.
Reynar and Ratnaparkhi19 create an SBD system using maximum entropy learning in a supervised setup. They consider candidate sentence boundaries to be any token containing a question mark, exclamation point, or period. They highlight the importance of training data for generalization and discuss two models: a domain-specific model trained for financial newspaper text, and a portable model for generic use. The models are tested on WSJ text and the Brown corpus20. The domain-dependent system achieves accuracies of 98.8% and 97.9%, respectively, on the two datasets, and the portable system achieves accuracies of 98.0% and 97.5%. The SBD module in OpenNLPb is based on this implementation. Palmer and Hearst21 present SATZ, a system that uses the part-of-speech distribution of the context surrounding candidate tokens for identifying sentence boundaries. Each word in the context is represented as a binary vector indicating plausibility of parts-of-speech for that word. Evaluation on WSJ text results in an error rate of 1.1% with a neural network and 1.0% using a decision tree. Gillick2 reviews supervised learning for SBD and discusses feature extensions using Naïve Bayes and Support Vector Machine (SVM) based models. Gillick focuses primarily on the period as a sentence separator. The best model uses an SVM, reporting an error rate of 0.25% on WSJ and 0.36% on the Brown corpus.
Mikheev22 proposes a rule-based system for the task of SBD. The system obtains a list of abbreviations and proper names in an unsupervised manner from a text corpus. Then, rules look for the presence of these terms among words to the left or right of a potential sentence boundary. The system achieves an error rate of 0.45% on WSJ and 0.28% on the Brown corpus. Extending this system with a part-of-speech tagger reduces the error rates to 0.31% and 0.20%, respectively. Kiss and Strunk10 treat SBD as a fully unsupervised task. Their Punkt system identifies abbreviations based on colocations between periods and sentence boundary words. They report an error rate of 1.02% on the Brown corpus and 1.65% on WSJ. Their system also shows promising results on corpora from ten other languages.
More recently, Read et al. 3 conducted a systematic analysis of SBD tools similar to ours, but with a focus on generalization towards user-generated Web content. They evaluated nine tools on five datasets. Specifically, they evaluated Stanford CoreNLP, Lingpipe, MxTerminator, OpenNLP, Punkt, RASP, Splitta, and tokenizer on the Brown Corpus, GENIA, WSJ, and the Conan Doyle Corpus23. They observe a performance degradation when moving from corpora with formal language to those that are less formal. They show that moderate interpretation of document structure contributes to an increase in the overall SBD performance.
As stated earlier, although the clinical domain is plagued with problems of text quality, SBD has not received much attention in this domain. Buyko et al. 24 present a study of SBD for the biology domain. They retrain components of the OpenNLP system with the GENIA and PennBiolE25 corpora. Using ten-fold cross validation, their retrained modules achieve accuracies of 99.0% and 97.4% respectively on the two corpora. Orosz26 present a hybrid system, a combination of rule-based, and unsupervised machine learning that achieves an F05-Score of 91.89%. Their corpus, a set of clinical notes in the Hungarian language, consists of 1,320 lines in the development set and 1,310 lines in the test set. The Punkt and OpenNLP systems yield F05 of 55.59% and 57.37% respectively on their test set. Recently, Kreuzthaler and Schulz27 presented an SBD system for German clinical narratives using an SVM and various handcrafted features. They report an Fl-score of 0.94 using ten-fold cross validation on 848 discharge summaries from the dermatology department.
To the best of our knowledge, the SBD module in cTAKES is the only system trained for clinical notes in English. It extends the SBD implementation from OpenNLP, which predicts whether a period, question mark, or exclamation mark is the end of a sentence. The best results are achieved with a model trained on data combined from the following datasets: GENIA, WSJ articles from the Penn Treebank (PTB), and a corpus of 273 clinical notes from the Mayo Clinic. This resulted in accuracies of 98.6%, 94.2%, and 94.9% on the three corpora, using 10-fold cross validation with an 80/20 split for each fold.
Our evaluation focuses on comparing performance between the general domain and the clinical domain: we use models trained on both types of text and evaluate them on both domains. Additionally, we evaluate only off-the-shelf models, with no additional training for a specific downstream task.
