Abstract
Hundreds of biological and biomedical ontologies have been developed to support data standardization, integration and analysis. Although ontologies are typically developed for community usage, community efforts in ontology development are limited. To support ontology visualization, distribution, and community-based annotation and development, we have developed Ontokiwi, an ontology extension to the MediaWiki software. Ontokiwi displays hierarchical classes and ontological axioms. Ontology classes and axioms can be edited and added using Ontokiwi form or MediaWiki source editor. Ontokiwi also inherits MediaWiki features such as Wikitext editing and version control. Based on the Ontokiwi/MediaWiki software package, we have developed Ontobedia, which targets to support community-based development and annotations of biological and biomedical ontologies. As demonstrations, we have loaded the Ontology of Adverse Events (OAE) and the Cell Line Ontology (CLO) into Ontobedia. Our studies showed that Ontobedia was able to achieve expected Ontokiwi features.
Introduction
A formal ontology is a set of terms and relations that represent entities in a specific domain and how they relate to each other. Hundreds of ontologies have been developed and have played a critical role in biomedical data and knowledge representation, integration, sharing and analysis. For example, the Gene Ontology (GO) [1] has been proven valuable in genome annotation and high throughput data analysis. The Ontology for Biomedical Investigations (OBI) has been used for representation of a wide range of investigations [2]. The Ontology of Adverse Events (OAE) has ontologically represented various adverse events (AEs) used in clinical vaccine and drug AE report and analysis [3]. The Cell Line Ontology (CLO) represents a large number of cell line cell types that are widely used in biological and biomedical research [4].
Community-based ontologies are typically designed for maximal community usages, and it would be beneficial to have wide community involvements in the entire ontology development process. For example, as a community- based ontology, the OAE is developed by following the best practice of the Open Biological and Biomedical Ontologies (OBO) development principles, such as openness, collaboration, and common format [3,5]. Compared to the classical AE classification terminologies, such as the Medical Dictionary for Regulatory Activities (MedDRA) [6], OAE has many advantages including its textual and logical definitions of various AEs, enhanced capability in robust AE classifications, and semantic framework in linking AEs and patients’ health backgrounds [3,7 –9]. However, one drawback of OAE is its low coverage of reported AEs. While MedDRA has over 65,000 classes, OAE only contains approximately 5,000 AE terms as of the end of 2015. It is time consuming to expand OAE while keeping its high quality standard. One possible strategy in future OAE development is to involve wide community support. Additionally, the development of CLO provides another example of the critical need of community involvements. CLO includes over 35,000 cell lines. In order to make CLO better serve the community, each of these cell lines in CLO requires more annotations and verifications. Such a goal is hardly achievable without a large voluntary community effort. To support community-based ontology development, efficient tools with community- editing, discussion, and version control are required.
While different software programs have been created to support community-based ontology development efforts, these programs either do not attract wide community participation or fail to provide critical ontological features. Wikipedia is a successful system that has attracted active community participation. Wikipedia is developed based on the MediaWiki software package. However, Wikipedia mainly uses free text editing, and does not support semantic definitions. To solve this problem, Semantic MediaWiki (SMW) is developed to extend the capability of MediaWiki to semantically annotate wiki pages and support semantic data storage and query [10]. SMW has been used in many applications, including LexWiki [11,12] and SNPedia [13]. OntoWiki is another open-source semantic wiki application developed to present different views on instance data [14]. Both SMW and OntoWiki are not designed to support ontology development since they treat normal pages as instances and do not display ontology classes as normal pages. SMW and OntoWiki also do not support logical ontological axiom definitions. WebProtege uses the Google Web Toolkit for the user interface and Protege for ontology supporting service [15]. WebProtege provides many good features to support ontology development and social discussion. However, its usage does not support non-ontological annotations, requires professional training, and does not attract wide domain expert and public involvements in comparison to Wikipedia.
Ontobee is an ontology browser and a linked ontology data server for dereferencing ontology terms [16]. To date, there are over 100 ontologies listed on Ontobee. Ontobee loads individual page for each class, property, or individual term in an ontology. All related information of a single term, such as label, definition, synonyms, superclass hierarchy, logical axioms, and term usage by other ontologies is displayed. Ontobee provides ontology term search and SPARQL query service supported by the He group Virtuoso RDF triple store1. Ontobee is a de facto linked ontology term server for most OBO Foundry [5] library ontologies. As a linked server for released ontology terms, Ontobee is not designed for wide community users to edit and update ontologies.
