Abstract
Responsiveness to drugs is an important concern in designing personalized treatment for cancer patients. Currently genetic markers are often used to guide targeted therapy. However, deeper understanding of the molecular basis for drug responses and discovery of new predictive biomarkers for drug sensitivity are much needed. In this paper, we present a workflow for identifying condition-specific gene co-expression networks associated with responses to the tyrosine kinase inhibitor, Erlotinib, in lung adenocarcinoma cell lines using data from the Cancer Cell Line Encyclopedia by combining network mining and statistical analysis. Particularly, we have identified multiple gene modules specifically co-expressed in the drug responsive cell lines but not in the unresponsive group. Interestingly, most of these modules are enriched on specific cytobands, suggesting potential copy number variation events on these loci. Our results therefore imply that there are multiple genetic loci with copy number variations associated with the Erlotinib responses. The existence of CNVs in these loci is also confirmed in lung cancer tissue samples using the TCGA data. Since these structural variations are inferred from functional genomics data, these CNVs are functional variations. These results suggest the condition specific gene co- expression network mining approach is an effective approach in predicting candidate biomarkers for drug responses.
Introduction
Cancer patients are highly heterogeneous1,2. Even patients with the same type of cancers often present different responses to drugs and therapeutic schemes3,4. Therefore, understanding and predicting the drug responses in cancer patients is critical to enable personalized treatment. Current methods to model drug effectiveness and resistance are limited to in vitro systems such as body-on-a-chip pharmacokinetic models5, tissue scaffolds6, or engineered tumor microenvironments7 –10; animal models such as genetically engineered murine systems11,12 have also shown promise. While these methods are effective at predicting general drug responsiveness to human cell lines, they fail to incorporate specific patient variability in a high-throughput manner. Single nucleotide polymorphisms (SNPs) are often used as measures of variance within a population and have proven invaluable for the development of personalized medicine13 –16. The problem with using SNP arrays as a basis for drug screening is that these microarrays often encompass all polymorphisms, including non-functional variations, between subjects. As nonfunctional polymorphisms do not directly correspond to genes, they are irrelevant to the determination of drug responsiveness17. Using gene expression data alleviates this issue by only surveying functional genomic data. One of the major efforts in understanding the molecular basis for drug responses in cancer is the Cancer Cell Line Encyclopedia (CCLE) project in which a large number (> 900) different cancer cell lines are treated with 26 different drugs including both chemotherapy drugs and targeted drugs18. The responses of the cancer cell lines to the drugs were recorded and the genome-wide gene expression profiles for these cancer cell lines before drug treatment were also generated. This dataset has hence become a valuable resource for characterizing the molecular basis of drug responses. In this paper, we take a systems biology approach to studying the CCLE by characterizing the gene co-expression networks (GCNs) specific to drug-responsive or unresponsive groups.
Gene co-expression is the phenomena wherein two or more genes tend to be expressed simultaneously across a large population19. Thus, in any one subject, two co-expressed genes will either both be highly or both lowly expressed comparing to other subjects in a cohort. There are multiple possible biological mechanisms leading to gene co-expression. For instance, genes co- regulated by the same set of transcription factors are often co-expressed. These co-expressed genes are often functionally related20 –25. In addition, genes located on the same cytoband may co-express in a cohort in which some of the patients have copy number variations (CNVs) on this cytoband26,27. Therefore co-expression analysis can reveal important structural and regulatory relationships in biological systems among a cohort.
Using high throughput gene expression algorithms, gene co-expression data is often measured by calculating the correlation between expression profiles of the two genes20,28. When co- expression analysis is expanded to all the genes in the genome, a network model called a gene co- expression network (GCN) is often adopted where genes are represented nodes29,30. For an unweighted GCN, the correlation coefficient value between two genes is used to determine if the two genes (nodes) are connected (often based on some threshold). For a weighted GCN, the correlation coefficient of its transformation is used as the weight for the edge linking the two genes28 –31. Gene co-expression network analysis (GCNA) can reveal functionally or genetically related gene clusters, which can subsequently lead to discovery of new gene functions and regulatory relationships19 –21,26. Such discoveries can bring to light new understandings of disease progression and therapy, as well as predicting new gene functions and even discovering new disease biomarkers. Compared to previously developed clustering algorithms, GCNA allows overlap between the modules; this overlap is particularly useful to visualize since genes can involve in multiple biological functions.