Methods
Datasets
We evaluate the selected toolkits on SBD in four datasets, covering a variety of sources and types of text: the British National Corpus, the Switchboard telephone corpus, the GENIA corpus of MEDLINE citations, and a set of clinical notes from the 2010 i2b2 challenge. Table 1 describes the text in each corpus, and shows the number of documents and sentences present in each. We note that each corpus defines a “sentence” differently; we describe the annotation choices and background of each dataset below.
Table 1.
Summary of datasets
| Corpus | Type of Data | # of Documents | # of Sentences | Avg. Sentence Length (# Tokens) |
|---|---|---|---|---|
| BNC | General text, mixed-domain | 4,049 | 6,027,378 | 16.1 |
| Switchboard | Telephone conversations | 650 | 110,504 | 7.4 |
| GENIA | MEDLINE abstracts | 1,999 | 16,479 | 24.4 |
| i2b2 | Clinical Notes | 426 | 43,940 | 9.5 |
The British National Corpus (BNC) 16 is a large corpus of British English written and spoken texts, sampled from academic writing, fiction, newspaper text, and spoken conversations, among other domains. It exhibits a concomitant variety of writing styles, document structure, and sentence formation. Documents are annotated for layout (e.g. paragraph breaks, section headers, etc.), segment boundaries, and tokens, including both words and punctuation. Individual tokens are annotated with their lemmatization and part of speech (POS) tag. Annotations were performed automatically28,29. The corpus is broken into segments, described in the documentation as “a sentence-like division of textc.” Though these do not always align with human judgments of sentence boundaries, we consider each segment to contain precisely one sentence.
The Switchboard (SWB) corpus17 consists of transcripts of spontaneous telephone conversations on a variety of topics. Switchboard has been heavily used in automatic speech recognition systems and related tasks30,31, and has previously been used to evaluate SBD in spoken text32. Switchboard exhibits markedly different linguistic structure from written text: sentences are left incomplete or change topic partway through, and speakers interrupt one another and add interjections and acknowledgments. These disfluencies, along with complete constituency parses, were hand-annotated by the Linguistic Data Consortium in the PTB style33,34. While the disfluencies present in normal speech complicate determining appropriate reference boundaries, we consider each root S node in the tagged documents to represent one sentence for the SBD task; we ignored all editorial marks and syntactic placeholders.
The GENIA corpus15 is a collection of 1,999 MEDLINE abstracts related to transcription factors in human blood cells. It is a popular resource for biomedical NLP35, and has been used as a training corpus for off-the-shelf language models in multiple toolkits, including cTAKES, CoreNLP, and LingPipe. The language in GENIA tends to be well-formed with long sentences, and the corpus has a high frequency for biomedical terms, phrases, and abbreviations. The abstracts are also all unstructured, with no headers for Materials, Methods, or Conclusion. We used GENIA Treebank version 1.036, which was hand-annotated with constituency parses37.
Our clinical notes corpus (referred to in this paper as i2b2) consists of all labeled clinical documents (n=426) from the i2b2 2010 shared task on identifying concepts, assertions, and relations in clinical text6. The documents are of two varieties: discharge summaries and progress reports. In contrast to the well-formed text found in GENIA and much of the BNC, text in clinical documents is often short, ungrammatical, and full of abbreviations and other forms of shorthand5. Many of the text segments considered as sentences for clinical purposes would not be treated as such in general-domain settings, e.g. topical headers and other short, distinct chunks of information. We obtained the corpus in plaintext format, hand-annotatedd with one sentence per line in each document.
Toolkits
We evaluate five off-the-shelf NLP toolkits on their performance of SBD across our corpora: the Stanford CoreNLP tools, LingPipe, Splitta, the SPECIALIST NLP tools, and cTAKES. It is worth noting that these toolkits perform a wide variety of NLP tasks beyond SBD, and each takes different approaches to its set of tasks. However, each toolkit has been used for SBD in the literature, and provides comparable functionality for the task. Table 2 lists where we obtained each toolkit, and what training data was used for the pre-trained models.
Table 2.