In this report, we represent our development of Ontokiwi, a new extension of MediaWiki that also incorporates many Ontobee-like features, with an aim to support community-wide ontology annotations and development. Ontokiwi integrates popular features from MediaWiki, Ontobee, and SPARQL-enabled semantic technologies, and generates new ontology development features unseen in SMW and other programs. Based on the Ontokiwi technology, we developed the Ontobedia web program for the community-wide biological and biomedical ontology development. Ontobedia has now included the OAE and CLO. In this manuscript, we demonstrate the features of Ontokiwi and Ontobedia primarily using the examples of community-based OAE development.
Methods
Development of ontokiwi:
Ontokiwi is developed as an extension of MediaWiki. Since MediaWiki is developed with PHP, we have also chosen PHP as our default programming language for Ontokiwi development. Ontology modification is achieved by editing the Wikitext source of the corresponding page. The MediaWiki magic word and Wikitext parsing system is utilized and extended to support update ontology into MySQL and RDF triple store. The general Wikitext data is stored in MySQL by MediaWiki build-in system. The parsed ontology data from Wikitext is stored in a RDF triple store through SPARQL queries. For the RDF triple store development, we have used Virtuoso Open source version2for our testing and evaluations. SPARQL is used as the database query language to query and update data stored within the RDF triple store.
Development of ontobedia:
Developed by deploying MediaWiki with Ontokiwi extension, the Ontobedia web server provides a platform supporting community development of biological and biomedical ontologies. Ontobedia uses Two HP ProLiant DL380 G6 servers each with quad core Intel Xeon CPUs and 3 TB hard drives. Red Hat Enterprise Linux Server release 6.7 is used as the server operating system. The MySQL database version 5.1 is used to store the default MediaWiki data and Ontokiwi-specific local data. The Ontobedia ontologies are stored in a RDF triple store generated using Virtuoso open source version 7.2.0. The default SPARQL program in the Virtuoso system was used for data access, query, and processing.
OAE and CLO loading to Ontobedia:
To provide the first demonstration of Ontobedia, the most recent versions of OAE and CLO were stored in a Virtuoso triple store generated in the Ontobedia server. The OAE and CLO ontologies were then loaded to Ontobedia by using Ontokiwi importing function. Features of Ontokiwi were tested and evaluated by the loaded OAE within Ontobedia.
Ontokiwi license and download:
The Ontokiwi source code is stored in the Github repository: https://github.com/e4ong1031/Ontokiwi. Its usage follows the open Apache License Version 2.0.
Results
Overall Ontokiwi system design
Fig. 1 illustrates how Ontokiwi is designed and integrated with the existing MediaWiki. Ontokiwi is the core technology we developed. The Ontokiwi system (enclosed in blue box in Fig. 1) extends the MediaWiki setup, language system, and parsing/rendering programs. In addition to the MediaWiki’s MySQL relational database, Ontokiwi includes a new Virtuoso RDF triple store that stores the ontology RDF triples including ontology classes, object and annotation properties, and ontological axioms. The ontology RDF triples are queried through SPARQL to support different programs including OWL ontology import and export. The information from both MySQL and RDF triple store can be queried and displayed in the Ontokiwi browser. A user can also edit the ontology contents using an Ontokiwi form or the source editing function. The edited results will be saved back to the Ontokiwi RDF triple store as well as the MediaWiki MySQL database.
Figure 1.
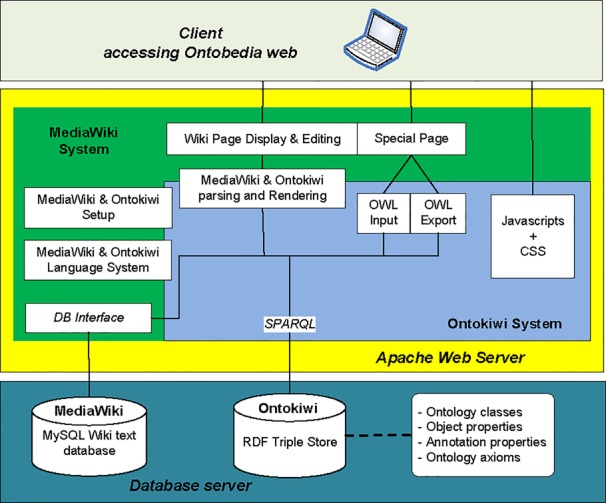
Overall Ontokiwi software system design. See the text for details.
Similar to the MediaWiki system being used in Wikipedia, the Ontokiwi/MediaWiki software package is used to develop Ontobedia (http://ontobedia.hegroup.org), a community-driven system for ontology annotation and development. Below we will use the examples found in Ontobedia to demonstrate how Ontokiwi works.