In this paper, we carry out weighted GCN (WGCN) analysis to identify highly co-expressed gene network modules in different groups of lung cancers. Specifically we compare the modules identified from lung cancer cell lines that are responsive to Erlotinib versus the ones identified from lung cancer cell lines that are not responsive to the drug. Erlotinib is a targeted cancer drug; specifically it is an ATP-competitive, tyrosine kinase inhibitor acting on EGFR, that blocks signaling cascades3. While it was originally used for lung cancers, not all patients are responsive to the drug4,32,33. Therefore, it is of great interest to identify the potential genetic factors associated with the drug response.
From our WGCN analysis, we have identified 73 gene modules for the drug responsive group and 51 gene modules for the unresponsive group. Interestingly, several of the condition-specific gene modules are highly enriched on specific cytobands, suggesting potential roles of the copy number alterations in drug responses. The existence of CNVs on these loci was further confirmed using cBioPortal (http://www.cbioportal.org). While further validation of the roles of the drug resistance of these loci requires experimental validation, our workflow nevertheless led to new hypotheses regarding the genetic basis for cancer drug resistance. While our focus is on the effects of Erlotinib on non-small cell lung cancer, specifically lung adenocarcinoma, our approach can nonetheless be applied to all other cancers and drugs.
Methods
Datasets
The gene expression dataset used for all analysis was downloaded from the Broad-Novartis’s database, the Cancer Cell Line Encyclopedia (CCLE). The file, “CCLE_Expression_Entrez_2012- 09-29.gct” contains gene-centric RMA-normalized mRNA expression data for 1037 cancer cell lines and 18987 genes. The file was converted to a Tab-Delimited-Text file in order to be read and processed by MATLAB. The genes and corresponding expression data were doubly filtered by low-value and low-variance MATLAB filters to limit the data to the top 10000 gene probes.
The drug responsiveness dataset used, ‘CCLE_GNF_data_090613.xls’, was also converted to a Tab-Delimited-Text file and the IC50 values from the data were used in order to determine responsiveness for 203 cancer cell lines to each of 26 drugs.
Workflow
As shown in Figure 1, our workflow contains multiple steps. The first three steps are data preprocessing steps and the following three steps are WGCN analysis steps. Below we describe each step in details.
Figure 1.
The workflow for our analysis.
Filter CCLE gene expression data: After the gene expression and drug response files were downloaded from the CCLE database and converted to‘.txt’ formats, the first step was to identify the common cell lines to both datasets and isolate the lung cancer cell lines. Within the gene expression dataset, the genes and corresponding expression data were doubly filtered by low- value and low-variance MATLAB filters to limit the data to the top 10000 unique genes. For this study, the lung cancer drug Erlotinib was chosen and the drug data was limited accordingly.
Isolate common lung cancer cell lines: The intersection between the two datasets described in the previous section yielded 195 common cancer cell lines. As the primary site of cancer was recorded along with the cell lines in each file, a simple search and comparison algorithm was conducted in MATLAB to isolate the 38 lung cancer cell lines of interest. These 38 lung cancer cell lines and their corresponding gene expression and drug responsiveness data were used for this study’s subsequent analysis.
Separate by drug responsiveness: Two threshold IC50 values were set to deem each cell line responsive or unresponsive to Erlotinib. As IC50 values determine the dosage concentration needed for 50% inhibition (in this case, of cancer cells), higher IC50 values correspond with less effective drugs or more resistant cells. For IC50 ≤ 5, the cell line and its corresponding gene expression data were sorted into the responsive data matrix. For IC50 ≥ 10, the cell line and its corresponding gene expression data were sorted into the unresponsive data matrix. The upper threshold value of 10 was chosen to reflect the spread of the data from 0≤IC50≤20 and thus term the upper half unresponsive. The responsive data was also limited by the lower threshold value of 5 in order to exclude cell lines that were neither responsive nor unresponsive to Erlotinib.
Correlation matrix per group: For both the unresponsive and responsive expression data, correlation matrices were created to quantify the co-expression between each pair of genes by calculating the Pearson correlation coefficient between the two expression profiles.
