Abstract
Clinical and Translational Science Award (CTSA) recipients have a need to create research data marts from their clinical data warehouses, through research data networks and the use of i2b2 and SHRINE technologies. These data marts may have different data requirements and representations, thus necessitating separate extract, transform and load (ETL) processes for populating each mart. Maintaining duplicative procedural logic for each ETL process is onerous. We have created an entirely metadata-driven ETL process that can be customized for different data marts through separate configurations, each stored in an extension of i2b2 ‘s ontology database schema. We extended our previously reported and open source Eureka! Clinical Analytics software with this capability. The same software has created i2b2 data marts for several projects, the largest being the nascent Accrual for Clinical Trials (ACT) network, for which it has loaded over 147 million facts about 1.2 million patients.
Introduction
A recent Institute of Medicine report on the CTSA program recommended that the consortium’s members engage more in consortium-level activities regionally and nationally1. The consortium’s members have begun initiatives in enhancing access to electronic health record (EHR) data for accrual into clinical trials and conducting comparative studies of treatment effectiveness in diverse populations2,3. Both of these use cases may require looking beyond one’s own institution to find study participants4. Leading technologies for implementing networks of EHR data for research are i2b25 and SHRINE6. They provide access to local data and mechanisms to query for aggregate information such as counts’ and the networking protocols and interfaces for sharing aggregate information across institutions, respectively. Different i2b2 projects within an i2b2 deployment can contain different datasets. Populating an i2b2 project from local EHRs and data warehouses remains a function that sites adopting i2b2 must implement separately due to the variety of data environments found at academic centers.
Loading data from local systems into an i2b2 project is achieved through ETL software7. Commercial and locally developed ETL solutions typically involve specifying the procedural steps needed to move data from one schema and representation to another. Even with commercial tools, for a typical academic health system several FTEs of local support are needed to build and maintain the procedural logic. Because research data networks use different data models and ontologies for representing data, it is typically necessary to maintain separate i2b2 databases and ETL processes for each network of interest. Reusing procedural logic between those ETL processes would substantially reduce the burden of joining a new network, but existing tools typically do not facilitate reuse8. While i2b2 does not have an ETL implementation built-in, it does implement an ontology cell that provides storage and interfaces for working with standard terminologies and custom data representations. I2b2 also has a simple star schema with a central fact table (observation_fact) and dimension tables for patients, visits, providers and concepts. We hypothesized that i2b2’s data schema design and ontology cell together provide most of the metadata needed to configure an ETL process that can be reused across i2b2 projects at an institution.
We previously developed Eureka! Clinical Analytics, an open source ETL system that is designed to create data marts from clinical data warehouses and other large clinical datasets9. Eureka!’s core data loading code is the Analytic Information Warehouse system, a tool we previously applied to processing large clinical datasets in analyses of hospital readmissions10. Eureka! supports extracting data from clinical data warehouses with a variety of data schemas and representations; optionally computing clinical phenotypes10,11 representing patterns in EHR data that signify disease, treatments and responses; and loading the data and computed phenotypes into i2b2. In support of this process, we originally implemented in Eureka! using the Protégé ontology editor12 an ontology containing a clinical data model and hierarchies representing standard terminologies. This ontology was the primary source of configuration information for controlling the data loading and phenotyping processes. Because the ontology was unique to Eureka!, keeping it up-to-date with current versions of standard terminologies became prohibitively resource intensive. Meanwhile, the database schema implemented by i2b2’s ontology cell has become a common format for sharing standard medical terminologies. We aimed to replace the Protégé ontology with support for reading the data model and terminology information from an i2b2 ontology cell’s database schema. This paper describes our technical implementation and discusses the extent to which we have achieved our goal of relying primarily on the ontology cell metadata to control our ETL process into i2b2.
Methods
Use cases
We are evaluating the flexibility of the metadata-driven ETL process implemented by Eureka! in three scenarios that are either under development or completed. While the sections below focus on a single representative project within each of the three use cases, the Results section more broadly shows statistics on all of the major projects in which Eureka! has been used to-date.