Unique Ontokiwi features with demonstrations in Ontobedia
In this section, we will introduce representative Ontokiwi-specific features that are not implemented in the original MediaWiki. These features will be explained with the OAE represented in Ontobedia.
(1) Ontology storage in RDF triple store and import to Ontobedia
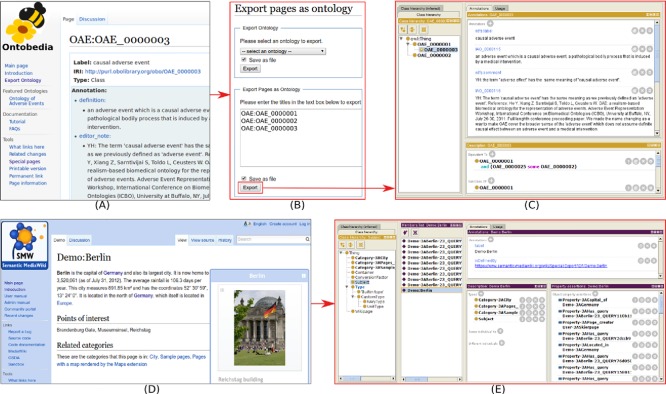
The new Ontokiwi feature “import pages from ontology” provides a convenient way to easily import an ontology from an existing RDF triple store to an Ontokiwi-based web system (e.g., Ontobedia). As shown in Fig. 2, this program requests related ontology information, and then processes the information based on a backend program.
Figure 2.
Ontokiwi automatic generation of wiki pages by importing ontology data from existing RDF triple store. Note that many items of information, including the term creation digit of 7, are requested in order for Ontokiwi to generate new ontology IDs for user-added ontology terms.
(2) Ontokiwi page display of ontology class information
For each ontology term page, we use the naming strategy of:
For example, for the OAE term OAE_0000003 (label: causal adverse event) and Basic Formal Ontology (BFO) term BFO_0000001 (entity) displayed in the Ontobedia OAE system, the following web URL naming strategy is provided for these terms: OAE:OAE_0000003 and OAE: BFO_0000001.
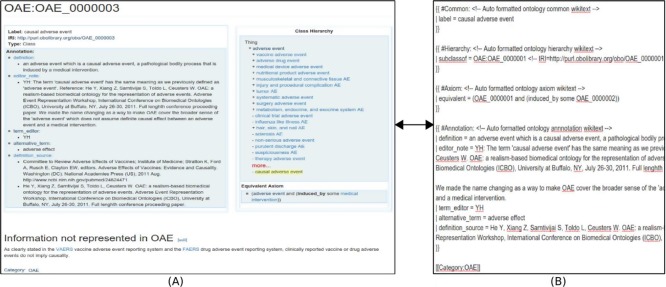
Ontokiwi represents each ontology term using Ontobee-like style (Fig. 3). Specifically, Ontokiwi first lays out the label, the Internationalized Resource Identifier (IRI), and the type of an ontology term. In the section of Annotation, Ontokiwi provides the detailed annotation information based on the annotation properties. Ontokiwi lays out the class hierarchy in the right side of the page as an embedded section (Fig. 3A).
Figure 3.
Display hierarchy, axioms, and annotations. (A) Display of ontology term information including label, IRI, type, annotations, class hierarchy, and axiom definitions. The bottom part shows the text information not represented in OAE. (B) The source of the ontology term page.
(3) Ontokiwi ontology term and axiom editing using source text or online forms
Ontokiwi provides convenient access for users to edit existing ontology terms. Fig. 3B shows the Wikitext source of the ontology term ‘causal adverse event’ (OAE_0000003). The Wikitext in original MediaWiki system is a markup language without any support for ontology editing. Therefore, we have embedded some specifically designed formats on top of the original syntax in Ontokiwi Wikitext parser. For example, we used the text “label =“, “subclassof =“ and “equivalent =“ to represent the type of information that is provided after the sign of “=“. If the text following “=“ sign is expected to be an ontology term, the information is automatically validated and reformatted using the data queried from the RDF triple store.
In addition to ontology related information, the ontology term can also have text descriptions or annotations that are not linked to the ontology annotations and axiom definitions. Users can add these texts using the original MediaWiki Wikitext format (Fig. 3B). Since the newly added information is unrelated to ontology, they will be only stored in MediaWiki MySQL relational database.