Summary of toolkits
| Toolkit | Version | URL (as of 1/7/2016) | Training Corpora |
|---|---|---|---|
| Stanford CoreNLP |
3.5.2 (4/20/15) |
http://nlp.stanford.edu/software/corenlp.shtml | PTB, GENIA, Other Stanford corpora |
| Lingpipe | 4.1.0 (6/27/2011) |
http://alias-i.com/lingpipe/ | MEDLINE abstracts, general text |
| Splitta | 1.03 (2/16/10) |
http://code.google.com/p/splitta/ | PTB |
| SPECIALIST | 2.4C (11/10/06) |
http://lexsrv3.nlm.nih.gov/Specialist/Home/index.html | SPECIALIST lexicon50 |
| cTAKES | 3.2.2 (5/30/15) |
http://ctakes.apache.org/ | GENIA, PTB, Mayo Clinic EMR |
The Stanford CoreNLP 12 toolkit contains several NLP tools used for tasks such as POS tagging, named entity recognition, coreference resolution, sentiment analysis, and pattern learning. Its components have been heavily used in general-domain NLP38,39, as well as clinical text mining40 –42. The parsing tool, which is most relevant to our evaluation, uses a rule-based system developed on the WSJ portion of the PTB, the GENIA corpus, and other general-domain English text.
LingPipe 13 is a general-domain NLP toolkit designed for a wide range of NLP tasks, from tokenization and parsing to POS tagging, named-entity recognition, sentiment analysis, and word sense disambiguation, among others. It comes with two pre-trained sentence models: one trained on MEDLINE abstracts (denoted here as LingPipeME), and the other trained for general Indo-European text (denoted as LingPipeGEN); we evaluate both models. LingPipe has been used previously in the biomedical informatics literature for NER, among other tasks43,44.
Splitta 2 is a dedicated toolkit for tokenization and SBD. It is provided with two classification models pre-trained on the WSJ and Brown corpus portions of the PTB; one model using a support vector machine (SVM) approach, and the other based on a Naive Bayes classifier. We evaluate both models on our task. It has seen prior use in SBD evaluations2,3 and clinical text mining45.
The SPECIALIST NLP tools 14 are a suite of tools created by the National Library of Medicine (NLM) for NLP tasks in the biomedical domain. The tools perform a range of tasks, from tokenization and parsing to identifying lexical variants and adding POS tags, and are used as part of the popular MetaMap software46. For the purposes of this evaluation, we used only the Tokenizer component of the SPECIALIST Text Tools, which tokenizes and provides phrase-level, sentence-level, and section-level boundaries. The tools are trained on the documents used to create the SPECIALIST lexicon.
The Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) 11 is an NLP system designed for information extraction from clinical text, building on the general-domain OpenNLP toolkit. cTAKES performs a rich variety of NLP tasks, including parsing, extraction and annotation of UMLS concepts, and coreference resolution, among others. It has seen increasing use in clinical research47 –49. The pre-trained models provided with the toolkit were created from a mix of documents including the GENIA corpus, documents from the PTB, and clinical documents sampled from the Mayo Clinic EMR corpus.
Experimental Method
For BNC, Switchboard, and GENIA, we extracted a plaintext version of each document, along with sentence boundaries calculated during the extraction. For i2b2, since we obtained the corpus in plaintext format, we only calculated sentence bounds using the implicit annotations in the documents. All sentence bounds were calculated as character offsets from the beginning of the document. We used whitespace to maintain document structure when necessary, substituting one or more newline characters between paragraphs, sections, etc.
We evaluated each toolkit’s performance on detecting sentence boundaries in each of the four corpora. We follow Read et al. 3 by evaluating SBD using character offsets from the beginning of a document to denote its bounds. Thus, we ran each toolkit on each dataset and extracted the sentence boundaries from the results, which we compared against gold standard bounds extracted from the annotated corpora. Sentence boundaries assigned by the toolkits that were not present in the reference bounds were treated as false positives, and reference bounds that were missing in the toolkit output were considered false negatives. We note that under this design, there are no true negatives for the purposes of evaluation; negative results are reflected in decisions not made in the SBD task. Additionally, each correct bound detection counts twice (once for the end of the sentence and once for the start of the next sentence), and each error does so as well.