WGCN mining of each group: Both correlation matrices were then passed through a weight network quasi-clique mining algorithm in order to mine through the network and identify modules of strongly co-expressed genes. Here, we applied our recently-developed local maximal quasi- clique merging (lmQCM) algorithm27. This algorithm was specifically designed to mine weighted graphs. Unlike the well-known WGCNA R-package developed by the Horvath group that uses hierarchical clustering algorithm to identify gene modules28, this algorithm takes a graph mining approach and thus allows overlap between gene modules. In addition, it uses an adaptive module density threshold instead of a global distance threshold, as in hierarchical clustering. The density of gene modules is guaranteed to be above a lower bound determined by the parameters. One of the key parameters of the algorithm is γ that defines the ratio between the weight of the first edge of a module and the maximal edge weight in the graph. The larger the value of γ, the more stringent it is to start a new module and the less number of modules will be identified. In this study we tested multiple values of γ ranging from 0.85 to 0.95. We finally chose 0.86 in order to constrain the maximum module to 50-500 genes and the number of modules to 10-100. These ranges were chosen to ensure that only highly co-expressed genes were clustered and limit the possibility of clustering genes together with only marginal correlation. Additionally the parameter of minimum cluster size determined the fewest number of co-expressed genes that could represent a gene module; this value was set at 10 genes.
Cluster intersection and hypergeometric tests: Every pair of responsive and unresponsive gene modules were compared to determine the intersection and union between the two gene lists. Using this information, the Jaccard Index (J.I) and Altered Jaccard Index (A.J.I) were calculated based on the following equations:
| [1] |
where ‘size’ indicates the number of clusters in each intersection or union, respectively. In addition, the statistical significance for the intersection was determined using a hypergeometric test. Since we are interested in the modules that are unique in each responsiveness condition, we selected the modules that do not have significant intersections with any modules in the other group. In order to be more stringent in this selection, we used a p-value cutoff as 0.05, instead of any lower threshold from multiple test compensation. Essentially, a module is selected if the hypergeometric test p-values between it and all modules in the opposite group is larger than 0.05.
Gene enrichment analysis: Cluster intersections and set differences larger than 5 genes with a Jaccard Index > 0.03 were analyzed using the website ToppGene’s functional annotation software, ToppFun, for gene enrichment analysis (https://toppgene.cchmc.org/enrichment.jsp). For each inputted gene list, information regarding the common biological processes, transcription factor binding sites, cytoband (chromosome band locus), molecular functions, and interactions that the genes are involved in were recorded along with p-values. In addition, information was collected on drugs that act to target these genes and diseases the genes are known to play roles in.
Results
After passing both unresponsive and responsive matrices through the clustering algorithm at the gamma value of choice, we obtained 73 gene modules for the Erlotinib responsive group and 51 modules for the unresponsive group. The Jaccard and adjusted Jaccard distances between every pair of responsive-unresponsive modules were calculated. In addition, hypergeometric tests were conducted to calculate the statistical significance of the intersections between the modules. Since our goal is to identify the conditional specific gene modules, we focused on the genes modules that had no significant intersection with any modules in the other group. Table 1 shows the lists of conditional specific modules and their sizes.
Table 1.
Condition specific modules and the lowest hypergeometric test p-values. N/I means that the module has no intersection with any module in the other group.
| Responsive Group | Module # | 16 | 25 | 29 | 37 | 62 | 66 | 67 | 68 | 69 | 72 |
| Module size | 46 | 27 | 26 | 24 | 22 | 21 | 21 | 21 | 21 | 21 | |
| Lowest p-value | 0.1012 | 0.0589 | 0.0568 | N/I | N/I | 0.2787 | N/I | 0.0960 | 0.3714 | N/I | |
| Unresponsive Group | Module # | 11 | 12 | 31 | 32 | 35 | 48 | ||||
| Module size | 18 | 17 | 11 | 11 | 11 | 10 | |||||
| Lowest p-value | 0.0563 | 0.0852 | 0.3106 | 0.3106 | N/I | N/I |
We then further carried out gene enrichment analysis for these gene modules and the highly enriched terms for both groups. The analysis for the responsive group-specific modules is shown in Table 2 while the analysis for the unresponsive group-specific modules are shown in Table 3.
Table 2.