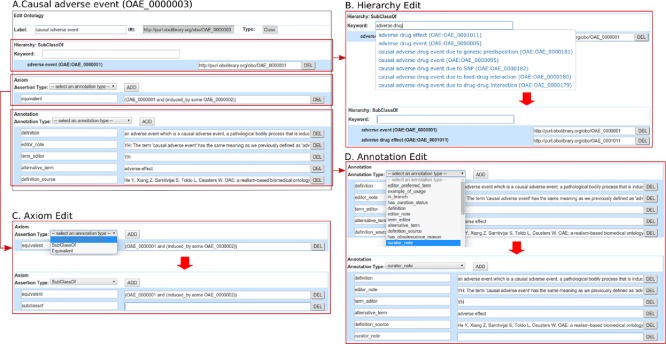
In addition to the Wikitext editor, a user can edit the ontology term information using a form-based Ontokiwi editing system, which includes four major sections (Fig. 4):
-
(i)
General information: This section includes labels, IRI and the type of current ontology term (Fig. 4A). Note that the editing of IRI and term type is restricted to a user with an administrative privilege because improper modification of these properties may introduce ontological conflicts.
-
(ii)
Hierarchical definition: This section defines the parent term of current ontology term with the World Wide Web Consortium (W3C) RDF standard sub-class-of property ‘http://www.w3.org/2000/01/rdf-schema#subClassOf (http://www.w3.org/TR/owl-ref/#subClassOf-def) (Fig. 4B).
-
(iii)
Annotation information: Annotation text can be added/edited with pre-defined annotation properties (Fig. 4C).
-
(iv)
Axiom definition: This section allows a user to define ontological axioms (Fig. 4D). Using the form, a user can first specify one of the two W3C RDF standard logical axiom types: sub-class-of property (see above), or equivalent property ‘http://www.w3org/2002/07/owl#equivalentClass’ (http://www.w3.org/TR/owl- ref/#equivalentClass-def). Next, a logical axiom definition can be generated using the W3C Manchester format (http://www.w3.org/TR/owl2-manchester-syntax/).
Figure 4.
Edit an existing ontology term and create a new term using Ontokiwi forms. (A) General term editing interface for editing the term ‘causal adverse event’ (OAE_0000003). (B) Hierarchical parent term definition. This form provides a way for users to define a parent term for existing term. (C) Axiom editing form. This form allows a user to generate ontological axioms. (D) Annotation editing form. A user can identify an annotation property and provide annotation text under the specific annotation property.
(4) Ontology export
The original MediaWiki export function only exports the page content as XML and does not contain ontology data. To overcome this shortcoming, Ontokiwi introduces a unique function to export the ontology data in the RDF/XML format, which correctly represents logical ontological definitions and axioms (Fig. 5). The Ontokiwi ontology data export feature is achieved through querying the backend RDF triple store through SPARQL. The queried results are then transformed to RDF/XML format. The exported ontology data can be either a whole ontology or a subset of ontology terms. The output file can be visualized in any ontology editor such as the Protege (Fig. 5C). In comparison, SMW can also export page results to RDF/XML format. However, the SMW’s regular pages are instance data even for ontology class terms, as demonstrated by the exported instance information shown in Protege (Fig. 5D-E).
Figure 5.
Ontokiwi ontology export and its comparison with SMW export. (B) The Ontobedia wiki page of a term to be exported. (B) Ontokiwi page export web interface. The top part allows the export of a whole ontology. The bottom part allows the export of pages as an ontology subset. In this example, three OAE terms were targeted for export. (C) A Protege display of the source file, which correctly represents OAE_0000003 (i.e., ‘causal adverse event’) as an ontology class, related annotations and axioms, and its parent class OAE_0000001 (i.e., ‘adverse event’). Note that this program currently does not export other ontology terms semantically associated with these three designated classes. (D) the SMW demo page of Berlin. (E) Protege display of an exported file from the SMW Berlin demo page. In this case, the ‘Demo:Berlin’ is an instance of ‘Subject’ and three other types of categories.
Inheritance of MediaWiki features to Ontokiwi for ontology community development
In addition to the new Ontokiwi-specific features, since Ontokiwi extends MediaWiki, the whole Ontokiwi/MediaWiki package also incorporates all the original MediaWiki features.
One basic MediaWiki feature frequently used in Ontokiwi is the Wikitext format, which is a predefined text format that shows the source of the web page (Fig. 3B). In addition to the traditional Wikitext usage, we have embedded our Ontokiwi-specific styles in Ontokiwi as demonstrated in Fig. 3B and explained in previous Ontokiwi specific feature section (3).
