Abstract
Prenatal and perinatal exposures vary seasonally (e.g., sunlight, allergens) and many diseases are linked with variance in exposure. Epidemiologists often measure these changes using birth month as a proxy for seasonal variance. Likewise, Genome-Wide Association Studies have associated or implicated these same diseases with many genes. Both disparate data types (epidemiological and genetic) can provide key insights into the underlying disease biology. We developed an algorithm that links 1) epidemiological data from birth month studies with 2) genetic data from published gene-disease association studies. Our framework uses existing data repositories — PubMed, DisGeNET and Gene Ontology — to produce a bipartite network that connects enriched seasonally varying biofactorss with birth month dependent diseases (BMDDs) through their overlapping developmental gene sets. As a proof-of-concept, we investigate 7 known BMDDs and highlight three important biological networks revealed by our algorithm and explore some interesting genetic mechanisms potentially responsible for the seasonal contribution to BMDDs.
1. Introduction and Background
Since antiquity [1], the relationship between disease and birth seasonality was described, pondered and studied. Of particular interest to modern researchers is the relationship between developmental seasonality (using birth month as a proxy) and disease risk. The recent expansion in Electronic Health Records (EHRs) usage throughout the United States of America has enabled researchers to conduct diverse high-throughput exploratory analyses [2 –5]. Recently, we developed and conducted a Season-Wide Association Study (SeaWAS), to systematically identify dependencies between birth month and disease risk using EHRs [6]. The initial study found disease-birth month associations [6], however it did not provide any molecular explanation for the dependency.
Recently, Dopico et al. demonstrated that gene expression can vary seasonally [7]. Additionally, we know that many biological compounds can vary seasonally in humans [8 –11]. Therefore, we decided to develop an algorithm that uses existing data repositories containing genetic information for various disease states and Seasonally Varying Biofactors (SVBs) to find genes potentially responsible for reported birth month dependent diseases (BMDDs). We hope to use these genes to uncover mechanisms behind birth month-disease relationships.
One well-studied BMDD is asthma. Several studies have linked asthma risk to birth month [6,12] where birth month is a proxy for a perinatal environmental exposure. Environmental factors play a key role not only in asthma development but also in its progression. Others have demonstrated that asthma flare-ups are seasonally dependent [13,14] and that genetic mechanisms are involved in this seasonal dependency [15]. Despite all the knowledge behind asthma seasonality and the role of perinatal exposures in regards to disease risk, we cannot easily point to a biological mechanism behind the birth month association. In part, the difficulty lies in the fact that >1,200 genes have been implicated in asthma disease progression [16]. Because of the plethora of genes implicated in certain diseases, finding the potential genetic mechanisms underlying birth seasonality associations is non-trivial. This formed the motivation for our current study.
2. Materials and Methods
Our framework combines data from three public data repositories: PubMed (http://www.ncbi.nlm.nih.gov/pubmed), DisGeNET (http://www.disgenet.org) [16] and the Gene Ontology (GO - http://geneontology.org). Figure 1 illustrates the overall framework approach. Each step is described in more detail in the sections that follow.
Figure 1.
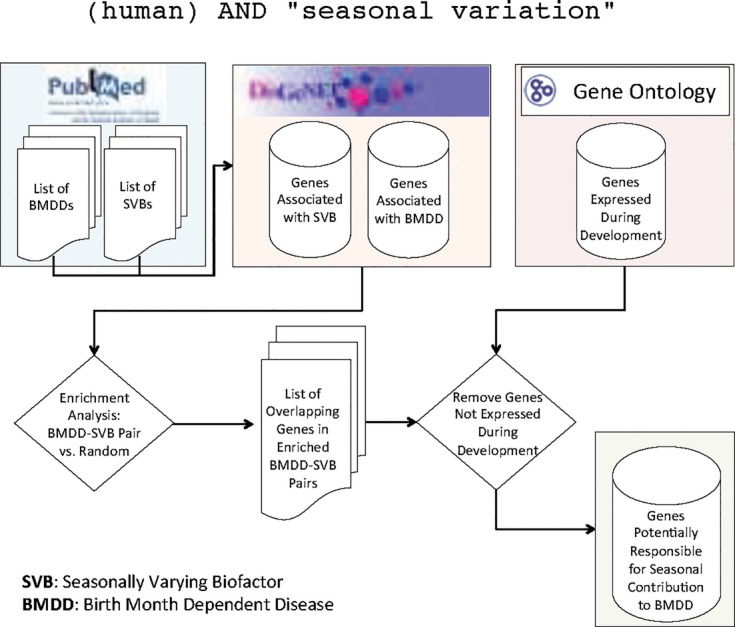
Overview off Our Method Designed to Locate Genes Potentially Responsible for Seasonal Contribution to BMDD.