Connecting to a national research data network
The Accrual for Clinical Trials (ACT) Network is a SHRINE-based network of over 20 CTSA hubs. It aims in part to enable investigators to query for patient counts from participating hubs using their web browser in a self-service fashion. The first implementation phase, which setup i2b2 and SHRINE components at each site and loaded them with data, was completed in Summer, 2015. Subsequent phases aim to use the network to facilitate accrual into high priority clinical trials. Our institution aimed to use Eureka! as the ETL engine for making local EHR data available to the network for query.
Local EHR data is available through the Emory Clinical Data Warehouse, an enterprise relational data warehouse architected with a dimensional modeling approach. It is implemented using the Oracle 11g database system (Oracle Corp., Redwood Shores, CA). It currently contains over 8 million patients and 35 million encounters from 5 hospitals and our institution’s clinics. It contains almost the entire contents of Emory’s EHR (Cerner Corp., Kansas City, MO) integrated with data from billing and other systems. It is refreshed nightly. To avoid heavy loads on the production data warehouse, we developed a cloning process using Oracle’s dump file export and import tools that clones tables of interest from production into a staging area for this project.
Major technical work for the project involved mapping local codes for laboratory tests and medication orders to the LOINC laboratory test13 and RxNorm medication14 codes required by the project, as well as mapping local visit and demographics value sets for gender, race and the like to the value sets in the network’s data model. We developed and configured a data adapter for Eureka! to generate SQL for querying demographics, visits, labs, medication orders, diagnosis codes and procedure codes from the staging area. We used Eureka! to load 3 ! years of data from January 2012 through May 2015 into an i2b2 project.
Providing clinical data access for quality improvement investigations
Many academic centers are adopting national data registries containing EHR data that enable them to benchmark their performance against their peers using common metrics such as length of stay, rate of hospital readmissions within 30 days and rate of mortality. We created a local copy of 5 years of data from one of these registries, the UHC Clinical Database 15, to support developing local metrics to perform deep dives into the patient populations that drive performance on standard metrics.
The UHC Clinical Database contains de-identified administrative data and limited clinical data from over 200 hospitals associated with US academic medical centers. Variables include demographics and visit details mapped by UHC to an UHC-specific coding system, and ICD-9 diagnosis and procedure billing codes. De-identified data files going back many years are available to UHC members for download. We created a database schema that mirrors the structure of and relationships between the data files, and we loaded 5 years of content from the files into the schema. We developed and configured a data adapter for Eureka! to generate SQL for querying the schema. We loaded the data into an i2b2 project.
Providing access to clinical data sets for biomedical informatics training and education
Biomedical informatics training programs typically do not provide access to informatics systems loaded with real clinical data. Technical barriers include availability of large de-identified clinical datasets with data represented using standards, and a low-cost solution for loading such data into a widely used clinical data warehousing system that does not require extensive data warehousing expertise to use.
The MIMIC-II Database 16 is a large de-identified dataset containing clinical and administrative data on over thirty- two thousand patients who were in an intensive care unit. It is publicly available with a signed data use agreement. The laboratory test codes in the dataset recently were mapped to the LOINC standard17, and ICD-9 diagnosis and procedure codes are in the dataset. Demographics and visit details are represented using custom codes.
We developed and configured a data adapter for Eureka! that generates SQL for querying MIMIC-II data from the PostgreSQL database that is available from the PhysioNet project as a virtual machine (http://physionet.org). The data adapter supports querying demographics, visits, labs, diagnoses and procedures. These data were loaded into i2b2, and the resulting i2b2 project was made available to a class of over 20 students for assignments in working with clinical data, formulating hypotheses and performing data analysis. This deployment is part of the Cardiovascular Research Grid, an initiative in developing cloud computing resources for data management and analysis for cardiovascular research18.