Highly enriched biological terms for gene modules specific to the responsive group. If a category contains no significantly enriched terms, it is left blank.
| Module # | Gene Ontology term | Cytoband | Transcription factor |
|---|---|---|---|
| 16 | BP: GO:0006310 DNA recombination (p=4.171E-8, 8 genes) | 19p13.3 (p=6.935E-7, 7 genes) | E2F (p=1.280E- 5, 6 genes) |
| 25 | - | 8q24.3 (p=2.414E-6, 4 genes),10 genes on 8q21-24 | - |
| 29 | BP: GO:0006479 protein methylation (p=2.636E-5, 4 genes) | 17q11.2 (p=4.093E-17, 9 genes); 16p13.3 (p=8.009E-7, 5 genes) | - |
| 37 | - | 3q27.2 (p=2.769E-5, 2 genes), 8 genes on 3q21- | - |
| 62 | BP: GO:1903047, mitotic cell cycle process (p=1.387E-5, 7 genes) | 12p13 (p=8.557E-6), 7 genes on 12p13 | - |
| 66 | - | - | - |
| 67 | BP: GO:0009057, macromolecule catabolic process (p=5.022E-6, 8 genes) | 15q24 (p=6.533E-5, 2 genes), 12 genes on 15q13-24 | - |
| 68 | MF: GO0003723, RNA binding (p=1.005E-5, 9 genes) | 6q13-q14.3 (p=5.773E-4), 5 genes on 6q11-26 | - |
| 69 | - | 17q22 (p=3.224E-4, 2 genes), 4 genes on 17q21-23 | - |
| 72 | MF: GO:0044822, poly(A) RNA binding (p=1.405E-6, 9 genes) | 6p21.3 (p=4.184E-4, 3 genes) | - |
Table 3.
Highly enriched biological terms for gene modules specific to the unresponsive group. If a category contains no significantly enriched terms, it is left blank.
| Module # | Gene Ontology term | Cytoband | Transcription factor binding site |
|---|---|---|---|
| 11 | - | 3 genes on 12q24 | - |
| 12 | GO: BP:GO0045333, cellular respiration (p=3.831E-9, 6 genes); CC: GO0044429, mitochondrial part (p=1.397E-8, 9 genes) | - | - |
| 31 | - | 3 genes on 2q24-33 | - |
| 32 | - | - | - |
| 35 | - | 3 genes on 2p11-13 | - |
| 48 | - | 19q13.43 (p=8.438E-9, 4 genes), 10 genes on 19q13.4 | - |
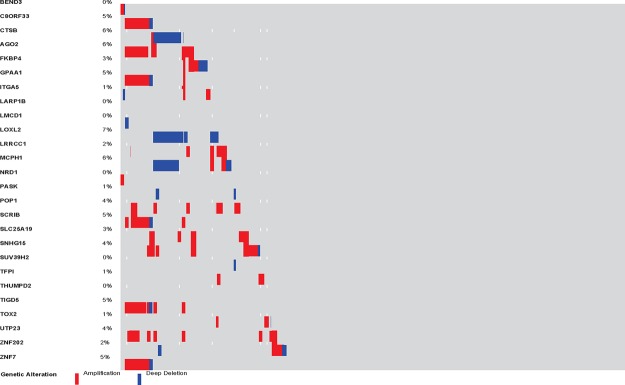
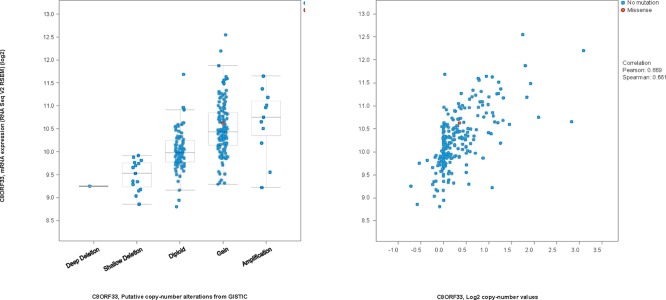
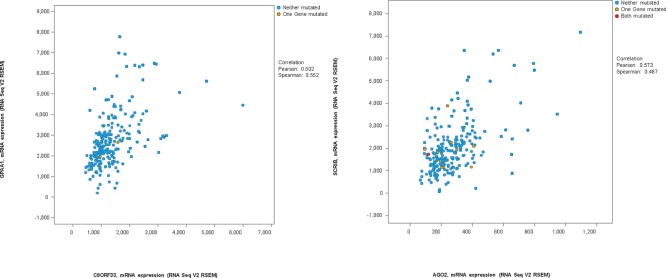
From Table 2, it is clear that many of the responsive group specific modules are enriched on different cytobands. While the trend for the unresponsive group is not clear, at least one module (# 48) is entirely on a single cytoband. This suggests potential involvement of copy number variations (CNVs) in the drug responses. While understanding roles of CNVs on these regions in drug responses requires further experimental study beyond the scope of this paper, we examined if CNVs on these regions were indeed present in the CCLE data and possibly other cancer data using cBioPortal. As shown in Table 4, the prevalence of CNVs in the modules with highly enriched cytobands in CCLE and lung cancer TCGA data are listed. An example of the patients with adenocarcinoma is shown in Figure 2. Specifically, among the 27 genes from module 25 of the responsive group, 10 are known to be on cytobands 8q21-24. Interestingly, among the 10 genes, seven show consistent amplification/gain in patients while the other three show consistent deletion/loss and the two types of alterations are mutually exclusive in patients. In addition, the expression levels of the genes with CNVs are indeed correlated with the copy number variance levels (Figure 3 shows an example). Furthermore, the genes with consistent CNV often co- express (two examples are shown in Figure 4).