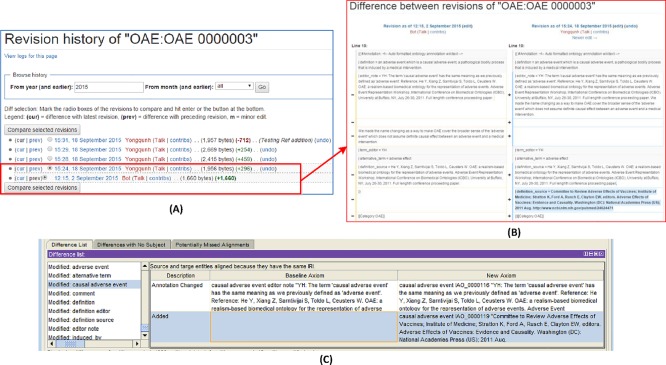
Wiki page version control is an important feature in MediaWiki. As an example, Fig. 6 shows how the Ontokiwi/MediaWiki package is able to support history checking and version control. After we make any text changes, the history of the changes is stored in the MediaWiki MySQL database and the information can be displayed in a user-friendly fashion. A user can also select two time points and compare the text differences between the two time points (Fig. 6B). In addition to the internal checking of version differences, ontology term pages can also be exported as a RDF/XML file, and compared with the original ontology document in another software program, for example, the Protege OWL editor (Fig. 6C).
Figure 6.
Ontobedia supported version control. (A) Revision history page of an edited term. (B) The detailed difference comparison between two versions. (C) Newly edited term information can be exported to a RDF/XML file, and displayed and compared with the original OAE file in Protege.
MediaWiki provides very good features to support social editing and discussion. Such social discussion features are available in Ontokiwi for the community to discuss the ontology terms in different areas with various opinions.
Furthermore, the MediaWiki Template mechanism can be used to generate different templates for saving duplicating work already done. Such templates (e.g., RefList for reference formatting) can be obtained from another wiki system such as Wikipedia. Such a feature enables Ontokiwi-based programs (e.g., Ontobedia) to easily integrate reusable templates to improve performance.
Ontobedia development and features
Ontobedia (http://ontobedia.hegroup.org) is an Ontokiwi-based web application targeted to support community- based biological and biomedical ontology development, non-ontological annotations, and discussions.
As the first demonstration, we have uploaded OAE in Ontobedia. OAE defines ‘adverse event’ (AE) as a pathological bodily process that occurs after a medical intervention (e.g., vaccination) [3]. OAE has been used in different applications, for example, the OAE AE classification system has been used to classify and analyze the AEs associated with live attenuated and killed inactivated influenza vaccines using the clinically reported VAERS vaccine AE data [8]. OAE has also been used to classify and analyze tyrosine kinase inhibitor (TKI) cardiovascular AEs associated with TKI chemical drugs [17]. In addition, OAE is reused in development of other ontologies. For example, The OAE and the Vaccine Ontology [18] have been extended to generate the Ontology of Vaccine Adverse Events (OVAE) [9] as a knowledge base of AEs induced by licensed vaccines. An extension of the OAE and the Drug Ontology (DrON) [19] has been implemented to generate the Ontology of Drug Neuropathy Adverse Events (ODNAE) as a knowledge base of neuropathy adverse events induced by chemical drugs [20]. The Drug- drug Interactions Ontology (DINTO) (http://pubs.acs.org/doi/abs/10.1021/acs.jcim.5b00119) and eNanoMapper project [21] also use OAE to represent AEs in their domains of representations. With the popular usage of OAE and the importance of studying AEs to advance drug and vaccine safety, it is a good option to use OAE as our first ontology to evaluate in Ontobedia.
The features of Ontobedia were evaluated using the OAE use case. First of all, Ontobedia was able to import all the OAE ontology terms. Currently, Ontobedia holds 5,027 OAE ontology classes and their corresponding WikiPages, and supports 111 ontology annotation properties, and 158 object properties. It is noted that OAE [3] imports many terms from other reliable ontologies, for example, BFO [22], UBERON [23], and OBI [2]. Ontobedia is able to correctly display these terms dynamically by querying the Ontobedia RDF triple store. In our evaluation, we randomly selected 50 Ontobedia web pages, each corresponding to one ontology term available in OAE. By manually comparing the ontology display information in Ontobedia and the information shown up using the Ontobee (http: //www.ontobee.org/ontology/OAE) and Protege OWL-editor, we found that Ontobedia is able to display all the information of selected OAE ontology terms as shown in Fig. 3 and Fig. 5. Furthermore, we have demonstrated in Fig. 4 that Ontobedia can be used to edit an existing ontology term and create a new term using Ontokiwi forms. As shown in Fig. 6A, Ontobedia was able to list all the revision history of term editing. The difference between edited version and the original version can be easily compared using the Ontobedia comparison feature (Fig. 6B) or using the Protege OWL editor after the Ontobedia output and display in Prote´ge´ (Fig. 6C).