Eureka! Clinical Analytics
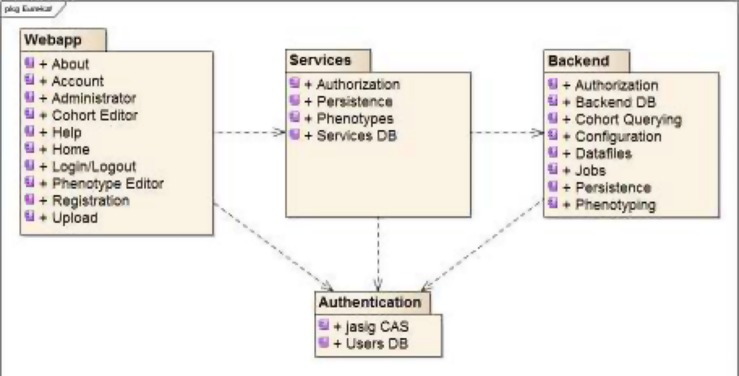
Eureka! has a three-tiered architecture, shown in Figure 1, with web application (user interface), services and backend layers that communicate via representational state transfer (REST) APIs. The server side is implemented in Java (https://www.oracle.com/java/), and the client side is implemented using modern web client programming languages and technologies. The backend implements ETL job processing and tracking. The web application (webapp) and services layers implement a user interface, file upload (for ETL jobs involving extracting data from files), and phenotype storage. A customized JA-SIG CAS (http://jasig.github.io/cas/4.1.x/index.html) server authenticates users in all three layers and facilitates audit trailing. It supports “local” accounts with usernames and hashed passwords stored in Eureka!’s database, or Lightweight Directory Access Protocol (LDAP) or OAuth (http://oauth.net) provider accounts.
Figure 1.
Eureka! Clinical Analytics component diagram.
Data Flow
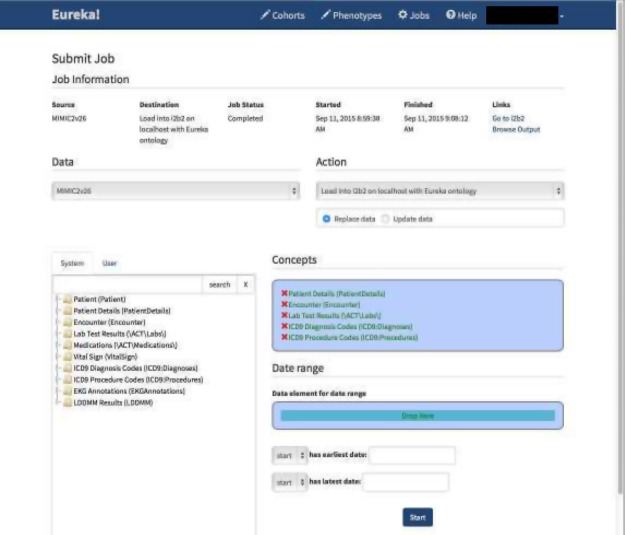
An engineer creates an ETL process using the webapp’s job submission screen, shown in part in Figure 2. The user specifies a data source adapter, an action to perform on the data, a set of concepts representing the data to load, and an optional date range. Data source adapters configure Eureka! to extract data from a specified database or data file. Actions load data into a target i2b2 project or other database or data file. Separate screens support specifying a data source adapter with database connection information, and mappings from the unique structure and semantics of the data source into Eureka!’s internal representation of EHR data. These configuration capabilities assume the existence of a central data warehouse or research database from which to extract data. Eureka! does not currently implement the substantially more diverse data transformations that would be needed to extract data directly from a transactional system like an EHR.
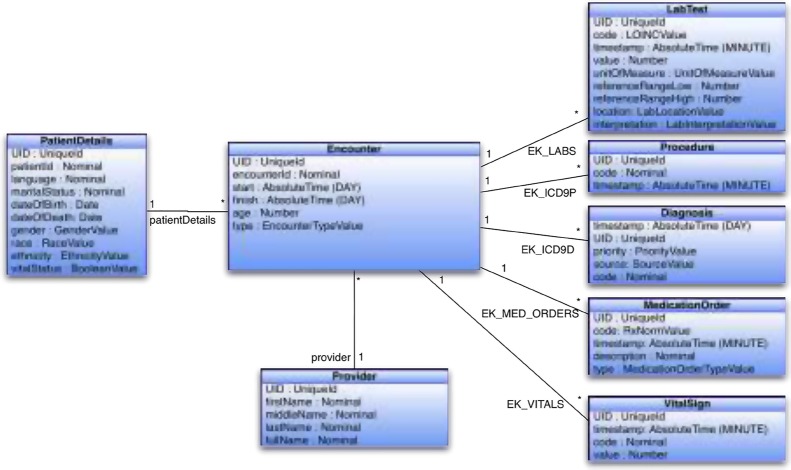
Part of Eureka! ’s internal data model is shown in Figure 3. While the model predates standards for modeling EHR data such as OMOP19, the PCORnet common data model20 and the ACT data model21, it mirrors their structure and semantics closely. While historically Eureka!’s internal model has used our institution’s local laboratory and medication order codes, we have moved in the past year to representing labs with LOINC and medication orders with RxNorm. Data transformations such as clinical phenotyping occur on data represented in the internal model.
