Abstract
The Quality Data Model (QDM) is an established standard for representing electronic clinical quality measures on electronic health record (EHR) repositories. The Informatics for Integrated Biology and the Bedside (i2b2) is a widely used platform for implementing clinical data repositories. However, translation from QDM to i2b2 is challenging, since QDM allows for complex queries beyond the capability of single i2b2 messages. We have developed an approach to decompose complex QDM algorithms into workflows of single i2b2 messages, and execute them on the KNIME data analytics platform. Each workflow operation module is composed of parameter lists, a template for the i2b2 message, an mechanism to create parameter updates, and a web service call to i2b2. The communication between workflow modules relies on passing keys ofi2b2 result sets. As a demonstration of validity, we describe the implementation and execution of a type 2 diabetes mellitus phenotype algorithm against an i2b2 data repository.
Introduction
The widespread adoption of electronic health records (EHR), driven in part by Meaningful Use regulations,1 has been a key enabler in the ability to measure clinical quality and population health on a large scale.2 In order to help achieve the full promise of these goals, the Office of the National Coordinator has promoted the development of the Quality Data Model (QDM), and the Health Quality Measure Format (HQMF), a Health Level Seven (HL7) standard for expressing electronic clinical quality measures.3 Leveraging these quality standards and the tools developed to support them, we have proposed in previous work to extend the use of the QDM to encompass EHR-driven clinical research, in particular for the creation of phenotype algorithms.4 Phenotype algorithms typically select cohorts of patients based on logical criteria expressed over EHR data such as diagnoses, medications, procedures, and encounters. Such algorithms have taken on an increasingly important role in clinical research as EHR data have become more widely available.5,6 As part of our efforts to achieve this goal, we have investigated ways to map QDM-based algorithms into executable artifacts, to reduce the degree of manual effort required to implement a phenotype algorithm once it has been specified.7,8
Mapping a QDM algorithm to an executable form inevitably raises the issue of how to link the algorithm logic (expressed using QDM-based concepts) to actual data stored in an individual healthcare institution’s local data repository. This issue has motivated us to leverage the data warehousing platform developed by the Informatics for Integrating Biology and the Bedside (i2b2), an NIH-funded National Center for Biomedical Computing.9 The i2b2 platform is widely adopted as a research data repository for EHRs at over 80 sites nationwide.10 This leads us to target the translation of QDM-based algorithms into executable artifacts that query i2b2 repositories.
The i2b2 platform has a modular architecture, where communications between modules use XML messages and RESTful web services. In particular, an i2b2 web client posts XML messages representing data queries to an i2b2 server containing patient data, and receives in return XML messages that contain keys for result sets. The web client can also use these result keys to construct next-step queries or to request patient or encounter lists. A previous effort to execute QDM-based queries on i2b2 mostly focused on creating a single i2b2 XML query message from a single HQMF document.11 This strategy is effective for many algorithms, but it is difficult to scale this approach to the full complexity of the QDM. This limits its potential with respect to many publicly available clinical quality measures12 and phenotype algorithms.13 Here, we propose a flexible, decompositional approach to break down a QDM algorithm into a workflow of unit i2b2 messages, where each message implements one step of a well-defined QDM operation.
In previous work,8 we have demonstrated the ability to implement QDM-based phenotype algorithms as executable workflows for the open source KNIME data analytics platform (https://www.knime.org/). This platform enables the graphical creation and execution of data workflows that can read, transform, visualize, and write data in various formats. KNIME workflows are persisted as sets of text file descriptors, which can be bundled together, exported and shared with other users. To support the generation of KNIME workflows from QDM algorithms, we have previously developed a software tool to automatically translate QDM into KNIME workflow descriptors,14 that can be subsequently manually edited to link to local data repositories. In the work described here, we build upon these previous efforts, using KNIME workflows in order to orchestrate the execution of sequences of i2b2 querying messages.
Methods
We manually decomposed a QDM algorithm into basic units of functionality, as defined in a recent formalization of the QDM.15 Each of these units of functionality is implemented in the KNIME workflow as a group of nodes that are bundled together into a single unit of workflow functionality. We call this bundle of nodes a QDM Operator Module. We link these modules together into an executable KNIME workflow to implement a complete QDM algorithm. The i2b2 query results from each QDM Operator Module are passed to subsequent modules in the form of i2b2 result set keys.
