Abstract
Recommendation of related articles is an important feature of the PubMed. The PubMed Related Citations (PRC) algorithm is the engine that enables this feature, and it leverages information on 22 million citations. We analyzed the performance of the PRC algorithm on 4584 annotated articles from the 2005 Text REtrieval Conference (TREC) Genomics Track data. Our analysis indicated that the PRC highest weighted term was not always consistent with the critical term that was most directly related to the topic of the article. We implemented term expansion and found that it was a promising and easy-to-implement approach to improve the performance of the PRC algorithm for the TREC 2005 Genomics data and for the TREC 2014 Clinical Decision Support Track data. For term expansion, we trained a Skip-gram model using the Word2Vec package. This extended PRC algorithm resulted in higher average precision for a large subset of articles. A combination of both algorithms may lead to improved performance in related article recommendations.
Introduction
PubMed®/MEDLINE® provides access to over 22 million citations in the biomedical literture1. One feature of PubMed is the recommendation of related articles that may be of interest to users. When a user examines a MEDLINE citation, PubMed suggests five related articles and displays them at the right side of the abstract. More related articles, ranked by relevance, are available on demand. Lin and Wilbur2 developed the PubMed Related Citations (PRC) algorithm, a topic-based content similarity technique that underlies this feature. The PRC algorithm considers term frequency (modeled as a Poisson distribution), inverse document frequency and document length when computing the similarity between two articles. A brief introduction to the PRC algorithm is available in the methods section. For more details, please refer to Lin and Wilbur2 and PubMed Help3.
However, the PRC algorithm may not always recommend desired articles to the reader. In particular, when the distributions of term counts are similar, the PRC algorithm is likely to conclude that the articles are similar, even though they may be about different topics. For example, if two articles detailing the mechanisms of different diseases describe similar techniques and mention related genes, they may have a large number of terms in common. On the other hand, when two articles discuss the same topic but use different terms, the PRC algorithm is likely to miss this recommendation.
Our objective was to modify the PRC algorithm and improve the selection of articles related to the same research topic. This was not the first attempt to do so and much effort has been spent to improve the retrieval performance of related MEDLINE citations. For example, Fontaine et al. 4 developed MedlineRanker, a system that identified the most discriminative words in query articles and then used these words as query terms to retrieve related citations. Poulter et al. 5 developed a system named MScanner that trained a naïve Bayes classifier on MeSH® terms and on journal identifiers extracted from a set of user provided articles and then used the classifier to select and rank related citations. Both performed well when compared on nine topics, in terms of the area under the ROC curve4. However, these approaches were not very practical because they both required users to provide a set of articles related to a query topic rather than a few keywords or a short description. eTBLAST6 is a method similar to PRC but it determines similarity based on word alignment. Therefore, the length of the query text has significant impact on the retrieval performance. Boyack et al. 7 investigated the accuracy of five similarity metrics (PRC, BM258, topic modeling, latent semantic analysis, and term frequency-inversed document frequency) for clustering over two million biomedical articles. They concluded that PRC generated the most coherent and most concentrated cluster solution. Aside from suggesting related articles to PubMed users, the PRC algorithm is used for other purposes as well. For example, Huang et al. 9 collected Medical Subject Headings (MeSH terms) from articles recommended by the PRC algorithm for assignment of MeSH terms to a new article. Lu et al. 10 used the PRC algorithm to recruit negative instances and balance the positive/negative instance ratio in a protein-protein interaction prediction task. Our goal was to explore extensions to the PRC algorithm that could produce an accurate short list of related articles.
Methods
a. A brief review of the PRC algorithm
The PRC algorithm2 predicts the conditional probability that a reader will be interested in unseen article c given that this reader shows an interest in article d. Every term in the text is associated with a unique topic and vice versa. The weight of a term t in a particular article d is defined as
| [1] |
where ídft is the inverse document frequency of term t, k is the term frequency of t in article d, l is the word count of d. λ is the expected occurrence of term t when this term is the topic of article d. Additionally, μ. is the expected occurrence of term t when t is not the topic of article d. λ and μ. were calculated using an extensive tuning approach. Based on the exactly matched terms (i.e., the same term appears in two articles) and term weights, the similarity score of two articles c and d is defined as,
| [2] |
where N is the number of matched terms in c and d.