Figure 3.
Eureka! internal data model, showing various entities and their properties (blue boxes), and references between entities (black lines).
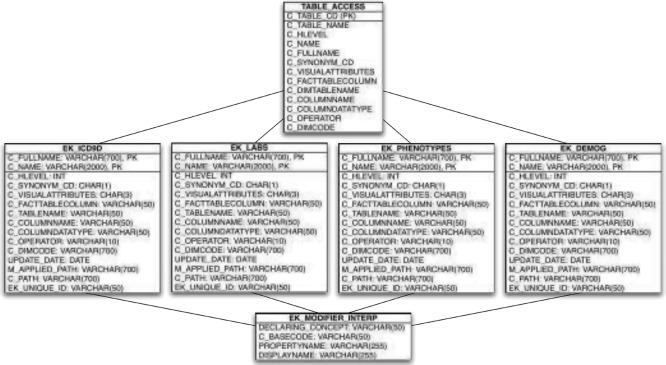
The terminologies used in the internal model are stored in an extended version of the i2b2 ontology cell’s schema, shown in Figure 4. We add one column to each ontology table, EK_UNIQUE_ID, to store a system-wide unique identifier for every concept. We also may store additional metadata about modifiers depending on how they are represented. Modifiers are attributes of a concept such as dose and route for a medication. They map to properties in the Eureka! internal model. A modifier’s value set can be represented as a list of values in the concept’s C_METADATA_XML fragment or as separate records in the concept’s ontology table. While the former provides sufficient metadata to map a modifier to a Eureka! property, the latter does not. Thus, we add to the ontology cell schema an additional EK_MODIFIER_INTERP table to annotate such modifier values with their value set membership. Finally, we add a EK_TEMP_UNIQUE_IDS temporary table for intermediate metadata processing during bulk retrieval. The extensions allow automated import of ontology cell tables as delimited files or SQL statements, which are increasingly common ways of sharing laboratory, medication, diagnosis and procedure codes. We have implemented simple scripts to populate the additional columns and table after importing ontology tables published by the ACT network21.
Figure 4.
Extended i2b2 metadata schema for use with Eureka!, showing four ontology tables that each have one additional column as compared with i2b2’s standard ontology tables, the standard TABLE_ACCESS table supporting concept lookup, and the Eureka!-specific EK_MODIFIER_INTERP table (see text for details).
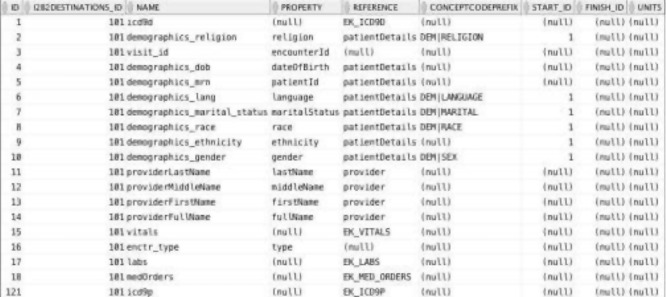
Accessed from the job submission page, screens allow configuring an action with the type of data target (i2b2 project), database connection information, and metadata for mapping from the Eureka! internal data model into the target database’s data representation. Example metadata for the “load into i2b2” action is shown in Figure 5. Processing one Encounter at a time, each record in this table specifies the location of facts or dimension column values in the internal data model (Figure 3). Mappings may be to a property of an Encounter or a path through a reference from the Encounter to the property of another entity in the model. For example, ICD-9 codes are found by traversing the EK_ICD9D reference from Encounter to Diagnosis. The patient’s date of birth is found by traversing the patientDetails reference from Encounter to PatientDetails. The patient’s race is found in the race property of the Encounter. A separate table, not shown, assigns records in this table to dimension table columns, otherwise records are assumed to specify facts. These tables, together with the extended ontology cell schema, provide sufficient metadata to load facts and dimension values into i2b2’s star schema.