To ensure the Ontobedia can be used for more than one ontology, we also uploaded the Cell Line Ontology (CLO) [4] to Ontobedia. CLO contains over 35,000 classes of specific cell line cells derived from over 200 in vivo cell types from various organisms. Our investigation shows that Ontobedia was also able to fully import CLO, and the community members can freely edit and discuss CLO terms using the Ontobedia platform (Data not shown).
Discussion
The major contributions of the paper include: (1) The first time introduction of Ontokiwi, a MediaWiki extension that supports community-based ontology development and annotations. (2) Description of new features of Ontokiwi and features inherited from MediaWiki. (3) The first time introduction of the Ontokiwi-based Ontobedia system that now includes OAE and CLO. Ontobedia provides a feasible decentralized mechanism and platform for possibly wide community involvements in ontology development, annotations, and discussions.
Since ontologies are typically consensus based and target for wide community usages, it is critical to involve more community efforts to know, edit, and discuss ontology details. However, currently most ontologies are developed by a small group of experienced developers. How to get more community involvements in ontology development has become a big challenge. Ontokiwi is developed with the purpose to support wide community-based ontology development. In addition to ontology development, the wiki features embedded in Ontokiwi allow users to provide additional Wikipedia-like annotations that would support better understanding and usage of individual ontology terms. Therefore, Ontokiwi also supports ontology annotation and distributions.
The Ontokiwi/Ontobedia aims to target wide range of users. Ontokiwi works as an integrative platform in the way that it simultaneously provides features for ontology experts/developers, domain experts who are unfamiliar with ontology development, and regular users who are just interested in searching for more information. For ontology experts and professional ontology developers, they can use the form editing function and benefit from the version control and discussion features. For those domain experts who know little about ontology but would like to contribute to ontology development, they can use the Wikitext editing features (same as those in regular Wikipedia) to edit the ontology or add annotations. For web users who are neither ontology experts nor domain experts, they can search for the interested terms and useful information using the Ontokiwi/Ontobedia platform, in the way similar to the regular Wikipedia usage. In addition to the support of collaborative community ontology development, the Ontokiwi tool can be installed on a private server to support close teamwork on private ontology development.
As briefly introduced in the Introduction section, Ontokiwi differs from other related tools, such as OntoWiki [14], SMW [10], and WebProtege [15], in many different ways. Table 1 provides more specific comparison among these tools. Specifically, Ontokiwi is similar to SMW in that both tools extend MediaWiki. The main difference between these two is that Ontokiwi focuses on displaying ontology classes as normal pages where SMW focuses on displaying instances under semantic structure. Due to such a main difference, Ontokiwi emphasizes ontological hierarchy and axioms. Unlike Ontokiwi, OntoWiki does not extend MediaWiki and focuses on different views of instance data [14]. Ontokiwi and WebProtege both focus on ontology community development. WebProtege incorporates features from the installable Protege OWL editor. Ontokiwi extends MediaWiki and also incorporates features from Ontobee and SPARQL-enabled semantics technologies. Due to the extension of MediaWiki, Ontokiwi obtains many wiki-based community involvement features (e.g., Wikitext editing, version control, social discussion, and web query). With the popularity of Wikipedia, these wiki features become more familiar to the public users.
Table 1.
Comparison of Ontokiwi, OntoWiki, Semantic MediaWiki (SMW), and WebProt´g´
| Feature | Ontokiwi | SMW | OntoWiki | WebProte´ge´ |
|---|---|---|---|---|
| Main program language | PHP | PHP | PHP | Java |
| MediaWiki based | yes | yes | no | no |
| Normal page saved as | Original ontology type | instance | instance | Original ontology type |
| Hierarchical display | yes | no | no | yes |
| Axioms with annotation properties | yes | yes | yes | yes |
| Axioms with object property | yes | no | no | yes |
| Object/Annotation property editing | no | yes | yes | yes |
| Version history | yes | yes | yes | yes |
| Version comparison | yes | yes | no | no |
| Social discussion | yes | yes | no | yes |
| Editing Type | Syntax, Form | Syntax, Form | Form | Form |
| Demo web site | ontobedia.hegroup.org | semantic-mediawiki.org | 3ba.se | webprotege.stanford.edu |
Unlike WebProte´ge´, Ontokiwi supports non-ontological annotations of ontology terms. For example, for the OAE term ‘causal adverse event’, in addition to ontology representation, we can also add annotation information that is not represented in OAE (see the bottom part of Fig. 3A). Such a Wikipedia-like feature supports free expression of information related to the ontology term, and the information can be searched and distributed on line by the public.