Results
We report precision, recall, and F1-scores from this comparison in Table 3. We do not report results for Splitta on BNC, because the toolkit implementation we used collapses consecutive whitespace characters, which are used to indicate document structure in our plaintext extraction of BNC. Our evaluation based on character offsets was therefore impractical for this pairing.
Table 3.
Precision (Pr), Recall (Re), and F1 score (FS) of sentence boundary detection task, evaluated for each tool on each dataset. The best results for each dataset are highlighted in bold.
| Toolkit | BNC | SWB | GENIA | i2b2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | FS | Pr | Re | FS | Pr | Re | FS | Pr | Re | FS | |
| Stanford | 0.89 | 0.77 | 0.82 | 0.59 | 0.37 | 0.45 | 0.98 | 0.98 | 0.98 | 0.58 | 0.34 | 0.43 |
| LingpipeGEN | 0.83 | 0.65 | 0.73 | 0.58 | 0.33 | 0.42 | 0.97 | 0.95 | 0.96 | 0.59 | 0.33 | 0.42 |
| LingpipeME | 0.82 | 0.65 | 0.72 | 0.59 | 0.34 | 0.43 | 0.99 | 0.99 | 0.99 | 0.54 | 0.34 | 0.41 |
| SplittaSVM | - | - | - | 0.55 | 0.30 | 0.39 | 0.98 | 0.96 | 0.97 | 0.59 | 0.34 | 0.43 |
| SplittaNB | - | - | - | 0.55 | 0.30 | 0.39 | 0.99 | 0.99 | 0.99 | 0.58 | 0.35 | 0.43 |
| SPECIALIST | 0.77 | 0.71 | 0.74 | 0.60 | 0.37 | 0.46 | 0.89 | 0.94 | 0.92 | 0.58 | 0.53 | 0.56 |
| cTAKES | 0.73 | 0.75 | 0.74 | 0.67 | 0.42 | 0.55 | 0.62 | 0.76 | 0.68 | 0.93 | 0.97 | 0.95 |
On a practical note, we found that the SPECIALIST and cTAKES tools could be run with no modification to the source code, but both required some post-processing to recover the character-level sentence bounds from the output. Splitta required some modifications to the source code in order to track sentence bounds at the character level, though this obviated the need for post-processing. LingPipe and Stanford CoreNLP required the most code-level work, needing API access to extract sentence bound information. The interested reader should refer to the source code for details.
Our experimental results show that all toolkits except cTAKES perform extremely well on the well-formed text in GENIA, and somewhat lower on the more mixed-domain text in BNC. The short, telegraphic sentences of Switchboard were extremely difficult for any of the toolkits to parse correctly: cTAKES performed the best, but still had a comparatively low F1-score (0.55). Finally, we saw that the clinical notes in the i2b2 corpus were even more difficult than Switchboard for every toolkit except cTAKES; its pre-training on clinical notes gave it an F1-score more than twice as high as that most of the other toolkits.
Analysis by terminal character
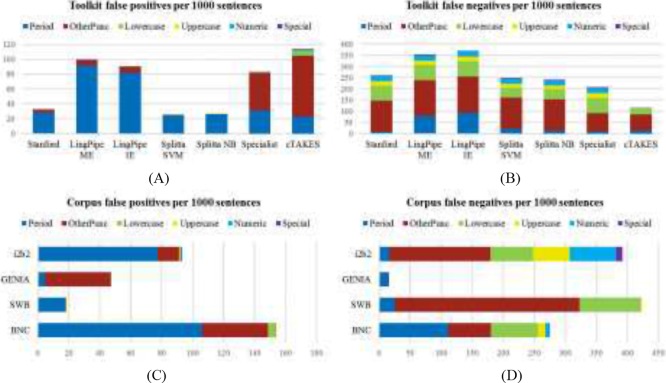
To get a more detailed picture of SBD performance for different kinds of sentences, we further break down our results by the type of character used to end a sentence in each of the four corpora. We group characters into periods, all other punctuatione, lowercase alpha characters, uppercase alpha, numeric characters, and all other characters (the “Special” category). Table 4 shows the distribution of these different terminal character types in the four corpora, and Figure 1 illustrates the distribution of false positive and false negative errors among them.
Table 4.