Figure 5.
Mappings from the Eureka! internal data model to facts and the dimension tables in i2b2’s star schema.
Thus, Eureka! extends the i2b2 ontology cell’s schema with a total of three tables and one additional column per ontology table. This is in addition to database connection information for the source and target databases, semantic mappings from source coding systems to the standard coding systems supported by the internal data model, and structural mappings from the source system’s physical schema to the structure of the internal data model.
System requirements
We typically install Eureka! on CentOS version 6 or 7 Linux systems (http://www.centos.org). For testing purposes we also have run it successfully on Ubuntu (http://www.ubuntu.com) and other distributions. Eureka! has been tested on Oracle Java 7 virtual machines. It runs in a Tomcat 7 servlet container (http://tomcat.apache.org) with the maximum heap size set to between 4 and 32 GB of RAM depending on the size of the datasets being loaded. Eureka!’s ETL implementation is pipelined, thus its performance benefits from multiple processors and cores.
We install i2b2 on CentOS version 6 or 7 Linux systems. While we currently are using i2b2 version 1.7.05, the projects described have used various releases of i2b2 1.5 and 1.7 over time.
We use Oracle 11g Express or Enterprise Edition database systems deployed on Linux except where indicated.
Results
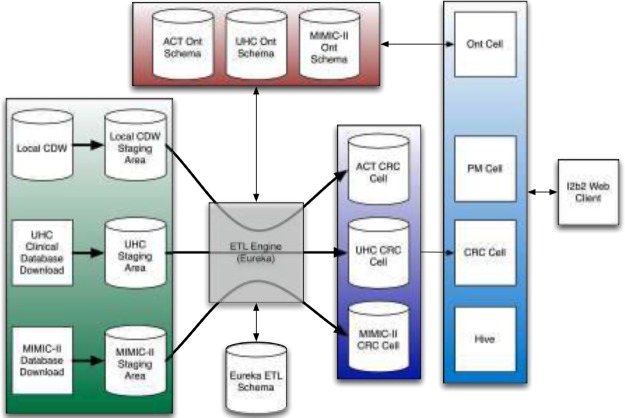
Eureka! is available as open source under the Apache 2 license or the GNU General Public License (GPL) version 3. The GPL-licensed version differs from the Apache-licensed version in supporting an additional action for writing data and phenotypes to the Neo4j graph database (http://www.neo4j.com). In addition to loading data into i2b2, both versions of the software also support loading data into a delimited file. Figure 6 illustrates Eureka! configured for three separate ETL processes for creating i2b2 projects containing a portion of our local clinical data warehouse, a local copy of 5 years of the UHC Clinical Database, and local copy of the MIMIC-II Database for the projects described in Use Cases above.
Data on various deployments of Eureka! are shown in Table 1, including data source, data target, data volume (count of patients loaded, number of phenotypes computed, and total number of facts loaded including phenotypes). While this paper has focused on loading data into i2b2 for hospital readmissions analyses and the CardioVascular Research Grid and NCATS ACT projects, the data target is pluggable as described above. Table 1 also shows projects that have involved exporting data to flat files and to Neo4j. The hospital readmissions analysis has been previously described10. The nascent Neo4j ETL capability is part of a grant in developing radiomics infrastructure for the Quantitative Imaging Network22. A team of three software engineers, a part-time project manager, and a project lead conducted the projects in Table 1, in many cases simultaneously.
Table 1.
Ongoing and completed project data (2011 - present).