QDM operator modules
We implemented a set of QDM Operator Modules that can handle a variety of QDM components, such as clinical data elements, logical operators (AND, OR, AND-NOT), temporal operators (point to point such as STARTS BEFORE START OF, and point to duration such as STARTS DURING), and a patient list retriever.
Although a single i2b2 message can represent a query with combined aggregative, logical, and temporal operations, in our implementation we restrict each QDM operator module to perform only a single unit of QDM operation as defined by the QDM formal model. We then compose these single steps into larger operations programmatically using KNIME workflows, allowing us to build queries of arbitrary complexity. Each QDM Operator Module contains the following core components:
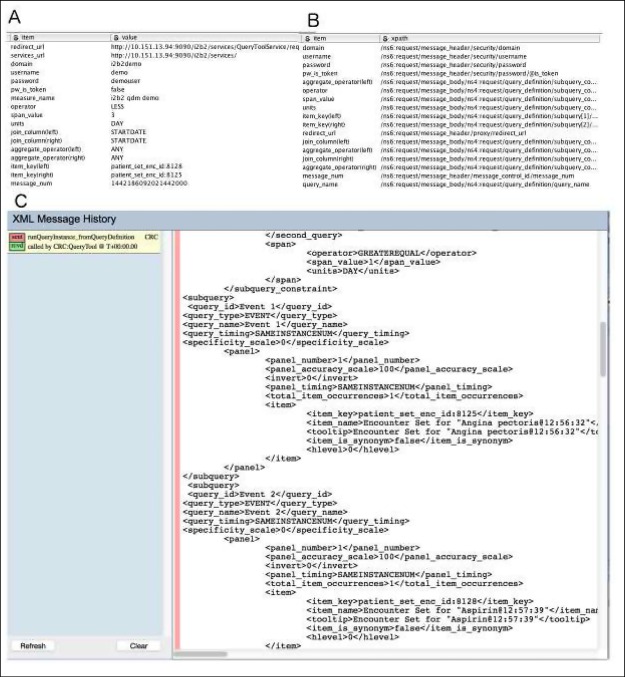
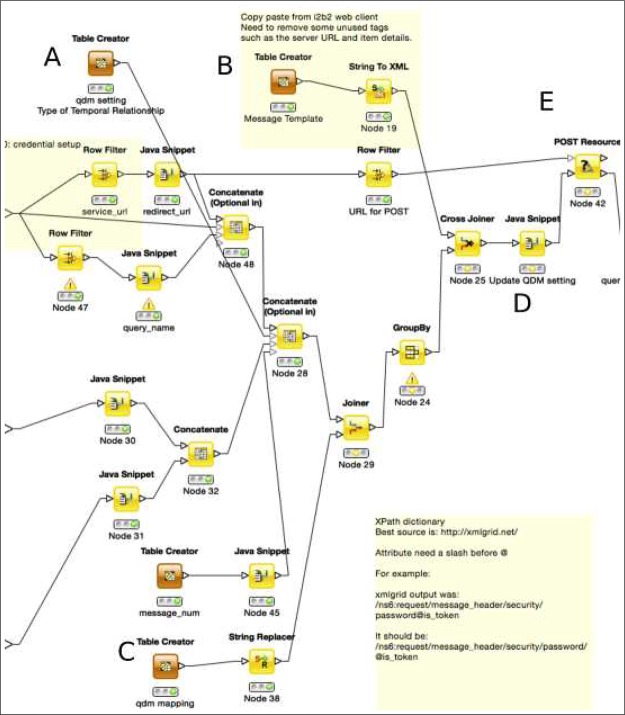
Parameter List: This list contains account credentials, keys for patient or encounter lists from upstream operations, and attributes of an operation (e.g., “starts less than 3 days before start of”). Each parameter has a name and a value (Figure 1A). The Table Creator nodes (Figure 2A) that store these parameters are designed to be updatable with external KNIME workflow constructing applications, such as our QDM translation tool.
Template of i2b2 Messages: We generated i2b2 XML message templates by performing related queries on an i2b2 Web Client, obtaining the resulting XML messages (Figure 1C), and placing abstractions of these messages as templates in KNIME nodes (Figure 2B).
XPath Maps and Update of XML Messages: For each parameter that requires an update on the XML message template, we assign an XPath to indicate the destination (Figure 1B, Figure 2C). In KNIME, we use a node to update the XML message with the guidance of XPath (Figure 2D).