Distribution of terminal sentence characters among the four corpora, by character type.
| Corpus | Period | Other Punctuation | Lowercase | Uppercase | Numeric | Special |
|---|---|---|---|---|---|---|
| BNC | 74% | 15% | 9% | 1% | <1% | <1% |
| Switchboard | 56% | 33% | 10% | <1% | 0% | <1% |
| GENIA | 99% | <1% | <1% | <1% | 0% | <1% |
| I2b2 | 51% | 22% | 9% | 7% | 9% | 1% |
Figure 1.
Average errors per 1000 sentences, by the type of terminal character in the sentence. A and B show errors created by each toolkit, calculated as a sum of the errors on each corpus weighted by the number of sentences in that corpus. C and D show errors that occurred in each corpus, averaged across the toolkits used.
Runtime
Figure 2 displays the compute time required to run each toolkit on the i2b2 corpus on an 8-core 64-bit server; note that none of the tools is very parallelized. The speed differences are stark, and only grow in impact with the size of the dataset: running the Naive Bayes Splitta on BNC requires over 19 hours, and cTAKES clocks in at just over 6.5 hours, while LingPipe processes the corpus in approximately 90 seconds. While each toolkit performs more operations than strictly necessary for the SBD task, the amount of extra processing does not directly correlate with the increase in runtime. Most significantly, we note that cTAKES, running in a minimal configuration to only get chunking results, is nearly 100x slower than LingPipe running with the MEDLINE model.
Figure 2.
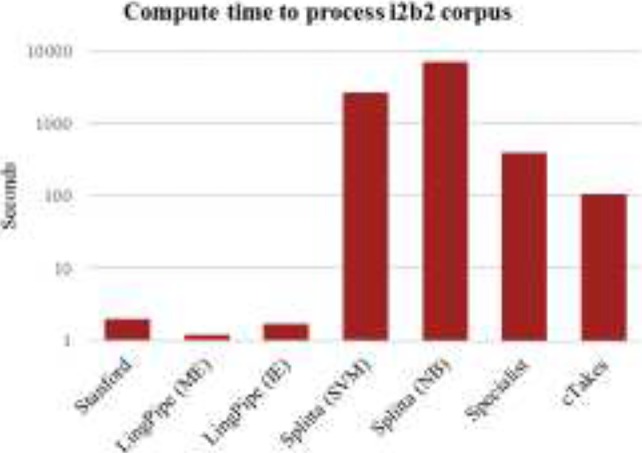
Runtime to process clinical notes corpus.
Discussion
We found several trends in the errors from our evaluation. One of the primary errors from every toolkit on every corpus was an oversensitivity to semicolons and colons: regardless of context, these marks were treated as strong indicators of the end of a sentence. Other common errors included treating less-common punctuation marks such as ellipses and repeated hyphens as sentence terminators. Some of these punctuation errors stem from the different definitions of a sentence, as discussed in the Methods section. For example, in the i2b2 corpus, colons are often annotated as sentence terminals in section headers and labels, whereas in GENIA, colons never denote sentence boundaries. Interestingly, in the BNC, colons are quite common as sentence terminals; this may reflect different usage in British English than in American English.
We also noted that the off-the-shelf models in the various toolkits showed different segmentation preferences based on their training domain; the shorter chunks marked as distinct segments in clinical text meant that cTAKES preferred to treat a colon as a sentence terminal, but the Stanford CoreNLP tools (trained on well-formatted text) skipped right past several true colon boundaries. These preferences were also reflected in the toolkits’ ability to detect short, non-terminated sentences, as are common in the i2b2 and Switchboard corpora. CoreNLP, LingPipe, SPECIALIST, and Splitta all tended to concatenate sequences of non-terminated sentences until they found a period; while cTAKES did slightly better in these cases, it was not immune to this error.
The period plays a central role in SBD, and a large number of the errors we encountered stemmed from periods. One of the most common types of false positive, especially for the clinical domain, came from unfamiliar or nonstandard abbreviations, including case-sensitivity. For example, while “Dr.” is a known abbreviation for all of the toolkits and only rarely treated as ending a sentence, the uppercase “DR.” or lowercase “dr.” tokens, both common in the i2b2 corpus, are frequently considered sentence-terminal. Initials are also handled poorly: “J. Smith” is frequently marked as ending one sentence and beginning another; this was a recurring error in both the i2b2 notes and the academic and news writing sections of the BNC.