| Project name | # of users | Data source | Data target (and software version, where applicable) | Count of phenotypes computed | Count of patients loaded | Count of facts loaded (including phenotypes) | Project status |
|---|---|---|---|---|---|---|---|
| Hospital readmissions analysis | 11 | Emory Clinical Data Warehouse (inpatient discharges from 10/2006-3/2011) | Tab-delimited file | 7,886,868 | 149,514 | 41,031,5151 | Completed |
| Hospital readmissions analysis | 11 | UHC Clinical Database (inpatient discharges from 10/2006-3/2011) | Tab-delimited file | 593,428,440 | 11,794,310 | 1,685,673,6831 | Completed |
| Lymphoma data registry | 4 | Emory Clinical Data Warehouse | i2b2 1.7.05 | 0 | 4,870 | 13,086,0712 | Completed |
| Lung cancer data registry | 4 | Emory Clinical Data Warehouse | i2b2 1.7.05 | 0 | 1,554 | 310,3302 | Completed |
| Cardiovascular Research Grid | 34 | MIMIC-II (all encounters) | i2b2 1.7.05 | 13,196 | 32,074 | 6,446,4132 | Production |
| Quantitative Imaging Network | 6 | NLST data files (all patients) | Neo4j 2.2.2 | 0 | 53,452 | 374,1643 | Development |
| NCATS ACT | 42 | Emory Clinical Data Warehouse (all discharges 1/2012-5/2015) | i2b2 1.7.05 | 0 | 1,153,320 | 147,345,6592 | Beta testing |
number of non-null values in the output file,
number of rows in the i2b2 project’s observation_fact table,
number of nodes in the graph database
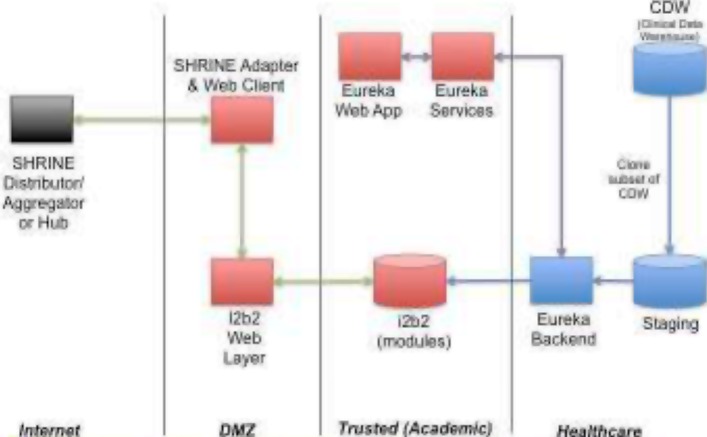
We make available upon request a self-contained VMWare (http://www.vmware.com) virtual machine containing integrated Eureka!, Oracle Express Edition and i2b2 installations on CentOS Linux. This virtual machine is deployed at Johns Hopkins University, where it has been used in the classroom setting described above. Local installations of Eureka! for the ACT Network and UHC projects, among others described in Table 1, involve a dedicated Oracle database server and run the i2b2 web services, the i2b2 web client, the ETL component of Eureka!, and the webapp and services components of Eureka! on separate application servers as shown in Figure 7.
Discussion
Using the previously developed Eureka! Clinical Analytics9,10 as a starting point, we successfully implemented an ETL process7 for populating i2b2 that we have reused for multiple projects. These projects differ in the source of clinical data (local clinical data warehouse, MIMIC-II16, UHC Clinical Database15). While these data sources implement typical data warehouse-style schemas7, they differ substantially in structure and semantic data representations. These projects also differ in data volume (Table 1) and the terminologies supported in the target i2b2 project (LOINC, RxNorm, and ICD-9; custom demographics and visit details value sets). Based upon these experiences, we believe this ETL process is general-purpose in its support for a variety of data sources and formats.
Our experiences also indicate that Eureka!’s highly metadata-driven ETL processing supports the architecture in Figure 6 in which procedural logic is encapsulated in a single ETL system, and a wide variety of ETL processes may be implemented through separate configurations of that system. With traditional procedural ETL systems, processes for populating i2b2 with local data may be time-consuming and expensive to maintain. Moving to metadata-driven systems such as Eureka! are likely to enable institutions’ efforts in joining research data networks, participation in which is becoming a national mandate as well as important infrastructure in support of multicenter clinical research.
Figure 6.
Eureka! data flow illustrated for three ETL processes for loading data into i2b2 (ACT Network, local copy of UHC Clinical Database, and local copy of MIMIC-II Database). Ont=Ontology; CRC=Clinical Research Chart; PM=Project Management.