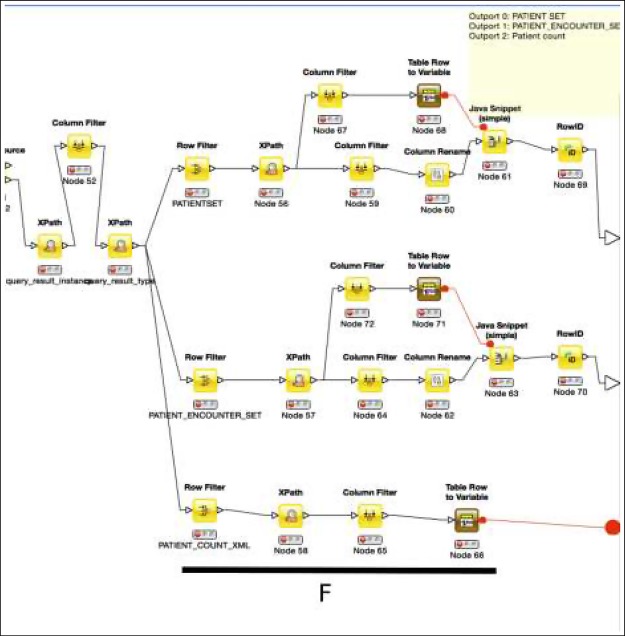
RESTful Communications: After an XML message is issued, the module will POST the message to the i2b2 server (Figure 2E). The module then will extract the result set keys for the next operations to use (Figure 3F). To retrieve patient lists, the module will POST an XML message with the result key as patient_set_coll_id to the i2b2 server.16
Figure 1.
Update Parameters on an i2b2 Message for an QDM Operator Module. A: a parameter list; B: An XPath map for the parameters; C: a template i2b2 message generated in i2b2 web client.
Figure 2.
An example of a QDM Operator Module. This module is for temporal operators. The three input ports are (from up to down): connection information and log-in credentials, left side input of the key for encounter list from the previous operation, the right side one. A: the list of operation specific parameters; B: the XML message template; C: XPaths for parameters; D: the Java Snippet to update the parameters on the XML message; E: RESTful communication with the i2b2 server. The rest of the module is shown in Figure 3
Figure 3.
An example of a QDM Operator Module, continuation from the one shown in Figure 2. F: Extracting information from the response message from an i2b2 server. The three output ports are: key for result patient set, key for result encounter set, count of patients.
A test case
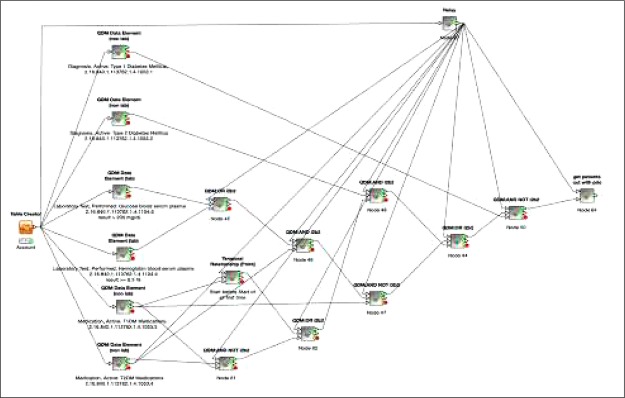
As a demonstration test case, we use a version of the type 2 diabetes mellitus (T2DM) phenotype algorithm created as part of the Electronic Medical Records and Genomics (eMERGE) network17 (Figure 4). This version of the T2DM algorithm was also previously used in the AMIA 2015 Demonstration S32 (PhEMA: Phenotype Modeling, Sharing and Execution Architecture).18 The T2DM algorithm requires a set of complex nested Boolean logical operations over diagnoses, labs, and medications. It also includes a point-to-point temporal operation on timing of medication use.
Figure 4.
High-level overview of the type 2 diabetes mellitus (T2DM) phenotype algorithm. The T2DM algorithm contains multiple nested Boolean operators expressed over diagnoses labs, and medications. It also includes a point- to-point temporal operation on timing of medication use. Use of disjunctive OR operator allows for mulitple paths to be selected into a T2DM cohort. The algorithm uses corresponding ICD9 codes for Diagnosis, Active, RxNorm codes of oral hypoglycemics for T2DM medications, and those of insulins for T1DM medications.
This level of complexity provides a good test of the composition of various QDM Operator Modules into a single executable workflow.