Furthermore, all of the toolkits were highly sensitive to unusual patterns around periods. In cases where a sentence began with a lowercase word, either due to typographical error (very common in i2b2, but also present in BNC) or because the first token is something normally written in lowercase, such as a gene name (common in GENIA), we found that the sentence boundary preceding it was missed more often than not. Similarly, when either a sentence began or ended with a number, the toolkits often missed its boundary.
Abbreviations and acronyms were also problematic. While the various off-the-shelf models were trained to recognize some abbreviations, unfamiliar abbreviations (e.g., “ibid.”) or unfamiliar forms of known abbreviations (e.g., “t.i.d.” vs “t.i.d”) were repeatedly classified as ending a sentence. This was a particular issue in the i2b2 corpus, which exhibits a wide variety of acronyms, often used in succession (e.g., “mg. po. b.i.d.”).
While cTAKES avoided some of the errors exhibited by the other toolkits on the i2b2 corpus, we note two unique kinds of errors from it. The first is that when a number is followed by a non-whitespace character, e.g., “12.3*”, as occurs in several notes describing patient measurements, cTAKES is unable to recognize the number and treats the decimal point as a bounding period. The second is that cTAKES is prone to treat any sequence of all-uppercase letters like a header; this becomes an issue in text with acronyms, e.g., “Natural Language Processing (NLP).” The problem is exacerbated when the text occurs in parentheses; several other false positives were a result of mixed alphanumeric text in parentheses, e.g., “(50 mg per day).”
Whitespace
One significant issue we encountered in running the different toolkits was inconsistent handling of whitespace. Given a text segment like:
“Sentence A. Sentence B.”
with two spaces between the sentences, CoreNLP will assign the spaces as both the ending of sentence A and the beginning of sentence B, LingPipe and SPECIALIST consider the first space as part of sentence A, cTAKES does not incorporate either space into either sentence, and Splitta reduces the two spaces to one in its output, with that space not being assigned to either sentence. Additionally, SPECIALIST considers two consecutive newlines to signify a sentence boundary, and three or more newlines indicates the end of the document. Thus, while the whitespace does not affect the tokens that are included in each sentence, these issues suggest that careful testing is needed any time the output of one of these toolkits will be cross-referenced with the original text downstream.
Mitigating errors
Unfortunately, avoiding many of the kinds of errors we observed via preprocessing alone is difficult. Some problems, such as sensitivity to punctuation, acronyms, and alphanumeric parenthesized text, are unpredictable and may require complex regular expressions to identify. Fixing others, such as lower-case gene names and numeric formatting of patient measurements, risks changing the meaning of the text or potentially impacting downstream NLP applications. However, the issues of whitespace and unknown acronyms and abbreviations can be largely fixed by automated formatting and improved abbreviation dictionaries that have been adapted to the target domain.
Conclusion
The task of identifying sentence boundaries is integral to many NLP applications. However, SBD has largely been treated as a solved problem in the biomedical domain, as it has been common practice to use off-the-shelf models to split sentences with minimal correction or adaptation for the specific task at hand. We describe and quantify the kinds of errors that arise from using several popular off-the-shelf SBD models on various domains, including clinical text. We find several interesting trends, primarily around domain-specific use of punctuation. In our clinical data, semicolons, colons, and newlines are heavily used and error prone, while periods caused errors in multiple corpora when used in unknown abbreviations, names, and numbers. Additionally, we note that both the ease of use of each toolkit and the additional work it performs on top of SBD varies widely, as does its runtime. Our observations indicate that SBD remains a difficult problem in the biomedical domain, and that the field will benefit from renewed effort to create or train efficient, domain-adapted models for this fundamental task.