This architecture complements traditional data warehouse architectures that have a single database that represents the source of truth for reporting on an institution’s operations. The principal benefit of a data warehouse system is integration of data from disparate transactional systems for reporting. As healthcare institutions pool data across institutions for research and quality improvement, maintaining a single data representation that is compatible with every cross-institution initiative is unlikely to succeed. Even with standards, different data networks may justifiably have different data models and representations that are optimized for their use cases. It is theoretically possible to represent synonyms in an i2b2 ontology that assign an observation multiple terms from different terminologies, thus allowing data to be stored once and queried using multiple data representations. However, in practice, differences in the granularity of terminologies make using synonyms for this purpose difficult. It appears increasingly common for institutions to build data marts off of a central warehouse for each network in which they participate.
Using the i2b2 ontology cell as the primary source of metadata for Eureka! has the principal benefit of leveraging the work of national projects in developing and maintaining hierarchies of standard codes. Typical coding systems are updated quarterly to annually, and standard coding systems still frequently lack hierarchies that group the codes in semantically meaningful ways. Thus, maintaining coding hierarchies is very resource intensive. A pitfall in using the i2b2 ontology cell is representing modifier values as separate records in an ontology table, which may result in incomplete metadata for the purpose of building an ETL process (see Methods). As we have demonstrated, a relatively small amount of additional metadata is sufficient to overcome this problem.
Eureka! is available as open source. Beyond several internal deployments, we support a deployment at Johns Hopkins University for the Cardiovascular Research Grid (see educational use case in Methods). We also recently began a deployment at Medical University of South Carolina as part of a southeastern regional CTSA consortium.
These efforts are enabling the development of installation tools for Eureka! that we expect will support dissemination to interested institutions. Additional work required to support dissemination will include more complete graphical screens for specifying the schema of a source system for Eureka!’s query generation, and development of solutions to sharing configurations for accessing frequently used public datasets, versions of standard terminologies in Eureka!’s extended i2b2 ontology schema format, common target data systems like i2b2, and the specification of clinical phenotypes.
Limitations of Eureka!’s implementation at present include lack of a capability to compose metadata describing data integration steps. Eureka supports integration of data from multiple source systems into a single target database, but it requires executing sequential data loads a data source at a time. Eureka lacks support for expression of negated concepts and inclusion and exclusion criteria, though in many cases phenotypes can be constructed that represent mutually exclusive states implicitly. Another limitation is lack of support for emerging standards for representing phenotypes such as the Health Quality Measure Format (HQMF)23. These are all areas of future work. Filling in these feature gaps will enable us to conduct more detailed evaluations of the metadata-driven ETL architecture.
Conclusion
Eureka! Clinical Analytics has supported deployment of clinical data from multiple sources to multiple data marts with different data model and ontology requirements with few technical resources. This technology provides a low- cost and sustainable technical infrastructure for joining research data networks and otherwise building research data marts that enhance access to EHR data for the research community.
Figure 2.
The Eureka! job submission screen, showing the submission of a job loading the MIMIC-II dataset (see Methods) into an i2b2 project. Data to load is specified by dragging concepts from the hierarchy on the left into the blue Concepts box on the right. Users also may specify a date range.
Figure 7.
Deployment diagram showing Eureka! receiving a feed from an enterprise clinical data warehouse. The diagram shows the database and application server deployments, and it shows the network zones in which those systems are deployed (DMZ, Academic, Healthcare).
Acknowledgments
This work has been supported in part by PHS Grant UL1 RR025008 from the CTSA program, NIH, NCATS; PHS grant UL1 TR000005 from the CTSA program, NIH, NCATS; NHLBI grant R24 HL085343; NCI grant U01 CA187013; Emory Healthcare; and Emory Winship Cancer Institute.
References
- 1.Committee to Review the Clinical and Translational Science Awards Program. The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research. In: Leshner AI, Terry SF, Schultz AM, Liverman CT, editors. Washington (DC): National Academies Press (US), National Academy of Sciences; 2013. [PubMed] [Google Scholar]
- 2.National Center for Advancing Translational Sciences. CTSA Consortium Tackling Clinical Trial Recruitment Roadblocks. 2015. [updated Aug 21, 2015; cited 2015 Sep 22]; Available from: http://www.ncats.nih.gov/pubs/features/ctsa-act.