Results
We successfully used the QDM Operator Modules on KNIME to implement the test T2DM phenotype algorithm (Figure 5), and performed the query on an i2b2 Virtual Machine (VM) server instance. Among the total 133 demonstration patients provided by the i2b2 repository, none of them were found to be qualified for the proposed inclusion criteria. There are 11 patients with T2DM ICD-9 codes, but only one patient (without a T2DM diagnosis) takes oral hypoglycemic (Table 1). This result is in accordance with the logic of the algorithm, which properly excluded all test patients from inclusion in the T2DM cohort.
Figure 5.
The KNIME i2b2 implementation of the eMERGE Type 2 Diabetes Mellitus Phenotype Algorithm modeled with QDM.
Table 1.
Diabetes-Related Records of i2b2 Demonstrate Patients. T1DM: Type 1 Diabetes Mellitus; T2DM: Type 2 Diabetes Mellitus; Dx: Diagnoses, with ICD-9 codes; Gluc: plasma glucose measure; A1C: Hemoglobin A1c; T1DM Med: mostly insulin; T2DM Med: mostly oral hypoglycemic. 1 or 0 indicates present or absent
| patient_id | T1DMDx | T2DMDX | Gluc > 200 mq/dl | A1C ≥ 6.5% | T1DM Med | T2DMMed |
|---|---|---|---|---|---|---|
| 1000000011 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000013 | 0 | 1 | 0 | 1 | 0 | 0 |
| 1000000029 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000043 | 0 | 0 | 0 | 1 | 0 | 0 |
| 1000000045 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000070 | 1 | 1 | 0 | 0 | 0 | 0 |
| 1000000083 | 0 | 1 | 0 | 0 | 1 | 0 |
| 1000000087 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000096 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000108 | 0 | 1 | 0 | 0 | 1 | 0 |
| 1000000109 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000119 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1000000124 | 0 | 0 | 0 | 0 | 0 | 1 |
In the implementation, we met some difficulty in the QDM data element modules, because of the different designs between the code-based value set system in the QDM and the path-based ontology system in i2b2. We used an external KNIME workflow to query the i2b2 path sets of the QDM value sets directly through i2b2 metadata database of the Ontology Cell. Then we loaded these i2b2 path sets to the data element modules in the demonstration workflow.
The T2DM KNIME demonstration implementation is available for download at: www.ProjectPhema.org.
Discussion
We have described our solution for integrating the i2b2 platform with complex phenotype representation models such as the QDM. We adopted the strategy of decomposing a QDM algorithm into a workflow of granular i2b2 queries, each of which is represented in the KNIME workflow as a single QDM Operator Module. Each module executes a single i2b2 query, and passes the i2b2 result set key to subsequent modules in the workflow. This design potentially enables i2b2 to support phenotype algorithms with arbitrary complexity. Our implementation of the eMERGE T2DM algorithm demonstrates the validity of this decompositional design.
We used the open source KNIME platform as a proof of concept to offer an easy-to-test prototype. KNIME provides an excellent workflow management service and graphical representation of a pipeline of modules. As our previous work has demonstrated,8 KNIME users can easily modify these workflows to adapt to local EHR availabilities or secondary projects. With a standardized repository system like i2b2, this new effort eliminates the need to write local SQL queries. We are also developing EM2KN,14 a Java-based tool that will translate a QDM algorithm represented in HQMF format into an executable i2b2 workflow in the KNIME platform. We will integrate eM2Kn with this i2b2 solution. Future work will demonstrate this approach on an expanded set of clinical quality measures and algorithms.
However, our design is agnostic to a particular implementation platform. Each QDM Operator Module only requires a template of i2b2 XML message from the web client, a list of case specific parameters, a list of XPaths to indicate the destination of parameter values, a RESTful communication service, and an XML parser for response messages. The communication between different modules uses result sets keys. Alternative implementation platforms could be used with this same approach.
The i2b2 infrastructure utilizes a modular design, an open messaging system, and reusable result sets. These qualities facilitate the development of external applications to complement existing i2b2 client applications. Our implementation provides another demonstration of the flexibility and power of the i2b2 architecture. The ecosystem of third party applications enabled by this architecture make the i2b2 platform extremely useful as a common EHR repository. Our work provides an additional alternative to the i2b2 Web Client and Workbench, which are no longer the only two client applications available for consuming i2b2 services.