Acknowledgments
Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award number R01LM011116, the Intramural Research Program of the National Institutes of Health, Clinical Research Center, and through an Inter-Agency Agreement with the US Social Security Administration. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Dr. O. Uzuner, personal communication
Includes the following characters:? !: ; , () … “ - ‘ ‘ ‘ ‘— –”
References
- 1.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–6. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillick D. Sentence boundary detection and the problem with the U.S. Proc Hum Lang Technol 2009 Annu Conf North Am Chapter Assoc Comput Linguist Companion Vol Short Pap - NAACL ‘09; 2009. p. 241. [Google Scholar]
- 3.Read J, Dridan R, Oepen S, Solberg J. Sentence Boundary Detection: A Long Solved Problem?; In Proceedings of COLING; 2012. [Google Scholar]
- 4.Marcus MP, Santorini B, Marcinkiewicz MA. Building a Large Annotated Corpus of English: The Penn Treebank. Comput Linguist. 1993;19(2):313–30. [Google Scholar]
- 5.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 6.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2012;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc. 2012;19(5):786–91. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker DJ, Clements DE, Darwin M, Amtrup JW. Sentence boundary detection: A comparison of paradigms for improving MT quality; In Proceedings of the MT Summit VIII; 2001. [Google Scholar]
- 10.Kiss T, Strunk J. Unsupervised Multilingual Sentence Boundary Detection. Comput Linguist. 2006;32(4):485–525. [Google Scholar]
- 11.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning C, Surdeanu M, Bauer J, Finkel J, Bethard S, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit; Proc 52nd Annu Meet Assoc Comput Linguist Syst Demonstr; 2014. pp. 55–60. [Google Scholar]
- 13.LingPipe 4.1.0. 2008. Available from: http://alias-i.com/lingpipe/
- 14.McCray a T, Srinivasan S, Browne a C. Lexical methods for managing variation in biomedical terminologies; Proc Annu Symp Comput Appl Med Care; 1994. pp. 235–9. [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J-D, Ohta T, Tateisi Y, Tsujii J. GENIA corpus--a semantically annotated corpus for bio-textmining. Bioinformatics. 2003 Jul;19(Suppl 1):i180–2. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 16.The British National Corpus, version 3 (BNC XML Edition). Distributed by Oxford University Computing Services on behalf of the BNC Consortium. 2007. Available from: http://www.natcorp.ox.ac.uk/
- 17.Godfrey J, Holliman E. Web Download. Philadelphia: Linguistic Data Consortium; 1993. Switchboard-1. Release 2 LDC97S62. [Google Scholar]
- 18.Riley MD. Some applications of tree-based modelling to speech and language. Proc Work Speech Nat Lang. 1989;(2):339–52. [Google Scholar]
- 19.Reynar JC, Ratnaparkhi A. A Maximum Entropy Approach to Identifying Sentence Boundaries; Proceedings of the 5th Annual Conference on Applied Natural Language Processing. Association for Computational Linguistics; 1997. pp. 16–9. [Google Scholar]
- 20.Francis WN. A Standard sample of present-day English for use with digital computers. 1964.
- 21.Palmer DD, Hearst MA. Adaptive Multilingual Sentence Boundary Disambiguation. Comput Linguist. 1997;23(2):241–67. [Google Scholar]
- 22.Mikheev A. Tagging sentence boundaries; Proc 1 st North Am chapter …; 2000. pp. 264–71. [Google Scholar]
- 23.Morante R, Blanco E. * SEM 2012 Shared Task: Resolving the Scope and Focus of Negation; Proceedings of the First Joint Conference on Lexical and Computational Semantics-Volume 1: Proceedings of the main conference and the shared task, and Volume 2: Proceedings of the Sixth International Workshop on Semantic Evaluation. Association for Computational Linguistics; 2012. pp. 265–74. [Google Scholar]
- 24.Buyko E, Wermter J, Poprat M, Hahn U. Automatically Adapting an NLP Core Engine to the Biology Domain; Proc ISMB 2006 “Joint Link Lit Inf Knowl Biol 9th Bio-Ontologies Meet; 2006. pp. 65–8. [Google Scholar]