b. An extension of the PRC algorithm
Our approach extends the PRC algorithm by considering similar terms. In the PRC algorithm, a topic is associated with a single unique term. We relaxed this assumption in the modified algorithm and allowed a topic to be associated with multiple similar terms. Similar terms were considered as important as the original term. We prepared similar terms for the vocabulary of TREC data using Word2Vec11, a package based on the Skip-gram model12. We trained distributed vector representations of terms with Word2Vec (vector size 100, minimum word count 40, window size 10) on three million MEDLINE citations that are available from the 2014 BioASQ challenge13, and then derived similar terms by comparing cosine distances between associated vectors. Training takes a few hours on a computer with four 2.67 GHz processors and 16 GB of RAM. This trained model and derived similar terms can be reused for other PubMed article retrieval tasks.
We expanded terms in the query article to a set that includes the original term and the five most similar terms according to the trained Skip-gram model. The expansion allows approximate term matching: for a particular term in the article, if one of its similar terms occurs in a candidate related article, then the similar term is treated as the original one, and the contribution of this pair of terms is included in the similarity score. Therefore, articles that focused on the same topic but used different terms had a higher chance of being connected.
Given an article c for a particular query article d, in the term weight function wt,c we changed the term frequency k to pΣiki, where Σiki is count of term t and its similar terms in article c, and p is the ratio of the count of term t in article d over the count of all terms in article d.
| [3] |
The term weights in article d are not changed. Therefore, the similarity score is asymmetric, and it depends on the set of terms in query article d.
c. Experimental design
We evaluated the performance of our eXtended PRC algorithm (XPRC) on two datasets separately: (1) 4584 articles (utilizing only title, abstract and MeSH terms) from the TREC 2005 Genomics Track evaluation dataset (Genomics data)14 (http://skynet.ohsu.edu/trec-gen/data/2005/genomics.qrels.large.txt), and (2) 3034 articles (utilizing only title and abstract) from the TREC 2014 Clinical Decision Support Track evaluation dataset (CDS data) (http://trec-cds.appspot.com/qrels2014.txt). Among the 4584 Genomics articles, we identified 4234 valid ones for evaluation [1]. These articles were assigned to 50 TREC official topics (i.e. information needs); 3034 CDS articles were assigned to 30 topics; one article could be assigned to múltiple topics. If an article was labeled as “possibly relevant” or “definitely relevant” to a topic, we assigned it to that topic. If two articles had topics in common, we considered them “similar” in our evaluation (the similarity measure is described later in the text).
In the evaluation step, within each dataset, each article served as a query article and the remaining articles were ranked according to the PRC or XPRC similarity (Code available from https://github.com/w2wei/XPRC.git). For every query article, the PRC algorithm recommended articles that were assigned into the true positive (TP) group or the false positive (FP) group according to the TREC gold standard. If the recommended article and the query article shared the same topic, this was considered a TP result. If the recommended article and the query article had no topics in common, this was considered a FP result.
We processed all text following NLM Help3. For example, we split the abstract and title of each article into terms following the PRC tokenization rules. In addition, terms were stemmed using the Porter stemmer. To understand why the PRC algorithm had false positives, we compared the number of matched terms in the sets of TP and FP articles under multiple conditions, and measured the Kullback-Leibler divergence (K-L divergence) of normalized weight distributions between articles and corresponding recommended articles obtained using the PRC algorithm. The term matching was based on string comparison, and the weights were calculated using the formula described in the NLM fact sheet1. When comparing two articles, matched terms were kept and their associated weights were normalized.
d. Evaluation measures
We used precision at the threshold of five articles the same way as described for the development of the PRC algorithm. In addition, we also measured average precision (AP) and mean average precision (MAP) at the same threshold. Precision, macro-average precision, AP and MAP are defined as
| [4] |
| [5] |
| [6] |
| [7] |
where i is the index of an article, N is the total number of articles in the corpus; macro-average precision is the average of the precision for all the articles; AP is the average precision on an article; K is the number of retrieved articles (K=5 in this study); P(k) is the percentage of correct articles among the first k recommendations in this article; I(k) is an indicator function (I(k) = 1 if the k-th retrieval is correct, otherwise I(k)=0).