However, the i2b2 ontology system for queries is still designed specifically for the interface of these two i2b2 native clients. For example, all the queries have to go through the Ontology Cell, and have to use long concept paths as keys. This design makes dynamic value set based data element models (such as the QDM) difficult to adapt to i2b2. In the work described here, we map value sets to i2b2 concept paths externally. Therefore, we suggest that i2b2 query messaging system provide mechanisms to bypass the Ontology Cell to directly query base codes.
A potential limitation of the work described here is that we have not assessed the real world performance characteristics of our decompositional approach. All our tests are performed on i2b2 demonstration instances. Moving forward, a production instance of an i2b2 EHR repository will be required for this assessment.
Acknowledgements
This work has been supported by funding from PhEMA (R01 GM105688), iPGx (R01 GM103859), and eMERGE(U01 HG006379, U01 HG006378, U01 HG006388).
References
- [1].Blumenthal D, Tavenner M. The Meaningful Use Regulation for Electronic Health Records. New England Journal of Medicine. 2010 Aug;363(6):501–504. doi: 10.1056/NEJMp1006114. Available from: http://www.nejm.org/doi/abs/10.1056/NEJMp10 0 6114. [DOI] [PubMed] [Google Scholar]
- [2].Klann JG, Buck MD, Brown J, Hadley M, Elmore R, Weber GM, et al. Query Health: standards-based, cross-platform population health surveillance. Journal of the American Medical Informatics Association. 2014 Jul;21(4):650, 656. doi: 10.1136/amiajnl-2014-002707. Available from: http://jamia.oxfordjournals.org/cgi/doi/10.1136/amiajnl-2 014-0 02707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].eCQI Resource Center. The one-stop shop for the most current resources to support electronic clinical quality improvement. 2015. Available from: https://ecqi.healthit.gov/
- [4].Thompson WK, Rasmussen LV, Pacheco JA, Peissig PL, Denny JC, Kho AN, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms; AMIA Annual Symposium Proceedings; 2012. p. 911. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC354 0514/ [PMC free article] [PubMed] [Google Scholar]
- [5].Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS computational biology. 2012;8(12):e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nature Reviews Genetics. 2012 Jun;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- [7].Mo H, Thompson WK, Rasmussen LV, Pacheco JA, Jiang G, Kiefer R, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. Journal of the American Medical Informatics Association. 2015 Sep:ocv112. doi: 10.1093/jamia/ocv112. Available from: http://jamia.oxfordjournals.org.proxy.library.vanderbilt.edu/content/early/2015/09/03/jamia.ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mo H, Pacheco JA, Rasmussen LV, Speltz P, Pathak J, Denny JC, et al. A Prototype for Executable and Portable Electronic Clinical Quality Measures Using the KNIME Analytics Platform. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2015;2015:127–131. [PMC free article] [PubMed] [Google Scholar]
- [9].Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Research. 2009 Sep;19(9):1675–1681. doi: 10.1101/gr.094615.109. Available from: http://genome.cshlp.org/cgi/doi/10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].i2b2: Informatics for Integrating Biology & the Bedside. 2015. Available from: https://www.i2b2.org/software/#documents.
- [11].Klann JG, Murphy SN. Computing Health Quality Measures Using Informatics for Integrating Biology and the Bedside. Journal of Medical Internet Research. 2013 Apr;15(4):e75. doi: 10.2196/jmir.2493. Available from: http://www.jmir.org/2013/4/e75/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Centers for Medicare and Medicaid Services eCQM Library. 2015. Available from: https://www.cms.gov/regulations-and-guidance/legislation/ehrincentiveprograms/ecqm_library.html.
- [13].PheKB: a knowledge base for discovering phenotypes from electonic medical records. 2015. Available from: https://phekb.org/
- [14].PheMA/qdm-knime. 2015. Available from: https://github.com/PheMA/qdm-knime.
- [15].Jiang G, Solbrig HR, Kiefer R, Rasmussen LV, Mo H, Speltz P, et al. A Standards-based Semantic Metadata Repository to Support EHR-driven Phenotype Authoring and Execution. Studies in Health Technology and Informatics. 2015;216:1098. [PMC free article] [PubMed] [Google Scholar]
- [16].i2b2: Informatics for Integrating Biology & the Bedside. 2015. Available from: https://www.i2b2.org/software/#documents.
- [17].Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association: JAMIA. 2012 Apr;19(2):212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Type 2 Diabetes Mellitus | PheKB. 2015. Available from: https://phekb.org/phenotype/type-2-diabetes-mellitus.