Currently Ontokiwi does not support the editing of object and annotation properties. SMW, OntoWiki, and WebProtege all include this function. For example, a SMW can add an object property “Capital of’ in a sentence: ‘“Berlin”‘ is the capital of [[Capital of::Germany]]. However, SMW does not allow users to specify a unique identifier for any property, and any SMW page editor can generate a user-defined property like “Capital of’ without the obligation of providing a definition. The reason why Ontokiwi does not provide the function now is that in the OBO ontology development community, instead of generating new properties, it is recommended to reuse object and annotation properties in existing ontologies. However, we will consider adding such a feature in the future to meet the occasional community needs to generate new properties.
It is important to understand the relations among Ontokiwi, Ontobedia and Ontobee. Ontokiwi is the technology used to generate Ontobedia. Since Ontokiwi is open source, users can download Ontokiwi and customize it for other ontology-related applications including private installation and local ontology teamwork. Ontobedia is our first application of Ontokiwi. We have first tested Ontobedia for OAE community development. Ontobedia is also our testing case for Ontokiwi feature evaluation. It is noted that the Ontobedia RDF triple store is not the same as the Ontobee RDF triple store. The data contained in the Ontobee RDF triple store will not be changed by Ontobedia editorial manipulations. The difference between the updated ontology information and the official released version of the ontology stored in the Ontobee RDF triple store can be easily compared (Fig. 6). After expert scrutiny, the results out of Ontobedia can also be officially added to the ontology and then released to Ontobee. New software programs can be later developed to facilitate such ontology version comparison and updating.
We will continuously develop the Ontokiwi tool and the Ontokiwi-based Ontobedia system. Since we are the primary group developing the OAE and CLO, we plan to invite internal and external OAE/CLO ontology developers to test the Ontobedia system for community-wide OAE/CLO ontology development. We will also advertise the Ontobedia to the public and encourage public users to search and test Ontobedia. More ontologies, especially OBO Foundry library ontologies, are expected to be included in Ontobedia. For example, we plan to add the Human Interaction Network Ontology (HINO) [24] to Ontobedia with an aim to represent non-redundant and integrated human interactions and pathways. Through more case studies with Ontobedia, more requests will be identified, new features on Ontowiki/Ontobedia will be added, and more benchmark analyses will be generated and reported correspondingly. Meanwhile, we also expect to collaborate with any interesting parties on the further development of the Ontokiwi tool to better serve the ontology society.
Conclusion
Ontokiwi is a MediaWiki extension that targets to support community-wide ontology term annotation and ontology development. Ontokiwi has many unique features to support ontology hierarchy structure display, term annotation, and axiom editing. Ontokiwi also incorporates many MediaWiki features such as Wikitext editing, version control, and social discussion. Ontobedia deploys Ontokiwi to upload the OAE and CLO ontologies and demonstrates the features of Ontokiwi. Differing from other related tools in different aspects, Ontokiwi provides a new platform for community-based ontology development, annotations, and distribution.
Acknowledgements
This project was supported by a MCubed Diamond project funded in the University of Michigan.