2.1 Assembling Data Sources from Existing Data Repositories
2.1.1 Develop List of Seasonally Varying Biofactors (SVBs)
Using PubMed, we searched for SVBs in humans (Homo sapiens). All non-humans (e.g., rats, geese, and even non-human primates) were excluded. While we required that the studies involve humans, we ignored the human state (e.g., post/pre menopausal, old/young). Figure 2 contains an example of two SVBs extracted from a study by Meier et al. [17] demonstrating the seasonality of parathyroid hormone (PTH) and vitamin D (specifically calcifediol). In Figure 2, we can see that PTH tends to be higher in the winter months (Jan-Mar) while vitamin D is noticeably higher in the late spring/summer months (Jun-Aug).
Figure 2.
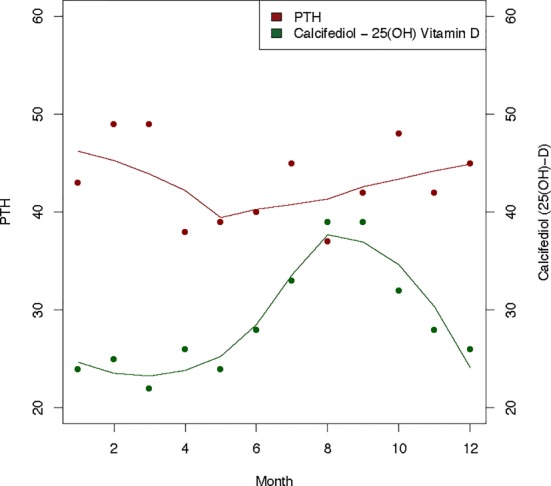
SVBs (parathyroid hormone and calcifediol) measured by Meier et al. 2004. Best-fit lines were applied and slight non-significant anti-correlation was observed (r=−0.303, p=0.338).
To develop a list of literature-backed SVBs, we first queried PubMed with the query: returning 4,091 articles. We then added a species filter (humans) and a language filter (English), which reduced the results set down to 3,627 articles.
We are interested in SVBs and not disease flare-ups (e.g., asthma exacerbations occur seasonally). Therefore, we decided to modify the query to also include the compound. We then ran this for a large variety of compounds (e.g., vitamin D, lactic acid, eosinophils, neutrophils, estrogen, testosterone) to retrieve articles related to their seasonality or lack thereof.
We then read through the resulting abstracts to determine if the compound was accurately found, and to remove any non-human studies that managed to pass the earlier filter. After these initial checks, we determined if seasonal variation was found or not found by the study.
2.1.2 Develop List of Birth Month Dependent Diseases (BMDDs)
Previously, a curated reference set of BMDDs was assembled to assess the quality of SeaWAS results [6]. To derive this reference set, we extracted all articles from PubMed using the term “birth month” and one additional article referenced from a located article (156 articles total). After manually reviewing all articles, we identified 92 relevant articles where birth month was used to study disease risk as a proxy for an environmental exposure. Each article was manually classified regarding whether they found or failed to find an association between the disease of interest and birth month. We then mapped these diseases to EHR-extractable conditions and found that 19 diseases reported in the literature could be mapped including 16 positively associated and 3 not associated with birth month (<50% literature support for an association with birth month). Because we wanted a list of BMDDs with at least 1 publication supporting the relationship between birth month and disease risk, we extended the original list to include the 12 novel findings from SeaWAS [6]. We provide this list with the supplemental information available on figshare. We will use the phrase ‘see supplement’ throughout this paper and we are referring to figshare accessible via: figshare.com/s/b47610ea62d111e5b56406ec4bbcf141
2.1.3 DisGeNET
We mapped each SVB to a disease involving dysregulation of a SVB because DisGeNET only contains genes associated with disease states. For example, vitamin D is an SVB. Hence vitamin D deficiency was used as one of the diseases involving the SVB vitamin D. All genes implicated via association studies in the disease of vitamin D deficiency were used as vitamin D genes. To match SVBs to diseases in DisGeNET, we performed substring matching to the SVB query term. In some cases, we had to modify the SVB search term used. We also did this to ensure that SVBs such as vitamin C (also known as ascorbic acid) were mapped properly. A list of SVBs and the exact query terms used for extracting genes from DisGeNET is included with the supplement. We include examples in Table 1 for explanatory purposes along with example diseases and example genes implicated in those diseases for each SVB in Table 1. However, these examples are not exhaustive.
Table 1.