- 3.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(4):576–7. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appel LJ. A primer on the design, conduct, and interpretation of clinical trials. Clin J Am Soc Nephrol. 2006;1(6):1360–7. doi: 10.2215/CJN.02850806. [DOI] [PubMed] [Google Scholar]
- 5.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMurry AJ, Murphy SN, MacFadden D, Weber G, Simons WW, Orechia J, et al. SHRINE: enabling nationally scalable multi-site disease studies. PloS one. 2013;8(3):e55811. doi: 10.1371/journal.pone.0055811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimball R, Ross M. The Data Warehouse Toolkit: The Complete Guide to Dimensional Modeling. 2nd ed. New York: Wiley Computer Publishing; p. 2002. [Google Scholar]
- 8.He T, Gudyka M. Build a Metadata-Driven ETL Platform by Extending Microsoft SQL Server Integration Services. 2008. [cited 2016 Jan 5]; Available from: http://download.microsoft.com/download/D/2/0/D20E1C5F-72EA-4505-9F26-FEF9550EFD44/Build%20a%20Metadata-9.Driven%20ETL%20Platform%20by%20Extending%20Microsoft%20SQL%20Server%20Integration%20Services.docx.
- 9.Post A, Kurc T, Rathod H, Agravat S, Mansour M, Torian W, et al. Semantic ETL into i2b2 with Eureka! AMIA Summits Transl Sci Proc. 2013:203–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Post AR, Kurc T, Cholleti S, Gao J, Lin X, Bornstein W, et al. The Analytic Information Warehouse (AIW): A platform for analytics using electronic health record data. J Biomed Inform. 2013;46(3):410–24. doi: 10.1016/j.jbi.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011:27483. [PMC free article] [PubMed] [Google Scholar]
- 12.Stanford Medical Informatics. The Protege Ontology Editor and Knowledge Acquisition System. 2012. [cited 2012 December 11]; Available from: http://protege.stanford.edu/
- 13.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(4):624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 14.Parrish F, Do N, Bouhaddou O, Warnekar P. Implementation of RxNorm as a terminology mediation standard for exchanging pharmacy medication between federal agencies. AMIA Annu Symp Proc. 2006:1057. [PMC free article] [PubMed] [Google Scholar]
- 15.Sutton JM, Hayes AJ, Wilson GC, Quillin RC, 3rd, Wima K, Hohmann S, et al. Validation of the University HealthSystem Consortium administrative dataset: concordance and discordance with patient-level institutional data. J Surg Res. 2014;190(2):484–90. doi: 10.1016/j.jss.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 16.Joon L, Scott DJ, Villarroel M, Clifford GD. Open-access MIMIC-II database for intensive care research. In: Saeed M, Mark RG, editors. Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE; 2011. Aug. 30 2011-Sept. 3 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abhyankar S, Demner-Fushman D, McDonald CJ. Standardizing clinical laboratory data for secondary use. J Biomed Inform. 2012;45(4):642–50. doi: 10.1016/j.jbi.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winslow RL, Saltz J, Foster I, Carr JJ, Ge Y, Miller MI, et al. The Cardiovascular Research (CVRG) Grid. Proceedings of the AMIA Summit on Translational Bioinformatics. 2011:77–81. [Google Scholar]
- 19.Observational Health Data Sciences and Informatics (OHDSI) OMOP Common Data Model. 2016. [cited 2016 Jan 5]; Available from: http://www.ohdsi.org/data-standardization/the-common-data-model/
- 20.PCORnet. Common Data Model (CDM) Specification. Version 3.0. 2015 [cited 2016 Jan 5]; Available from: http://www.pcornet.org/wp-content/uploads/2014/07/2015-07-29-PCORnet-Common-Data-Model-v3dot0-RELEASE.pdf.
- 21.ACT Data Harmonization Working Group. ACT Ontology and Data Dictionary v1.3. 2015. [cited 2016 Jan 5]; Availablefrom: https://ncatswiki.dbmi.pitt.edu/acts/attachment/wiki/DataHarmonization/ACT%20Ontology%20and%20Data%20Dictionary%20v1.3.docx.
- 22.Kurland BF, Gerstner ER, Mountz JM, Schwartz LH, Ryan CW, Graham MM, et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magn Reson Imaging. 2012;30(9):1301–12. doi: 10.1016/j.mri.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Health Level 7. Representation of the Health Quality Measures Format (eMeasure), Release 2. Ann Arbor, MI: 2012 V3_HQMF_R2_DSTU_2012SEP. [Google Scholar]