Results
a. Evaluation of the PRC algorithm
We recorded some characteristics of TP articles and FP articles. First, as expected, the average number of matched terms in TP articles is different from the number in FP articles. The average number of matched terms in TP was 29, and the average number in FP was 24. We used an independent two sample t-test on the 4234 Genomics articles to test the null hypothesis that the average number of matched terms in TP was equal to the number in FP. The p value was 9e-139 hence the hypothesis was rejected, as expected.
Next, we considered the normalized weight distributions of matched terms in TP and FP articles. The average K-L divergence between a query article and the TP articles from the PRC algorithm recommendations was 0.18, while this statistic was 0.21 between a query article and its FP recommendations. Using an independent two sample t-test and the Genomics dataset, we tested the null hypothesis that the average K-L divergence between query articles and their TP articles was equal to the average K-L divergence between query articles and the FP articles. The p value was 3e- 93 hence the hypothesis was rejected, as expected.
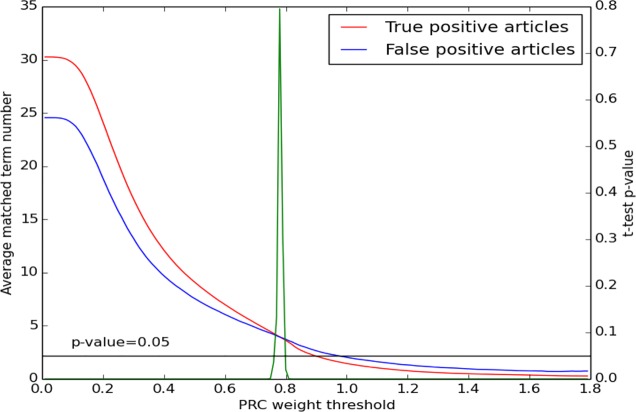
Finally, we analyzed PRC’s capability to match high-weight terms in TP and FP articles. We used a series of independent two sample t-tests to test the null hypothesis that the count of matched high-weight terms in the set of TP articles was equal to the count of high-weight terms in the set of FP articles at different weight thresholds on the Genomics dataset. As we increased the threshold from 0 to 1.8, there was a significant change in the counts of matched high-weight terms in the two groups (Figure 1), except for a small region in which the null hypothesis of equal counts could not be rejected. When the weight threshold was lower than 0.75, TP articles matched significantly more high- weight terms than FP articles. However, when the threshold was over 0.8 (i.e., only terms with weight over 0.8 were considered in computing the similarity score), FP articles matched significantly more terms than TP articles. This conflicts with our intuition that TP articles should share more meaningful and important terms with the query article than FP articles. In the experiments, we observed that PRC high-weight terms were not necessarily the critical terms (i.e., terms directly related to the focus of the article, such as disease names, gene names) in an article. High-weight terms were often general terms such as “gene”, “protein” and “disease”. When critical terms are missing from matched terms, terms that are less relevant to the focus of the article make major contributions to the similarity score. If there are a large number of such high-weight matched terms, a FP article is recommended. For example, the PRC algorithm recommends article PMID11480035 as related to article PMID10226605, although they are not under the same topic according to the TREC evaluation dataset. They match high-weight terms such as “mucosa”, and “mRNA”. But PMID11480035 lacks critical terms such as “APC”, “colon” and “colorectal”.
Figure 1.
A comparison of the number of matched term counts at different PRC weight thresholds. The red curve is the smoothed trend of matched terms in TP articles. The blue curve is the smoothed trend of matched terms in FP articles. The two curves cross between X= 0.76 and X=0.8. The green curve illustrates the p-value of the difference between TP and FP for the null hypothesis that the count of matched terms in TP is equal to the count of terms in FP above different weight thresholds. When 0.76<X<0.8, we cannot reject the null hypothesis of equal counts. Only 0.14% of all term occurrences have weights over 1.8. Therefore, we do not show these special cases in this figure.
b. XPRC: eXtended PRC algorithm
Term expansion is an effective approach to improve the performance of the PRC algorithm. The expansion helps the PRC algorithm recognize articles on related topics, even though they do not have matched critical terms. We wanted to understand in which situations XPRC could potentially enhance the results of PRC. First, we stratified the articles according to precision and AP of the PRC algorithm. After that, we ran XPRC on every stratum of data and compared its performance with PRC. The results of XPRC and the comparisons stratified by precision and AP are shown in Figures 2 and 3, and in Tables 1, 2, 3 and 4. The results show that the XPRC algorithm achieves better performance than the PRC algorithm for certain categories of cases.