- 25.Liberman M, Mandel M, White P. Web Download. Philadelphia: Linguistic Data Consortium; 2008. PennBioIE Oncology 1.0 LDC2008T21. [Google Scholar]
- 26.Orosz G, Novak A, Proszeky G. Advances in Artificial Intelligence and Its Applications. Springer; 2013. Hybrid text segmentation for Hungarian clinical records; pp. 306–17. [Google Scholar]
- 27.Kreuzthaler M, Schulz S. Detection of sentence boundaries and abbreviations in clinical narratives. BMC Med Inform Decis Mak. BioMed Central Ltd. 2015;15(Suppl 2):S4. doi: 10.1186/1472-6947-15-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leech G, Garside R, Bryant M. CLAWS4: The tagging of the British National Corpus; Proc 15th Int Conf Comput Linguist (COLING 94); 1994. pp. 622–8. [Google Scholar]
- 29.Garside R, Leech GN, McEnery T, editors. Corpus annotation: linguistic information from computer text corpora. Taylor & Francis; 1997. [Google Scholar]
- 30.Seide F, Li G, Yu D. Conversational speech transcription using Context-Dependent Deep Neural Networks; Proc Annu Conf Int Speech Commun Assoc INTERSPEECH; 2011. pp. 437–40. [Google Scholar]
- 31.Kotti M, Moschou V, Kotropoulos C. Speaker segmentation and clustering. Signal Processing. Elsevier. 2008;88(5):1091–124. [Google Scholar]
- 32.Stolcke A, Shriberg E, Bates R, Ostendorf M, Hakkani D, Plache M, et al. Automatic detection of sentence boundaries and disfluencies based on recognized words; Int Conf on Spoken Language Processing (ICSLP); 1998. [Google Scholar]
- 33.Meteer MW, Taylor AA, MacIntyre R, Iyer R. Dysfluency annotation stylebook for the switchboard corpus. University of Pennsylvania; 1995. [Google Scholar]
- 34.Bies A, Ferguson M, Katz K, MacInture R, Tredinnick V, Kim G, et al. Bracketing guidelines for Treebank II style Penn Treebank project. Univ …. 1995 [Google Scholar]
- 35.Simpson MS, Demner-Fushman D. Mining text data. Springer; 2012. Biomedical text mining: a survey of recent progress; pp. 465–517. [Google Scholar]
- 36.GENIA Treebank v1.0. Available from: http://www.geniaproject.org.
- 37.Tateisi Y, Yakushiji A, Ohta T, Tsujii J. GENIA Annotation Guidelines for Treebanking. 2006.
- 38.Hirschberg J, Manning CD. Advances in natural language processing. Science (80-). American Association for the Advancement of Science. 2015;349(6245):261–6. doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 39.Xue N, Ng HT, Pradhan S, Prasad R, Bryant C, Rutherford A. The CoNLL-2015 Shared Task on Shallow Discourse Parsing; In Proceedings of the Nineteenth Conference on Computational Natural Language Learning - Shared Task; Beijing, China: Association for Computational Linguistics; 2015. pp. 1–16. [Google Scholar]
- 40.Patra BG, Ghosh N, Das D, Bandyopadhyay S. Computational Linguistics and Intelligent Text Processing. Springer; 2015. Identifying Temporal Information and Tracking Sentiment in Cancer Patients’ Interviews; pp. 180–8. [Google Scholar]
- 41.Torii M, Fan J, Yang W, Lee T, Wiley MT, Zisook DS, et al. Risk factor detection for heart disease by applying text analytics in electronic medical records. J Biomed Inform. Elsevier Inc. 2015. [DOI] [PMC free article] [PubMed]
- 42.Kilicoglu H, Rosemblat G, Cairelli MJ, Rindflesch TC. A Compositional Interpretation of Biomedical Event Factuality. 2004.
- 43.Carpenter B. LingPipe for 99.99% Recall of Gene Mentions; Proc Second BioCreative Chall; 2007. pp. 2–4. [Google Scholar]
- 44.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput. 2008;663:652–63. [PubMed] [Google Scholar]
- 45.Li Z, Liu F, Antieau L, Cao Y, Yu H. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc. 2010;17(5):563–7. doi: 10.1136/jamia.2010.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. Nature Publishing Group. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 48.Pathak J, Bailey KR, Beebe CE, Bethard S, Carrell DC, Chen PJ, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. J Am Med Inform Assoc. 2013;20(e2):e341–8. doi: 10.1136/amiajnl-2013-001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browne AC, McCray AT, Srinivasan S. The specialist lexicon. Natl Libr Med Tech Reports. 2000:18–21. [Google Scholar]