Table 4.
Prevalence of CNVs of the genes in the modules in CCLE and lung adenocarcinoma samples from TCGA
| Responsive Group | Unresponsive Group | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Module # | 16 | 25 | 29 | 37 | 62 | 67 | 68 | 72 | 48 |
| % in CCLE (995 cell lines) | 51.9 | 53.4 | 22.8 | 41.7 | 27.5 | 28.7 | 41.4 | 39.7 | 5.6 |
| % in Lung cancer TCGA (LUAD, 230 cases) | 32.2 | 32.6 | 14.3 | 27.0 | 15.7 | 28.3 | 33.5 | 23.5 | 0.9 |
Figure 2.
OncoPrint for the CNV events of the 26 genes (the 27th gene could not be found in the database) in gene module 25 of the responsive group in 230 TCGA lung adenocarcinoma patients generated by cBioPortal.
Figure 3.
Relationship between expression levels and CNV measurements for C8ORF33.
Left: the x-axis indicates patients with different status of CNVs. Right: The scatterplot between the expression levels and the CNV measurements. The Pearson correlation coefficient is 0.669.
Figure 4.
Relationship between different gene pairs identified on the same cytobands.
Left: C8ORF33 versus GPAA1, the Pearson correlation coefficient is 0.502. Right: AGO2 versus SCRIB. The Pearson correlation coefficient is 0.573.
Discussion
In this project, we implemented a novel workflow for identifying condition-specific gene co- expression networks associated to drug response in lung cancer. Instead of taking the traditional approach on inferring phenotype-associated differentially expressed genes, the condition-specific network approach fills in a gap between gene and functional levels. Most interesting about our findings is the prevalence of gene modules enriched on multiple cytobands in the Erlotinib responsive group. These results predict the potential functional roles of the structural variants in drug response and thus provide new hypotheses of biomarkers for personalized treatment in cancers.
Our approach has two major advantages. First, while traditionally CNVs are detected using genetic approaches or microarrays (e.g., SNP array), it is often unclear if the detected CNVs are functional. Since the potential CNVs loci we detected are based on functional genomics data (ie., gene expression), they are clearly functional variations. Secondly, the direct connection to functions can help to elucidate drive genes in cancer. While currently single nucleotide variations and indels are widely used for personalized targeted cancer treatment, robust CNVs have been recently proposed to be potential biomarkers and our study supports such potentials4,32,34.
Future work will include detailed bioinformatics analyses at the network and systems level to delineate potential drive mutations from the co-expressed modules. In addition, we plan to expand the study to other targeted drugs in CCLE, as well as other cancer types. The commonality between different cancer types may also suggest possible drug repurposing options.
Acknowledgements
This work is partially supported by NCI ITCR U01 grant.