Figure 2.
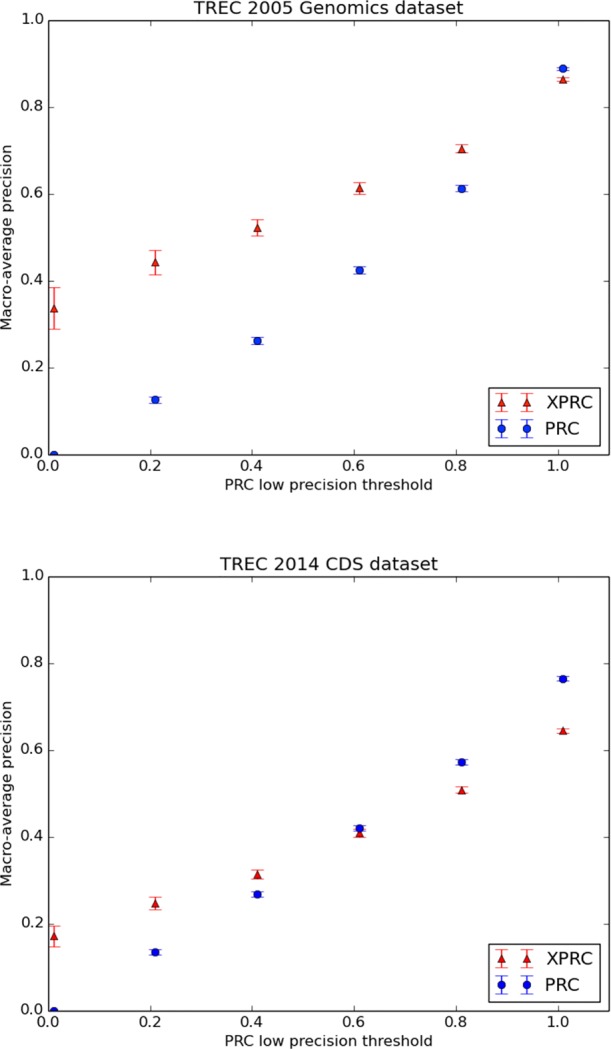
A comparison of PRC and XPRC at five precision levels determined by the PRC algorithm on the Genomics dataset and CDS datasets. For the Genomics articles in which PRC does not achieve perfect precision, XPRC has better overall performance in every group. For the CDS articles, XPRC achieved better performance in PRC’s low precision articles. Values associated with every data point are available in Tables 1 and 2. The error bars indicate standard errors.
Figure 3.
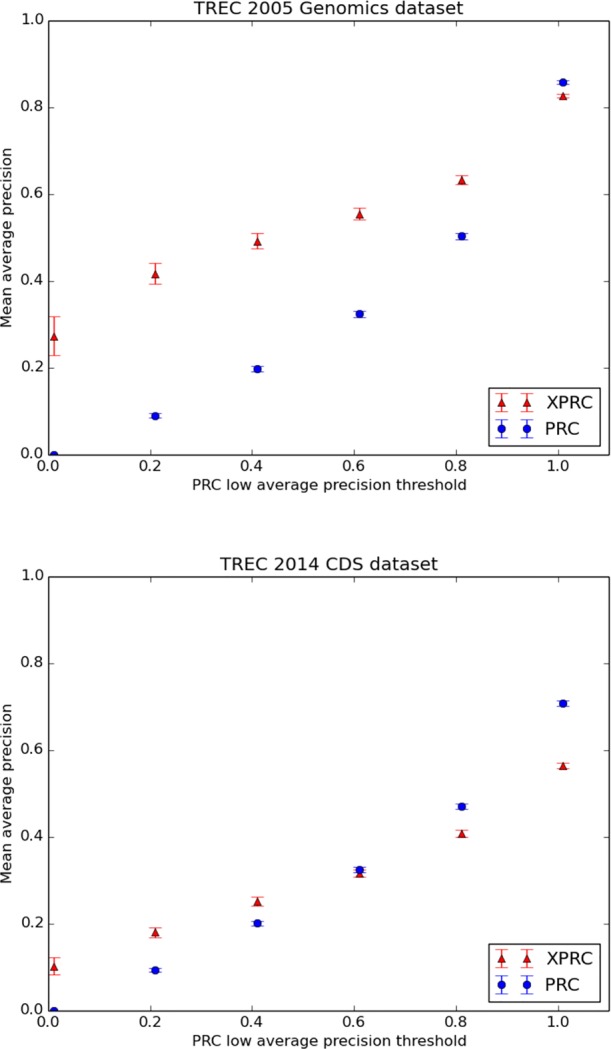
A comparison of PRC and XPRC at five average precision (AP) levels determined by the PRC algorithm on the Genomics dataset and CDS dataset. For the Genomics articles in which PRC does not achieve perfect AP, XPRC has better performance in every group. For the CDS articles, XPRC achieved better performance in PRC’s low precision articles. Values of every data point are available in Tables 3 and 4. The error bars indicate standard errors.