Footnotes
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, Ruttenberg A, Sansone SA, Soldatova LN, Stoeckert CJ, Jr., Turner JA, Zheng J, consortium, OBI Modeling biomedical experimental processes with OBI. Journal of biomedical semantics. 2010;1(Suppl 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Sarntivijai S, Lin Y, Xiang Z, Guo A, Zhang S, Jagannathan D, Toldo L, Tao C, Smith B. OAE: The Ontology of Adverse Events. Journal of biomedical semantics. 2014;5:29. doi: 10.1186/2041-1480-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, Schurer TC, Pang C, Malone J, Parkinson H, Liu Y, Takatsuki T, Saijo K, Masuya H, Nakamura Y, Brush MH, Haendel MA, Zheng J, Stoeckert CJ, Peters B, Mungall CJ, Carey TE, States DJ, Athey BD, He Y. CLO: The Cell Line Ontology. Journal of biomedical semantics. 2014;5:37. doi: 10.1186/2041-1480-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug safety. 1999;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y, He Y. The ontology of genetic susceptibility factors (OGSF) and its application in modeling genetic susceptibility to vaccine adverse events. Journal of biomedical semantics. 2014;5:19. doi: 10.1186/2041-1480-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarntivijai S, Xiang Z, Shedden KA, Markel H, Omenn GS, Athey BD, He Y. Ontology-based combinatorial comparative analysis of adverse events associated with killed and live influenza vaccines. PloS one. 2012;7(11):e49941. doi: 10.1371/journal.pone.0049941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcos E, Zhao B, He Y. The Ontology of Vaccine Adverse Events (OVAE) and its usage in representing and analyzing adverse events associated with US-licensed human vaccines. Journal of biomedical semantics. 2013;4(1):40. doi: 10.1186/2041-1480-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vo¨lkel M, Kro¨tzsch M, Vrandecic D, Haller H, Studer R. ‘Semantic Wikipedia’, in Editor (Ed.)^(Eds.): ‘Book Semantic Wikipedia’ (ACM Press, 2006, edn.), pp. 585–594. [Google Scholar]
- 11. https://wiki.nci.nih.gov/display/LexWiki/LexWiki.
- 12.Jiang G, Solbrig HR, Iberson-Hurst D, Kush RD, Chute CG. A Collaborative Framework for Representation and Harmonization of Clinical Study Data Elements Using Semantic MediaWiki. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2010;2010:11–15. [PMC free article] [PubMed] [Google Scholar]
- 13.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic acids research. 2012;40(Database issue):D1308–1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auer S, Dietzold S, Riechert T. OntoWiki - A Tool for Social, Semantic Collaboration, The Semantic Web - ISWC 2006. Lecture Notes in Computer Science. 2006;4273:736–749. [Google Scholar]
- 15.Tudorache T, Nyulas C, Noy NF, Musen MA. WebProtege: A Collaborative Ontology Editor and Knowledge Acquisition Tool for the Web. Semantic web. 2013;4(1):89–99. doi: 10.3233/SW-2012-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Z, Mungall C, Ruttenberg A, He Y. ‘Ontobee: A linked data server and browser for ontology terms’, in Editor (Ed.)^(Eds.): ‘Book Ontobee: A linked data server and browser for ontology terms’ (CEUR Workshop Proceedings, 2011, edn.), pp. Pages 279–281. [Google Scholar]
- 17.Sarntivijai S, Hur J, Ozgur A, Burkhart KK, He Y, Omenn GS, Athey BD, Abernethy DR. ‘Predicting gene interactions of tyrosine kinase inhibitor-induced cardiotoxicity with ontology of adverse events-assisted bioinformatics’, in Editor (Ed.)A(Eds.): ‘Book Predicting gene interactions of tyrosine kinase inhibitor-induced cardiotoxicity with ontology of adverse events-assisted bioinformatics’ (2014, edn.), pp. [Google Scholar]
- 18.Ozgur A, Xiang Z, Radev DR, He Y. Mining of vaccine-associated IFN-gamma gene interaction networks using the Vaccine Ontology. Journal of biomedical semantics. 2011;2(Suppl 2):S8. doi: 10.1186/2041-1480-2-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna J, Joseph E, Brochhausen M, Hogan WR. Building a drug ontology based on RxNorm and other sources. Journal of biomedical semantics. 2013;4(1):44. doi: 10.1186/2041-1480-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo A, Racz R, Hur J, Lin Y, Xiang Z, Zhao L, Jiang G, Zhu Q, Bai J, He Y. ‘Ontology-based collection, representation and analysis of drug-associated neuropathy adverse events ‘, in Editor (Ed.)A(Eds.): ‘Book Ontology-based collection, representation and analysis of drug-associated neuropathy adverse events ‘ (2015, edn.), pp. 1-6 pages. [Google Scholar]
- 21.Hastings J, Jeliazkova N, Owen G, Tsiliki G, Munteanu CR, Steinbeck C, Willighagen E. eNanoMapper: harnessing ontologies to enable data integration for nanomaterial risk assessment. Journal of biomedical semantics. 2015;6:10. doi: 10.1186/s13326-015-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grenon P, Smith B. SNAP and SPAN: Towards Dynamic Spatial Ontology. Spatial Cognition and Computation. 2004;4(1):69–103. [Google Scholar]
- 23.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome biology. 2012;13(1):R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Xiang Z. HINO: a BFO-aligned ontology representing human molecular interactions and pathways. arXiv, 2013, arXiv:1311.3355.