References
- 1.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeGrendele H. Epidermal growth factor receptor inhibitors, gefitinib and erlotinib (Tarceva, OSI-774), in the treatment of bronchioloalveolar carcinoma. Clin Lung Cancer. 2003;5(2):83–5. doi: 10.1016/S1525-7304(11)70324-2. [DOI] [PubMed] [Google Scholar]
- 4.Sahnane N, Frattini M, Bernasconi B, et al. EGFR and KRAS Mutations in ALK-Positive Lung Adenocarcinomas: Biological and Clinical Effect. Clin Lung Cancer. 2015 doi: 10.1016/j.cllc.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9(10):1385–94. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 6.Dunne LW, Huang Z, Meng W, et al. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials. 2014;35(18):4940–9. doi: 10.1016/j.biomaterials.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Infanger DW, Lynch ME, Fischbach C. Engineered culture models for studies of tumor- microenvironment interactions. Annu Rev Biomed Eng. 2013;15:29–53. doi: 10.1146/annurev-bioeng-071811-150028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedron S, Becka E, Harley BA. Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment. Adv Mater. 2015;27(9):1567–72. doi: 10.1002/adma.201404896. [DOI] [PubMed] [Google Scholar]
- 9.Villasante A, Vunjak-Novakovic G. Tissue-engineered models of human tumors for cancer research. Expert Opin Drug Discov. 2015;10(3):257–68. doi: 10.1517/17460441.2015.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DelNero P, Song YH, Fischbach C. Microengineered tumor models: insights & opportunities from a physical sciences-oncology perspective. Biomed Microdevices. 2013;15(4):583–93. doi: 10.1007/s10544-013-9763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usary J, Zhao W, Darr D, et al. Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res. 2013;19(17):4889–99. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakasone ES, Askautrud HA, Egeblad M. Live imaging of drug responses in the tumor microenvironment in mouse models of breast cancer. J Vis Exp. 2013;(73):e50088. doi: 10.3791/50088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 14.McLeod HL, Evans WE. Pharmacogenomics: unlocking the human genome for better drug therapy. Annu Rev Pharmacol Toxicol. 2001;41:101–21. doi: 10.1146/annurev.pharmtox.41.1.101. [DOI] [PubMed] [Google Scholar]
- 15.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–37. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 16.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145(10):749–57. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang GS, Wang J, Wang Z, Lee CG. Predicting potentially functional SNPs in drug-response genes. Pharmacogenomics. 2009;10(4):639–53. doi: 10.2217/pgs.09.12. [DOI] [PubMed] [Google Scholar]
- 18.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39(11):1338–49. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Lu K, Xiang Y, et al. Weighted frequent gene co-expression network mining to identify genes involved in genome stability. PLoS Comput Biol. 2012;8(8):e1002656. doi: 10.1371/journal.pcbi.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kais Z, Barsky SH, Mathsyaraja H, et al. KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol Cancer Res. 2011;9(8):1091–9. doi: 10.1158/1541-7786.MCR-10-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Y, Zhang CQ, Huang K. Predicting glioblastoma prognosis networks using weighted gene co-expression network analysis on TCGA data. BMC Bioinformatics. 2012;13 Suppl 2:S12. doi: 10.1186/1471-2105-13-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalluru V, Machiraju R, Huang K. Identify condition-specific gene co-expression networks. Int J Comput Biol Drug Des. 2013;6(1-2):50–9. doi: 10.1504/IJCBDD.2013.052201. [DOI] [PubMed] [Google Scholar]
- 24.Xiang Y, Zhang J, Huang K. Mining the tissue-tissue gene co-expression network for tumor microenvironment study and biomarker prediction. BMC Genomics. 2013;14 Suppl 5:S4. doi: 10.1186/1471-2164-14-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Xiang Y, Ding L, et al. Using gene co-expression network analysis to predict biomarkers for chronic lymphocytic leukemia. BMC Bioinformatics. 2010;11 Suppl 9:S5. doi: 10.1186/1471-2105-11-S9-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Ni S, Xiang Y, et al. Gene Co-expression analysis predicts genetic aberration loci associated with colon cancer metastasis. Int J Comput Biol Drug Des. 2013;6(1-2):60–71. doi: 10.1504/IJCBDD.2013.052202. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Huang K. Normalized lmQCM: an Algorithm for Detecting Weak Quasi-clique Modules in Weighted Graph with Application in Functional Gene Cluster Discovery in Cancer. Cancer Informatics. 2015 doi: 10.4137/CIN.S14021. (accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 30.Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard metaanalysis? PLoS One. 2013;8(4):e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46(11) [PMC free article] [PubMed] [Google Scholar]
- 32.Serizawa M, Takahashi T, Yamamoto N, Koh Y. Genomic aberrations associated with erlotinib resistance in non-small cell lung cancer cells. Anticancer Res. 2013;33(12):5223–33. [PubMed] [Google Scholar]
- 33.Platt A, Morten J, Ji Q, et al. A retrospective analysis of RET translocation, gene copy number gain and expression in NSCLC patients treated with vandetanib in four randomized Phase III studies. BMC Cancer. 2015;15:171. doi: 10.1186/s12885-015-1146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110(52):21083–8. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]