Table 1.
A comparison of PRC and XPRC at different precision levels determined by the PRC algorithm on the Genomics dataset. The cumulative article count is the number of articles with PRC precision below a given precision level. For example, there are 158 articles that result in PRC precisions lower than 0.2. PRC has precision 0.0 on all the 58 articles in the 0.0 group, so its macro-average precision and standard error are also 0. XPRC has better performance on these articles. p-value shows the significance of the difference between PRC and XPRC at different precision levels.
| Precision levels | 0.0 | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |
| Cumulative article counts | 58 | 158 | 314 | 603 | 1215 | 4234 | |
| PRC | macro-average precision | 0 | 0.127 | 0.262 | 0.424 | 0.613 | 0.889 |
| standard error | 0 | 0.008 | 0.009 | 0.008 | 0.007 | 0.003 | |
| XPRC | macro-average precision | 0.338 | 0.443 | 0.523 | 0.614 | 0.705 | 0.864 |
| standard error | 0.048 | 0.028 | 0.019 | 0.013 | 0.009 | 0.004 | |
| p-value | 3e-09 | 3e-21 | 3e-30 | 2e-31 | 2e-16 | 4e-07 | |
Table 2.
A comparison of PRC and XPRC at different precision levels determined by the PRC algorithm on the CDS dataset. The format of this table is the same as that of Table 1.
| Precision levels | 0.0 | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |
| Cumulative article counts | 87 | 268 | 539 | 1000 | 1670 | 3034 | |
| PRC | macro-average precision | 0 | 0.135 | 0.268 | 0.421 | 0.573 | 0.765 |
| standard error | 0 | 0.006 | 0.006 | 0.006 | 0.006 | 0.005 | |
| XPRC | macro-average precision | 0.172 | 0.248 | 0.315 | 0.410 | 0.509 | 0.645 |
| standard error | 0.024 | 0.015 | 0.011 | 0.009 | 0.007 | 0.005 | |
| p-value | 2e-10 | 2e-11 | 3e-4 | 0.294 | 4e-12 | 6e-57 | |
Table 3.
A comparison of PRC and XPRC at five average precision (AP) levels determined by the PRC algorithm on the Genomics dataset. The cumulative article count is the number of articles with PRC AP below the given AP level. XPRC has better performance at all levels except for 1.0. p-value shows the significance of difference between PRC and XPRC at different AP levels.
| Average precision levels | 0.0 | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |
| Cumulative article counts | 58 | 231 | 420 | 687 | 1215 | 4234 | |
| PRC | MAP | 0 | 0.09 | 0.198 | 0.324 | 0.504 | 0.858 |
| standard error | 0 | 0.005 | 0.007 | 0.007 | 0.007 | 0.004 | |
| XPRC | MAP | 0.274 | 0.417 | 0.493 | 0.555 | 0.633 | 0.827 |
| standard error | 0.045 | 0.024 | 0.018 | 0.014 | 0.01 | 0.004 | |
| p-value | 1e-07 | 8e-31 | 2e-45 | 2e-45 | 2e-25 | 1e-07 | |
Table 4.
A comparison of PRC and XPRC at five average precision (AP) levels determined by the PRC algorithm on the CDS dataset. The format of this table is the same as that of Table 3.
| Average precision levels | 0.0 | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |
| Cumulative article counts | 87 | 363 | 663 | 1079 | 1670 | 3034 | |
| PRC | MAP | 0 | 0.094 | 0.201 | 0.325 | 0.471 | 0.709 |
| standard error | 0 | 0.004 | 0.005 | 0.006 | 0.006 | 0.006 | |
| XPRC | MAP | 0.103 | 0.181 | 0.253 | 0.317 | 0.408 | 0.565 |
| standard error | 0.020 | 0.011 | 0.010 | 0.009 | 0.008 | 0.006 | |
| p-value | 2e-6 | 3e-12 | 8e-6 | 0.446 | 2e-10 | 6e-62 | |
c. The scalability of XPRC
We compared the time and memory usage for running queries using PRC and XPRC (Table 5). We ran queries on different sizes of corpora and recorded the time and maximum memory usage. The algorithm and its implementation can still be further optimized.
Table 5.
Time and memory usage of PRC and XPRC. The corpora were randomly selected from the Genomics dataset. For each algorithm, we ran 10 queries on every corpus. The time shown in this table is the average value of all queries on every corpus. The memory in this table is the maximum value for all queries on every corpus.
| PRC | XPRC | |||
|---|---|---|---|---|
| Corpus Size | Time (s) | Maximum Memory (MB) | Time (s) | Maximum Memory (MB) |
| 10 | 0.08 | 65 | 2.1 | 590 |
| 100 | 0.15 | 68 | 2.2 | 591 |
| 1000 | 0.7 | 144 | 2.6 | 601 |
Discussion
Expansion of terms in query articles using the five most similar terms found using the Skip-gram model significantly improved PRC performance in a subset of the articles. In addition to the XPRC algorithm, we explored other modifications and compared their performance against the PRC and the XPRC algorithms. One attempted solution was to filter out terms that were not in the Unified Medical Language System (UMLS) Metathesaurus15. Another attempted solution was to increase the weights of major (starred) MeSH terms and of terms that were present in titles. These solutions did not achieve better performance than XPRC.
The gold standard is critical in the measurement of model performance. In this study, the gold standards were provided in the 4584 annotated articles in the TREC 2005 Genomics Track data and the 3034 TREC 2014 Clinical Decision Support Track data. One issue with the gold standard is that there were a large number of articles under every topic. The average number of articles per topic in the Genomics dataset was 815 and this number was 1264 in the CDS dataset. This issue sometimes makes PRC and XPRC indistinguishable in terms of precision and AP: PRC and XPRC make different recommendations for the same query, but all of their recommendations are true positives. A second issue was found by our physician reviewers (RM and SS). They manually reviewed the PRC and XPRC outcomes and found cases in which similar articles were not annotated in a way that they could be categorized under the same topic, and this could cause the precision to appear lower than it actually was.
Our data-driven approach provided similar terms that could not be found in traditional synonym dictionaries. One limitation of the XPRC algorithm is that the expansion was applied to every term in the query article. This may introduce undesired expansion to non-critical terms. In addition, the parameters were not yet optimized for our experimental setting. We used the μ. and λ proposed by Lin and Wilbur2. To further improve the performance of the XPRC algorithm, we will develop targeted term expansion and optimize the parameters on the TREC evaluation dataset. However, the algorithm needs to be applied to more corpora so we can confirm our results and evaluate its scalability. From the analysis of the PRC algorithm, we confirm that TP articles and FP articles have different distributions of term weights, and that the majority of articles achieve perfect precision, but a significant number of them still result in low precision, leaving some room for improvement.
We are exploring heuristic methods to select when to use the PRC or XPRC results, but there is no simple solution. An empirical method16 achieved good performance on the Genomics dataset (i.e., better performance than PRC in all conditions) but it did not perform as well on the CDS dataset.
The principle of extending a set of terms to assess similarity can be utilized in other problems in which the goal is to find related objects. For example, XPRC can be used to find a set of articles that report on analyses on a particular data set of interest (i.e., articles that are related to the one that first described or utilized the data set). These articles may point to derived data or new and related data sets of interest. Term expansions could also be used for meta-data in the same way we used them for embedded terms in titles and abstracts. In future work we will investigate the use of extended algorithms to retrieve related data sets.
Conclusion
Term expansion is an effective approach to improve the performance of the PRC algorithm for finding related articles in PubMed for cases in which PRC precision is low. The Skip-gram model and the Word2Vec package provide a data- driven solution to term expansion. An extended PRC algorithm utilizing term expansion resulted in higher precision and higher average precision for cases in which there were low precision assignments by the original PRC algorithm.
Acknowledgement
This project was funded in part by grant 1U24AI117966-01 from the NIH. RM and SS were funded by grant T15LM011271 from the NIH. SW was supported by NHGRI R00HG008175 from the NIH.
Footnotes
Among the 4584 PMID in the TREC evaluation dataset, 92 PMID appear multiple times, 1 PMID no longer exists, 248 PMID have no abstract, 6 PMID have problems with PRC (i.e., the most similar article is not itself), 2 PMID has problems with Lucene, the indexing software (i.e., Lucene cannot retrieve similar articles for these two articles using the BM25 algorithm). Both PRC and XPRC were built on top of BM25 results. After removing all these articles, 4234 articles were used for the experiments.
References
- 1.NLM Fact Sheet. Available at < https://www.nlm.nih.gov/pubs/factsheets/medline.html>. Last visit 09/18/2015.
- 2.Lin J., Wilbur W. J. PubMed related articles: A probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.PubMed Help. Available at < http://www.ncbi.nlm.nih.gov/books/NBK3827/#pubmedhelp.Computation_of_Similar_Articl>. Last visit 09/18/2015.
- 4.Fontaine J.-F., et al. MedlineRanker: Flexible ranking of biomedical literature. Nucleic Acids Res. 2009;37:W141–W146. doi: 10.1093/nar/gkp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulter G. L., Rubin D. L., Altman R. B., Seoighe C. MScanner: A classifier for retrieving Medline citations. BMC Bioinformatics. 2008;9:108. doi: 10.1186/1471-2105-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Errami M., Wren J. D., Hicks J. M., Garner H. R. eTBLAST: A web server to identify expert reviewers, appropriate journals and similar publications. Nucleic Acids Res. 2007;35:W12–W15. doi: 10.1093/nar/gkm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyack K. W., et al. Clustering more than two million biomedical publications: Comparing the accuracies of nine text-based similarity approaches. PLoS One. 2011;6:e18029. doi: 10.1371/journal.pone.0018029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson S. E., et al. Okapi at TREC-3. NIST Spec. Publ. 1995:SP 109. [Google Scholar]
- 9.Huang M., Névéol A., Lu Z. Recommending MeSH terms for annotating biomedical articles. J. Am. Med. Inform. Assoc. 2011;18:660–7. doi: 10.1136/amiajnl-2010-000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krallinger M., et al. The Protein-Protein Interaction tasks of BioCreative III: Classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinformatics. 2011;12:S3. doi: 10.1186/1471-2105-12-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikolov T. Word2Vec. Available at < https://code.google.com/p/word2vec/>. Last visit 09/18/2015.
- 12.Mikolov T., Sutskever I., Chen K., Corrado G. S., Dean J. Advances in neural information processing systems. 2013. Distributed representations of words and phrases and their compositionality; pp. 3111–3119. [Google Scholar]
- 13.Balikas G., Partalas I., Ngomo A.-C. N., Krithara A., Paliouras G. Results of the BioASQ Track of the Question Answering Lab at CLEF 2014; Proceedings of the first workshop on Bio-Medical Semantic Indexing and question answering, a post-conference workshop of Conference and Labs of the Evaluation Forum 2014; 2014. pp. 1181–1193. [Google Scholar]
- 14.Hersh W., et al. TREC 2005 Genomics Track Overview; The fourteenth Text Retrieval Conference; National Institute for Standards & Technology; 2005. [Google Scholar]
- 15.Bodenreider O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004;32:267D–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu C.-N., et al. Integrating high dimensional bi-directional parsing models for gene mention tagging. Bioinformatics. 2008;24:i286–94. doi: 10.1093/bioinformatics/btn183. [DOI] [PMC free article] [PubMed] [Google Scholar]