Examples of SVBs and DisGeNET query terms used to extract SVB-related diseases and genes potentially involved in perturbation of SVBs
| SVB | SVB query term used | Example Disease Names | Example Genes Implicated |
|---|---|---|---|
| Vitamin D | “Vitamin_D” | Vitamin D Deficiency, Rickets Hereditary Vitamin D-Resistant | DHCR7, VDR |
| Parathyroid hormone | “Parathyroid” | Pseudo-hypoparathyroidism, Parathyroid Neoplasms | GNAS, CDC73 |
| Vitamin C | “ascorbic acid” | Ascorbic Acid Deficiency | GSTK1, HP, SLCO6A1 |
| Vitamin K | “Vitamin_K” | Vitamin K Deficiency, Vitamin K Dependent Clotting Factors Combined Deficiency | GGCX, VKORC1, F7 |
| Neutrophil | “Neutrophil” | Neutrophil Actin Dysfunction, Hereditary Neutrophilia | PARP1, CYBA, CSF3R |
| Eosinophil | “Eosinophil” | Eosinophilia, Hyper-eosinophilic Syndrome | IL5, FIP1L1, PDGFRA |
| Hemoglobin | “Hemoglobin” | Hemoglobinopathies, Methemoglobinemia | HBB, CYP1A2, HBA1 |
| Estrogen | “Estrogen” | Oestrogen deficiency, Estrogen Resistance | ESR1, RBBP4, BCAR1, PPP2CA |
Additionally, we mapped our list of BMDDs to diseases in DisGeNET. We did this using approximate string matching similar to above. As this is a proof-of-concept of our framework, we randomly chose 7 BMDDs for this analysis provided they spanned the distribution of number of distinct genes. The seven BMDDs and their query terms used in DisGeNET are given in Table 2. We only used 7 BMDDs because we wanted to test the feasibility of our framework and algorithm. Note the number of distinct genes involved in each disease varies largely from 21 genes (reproductive performance) to 1253 genes (asthma). We randomly selected the BMDDs with this one constraint.
Table 2.
BMDDs included in proof-of-concept along with query terms, example genes implicated and counts of genes involved in BMDD
| BMDD | BMDD query term used | Example Genes Implicated | No. Distinct Genes |
|---|---|---|---|
| Asthma | “Asthma” | SCGB1A1, TNF, CCL11 | 1253 |
| Attention Deficit Disorder | “attention_deficit_disorder” | COMT, LPHN3, GRM5 | 338 |
| Atrial Fibrillation | “Fibrillation” | ACE, NOS3, KCNE2, SELE, VWF | 318 |
| Reproductive Performance | “Reproductive” | BRCA1, BRCA2, TLR4, ESR1, MBL3P | 21 |
| Cardiovascular Disease | “Cardiovascular” | ACE, APOB, LPL, MTHFR | 775 |
| Cardiomyopathy | “Cardiomyopathy” | CSRP3, TTN, DES, TMPO, VCL | 717 |
| Mitral Valve Disorder | “mitral_valve” | FBN1, AGTR1, FBN2, NPPB, PLAU, COL3A1 | 82 |
2.2 BMDD-SVB Pair Enrichment Algorithm
We developed an algorithm to uncover BMDD-SVB pairs that were enriched using their respective gene sets. The first step was the creation of an empirical null distribution for each disease. For each BMDD, we randomly extracted genes from DisGeNET (of the same size as the number of genes for that BMDD). Therefore, 1,253 distinct genes would be randomly pulled from DisGeNET for the empirical null distribution for asthma. However, for reproductive performance only 21 genes would be randomly pulled. We calculated the overlap between the SVB and the random gene set. We iterated through this sampling protocol 100 times and then an overall average overlap was computed. Fisher’s exact test was performed between the true BMDD-SVB pair and the random average overlap computed above (Table 3). Bonferroni correction was applied to adjust for multiple comparisons since each SVB was compared to each BMDD.
Table 3.
The Structure of the Enrichment Algorithm: Each BMDD-SVB Pair was Compared Against a Randomly Generated BMDD-SVB Pair Specific for that BMDD
| No. of BMDD Genes Per SVB | No. Genes Per SVB - No. BMDD Genes Per SVB | |
|---|---|---|
| Actual BMDD-SVB Pair | A | B |
| Randomly Generated * | C | D |
Random was the average across 100 random gene set extractions from DisGeNET using the same number of genes as the BMDD
2.3 Restrict to Genes Involved in Developmental Processes
We aim to explore the contribution of birth month variance on lifetime disease risk. We are especially interested in genes involved in developmental processes, as these are most likely to contribute to a prenatal/perinatal contribution to increased disease risk due to environmental exposures around birth month. Using the Gene Ontology (GO), we restricted the gene set to only include those genes involved in developmental processes. We retained only those genes with at least one GO term containing ‘develop’ in its annotation term description. So for example, if a gene contained the term ‘positive regulation of hair follicle development’ or ‘embryonic placenta development’ it was retained in our analyses. This further reduced the gene set to about 30% of the size (for asthma, 439 asthma genes were enriched in asthma-SVB pairs and only 140 of those genes also had at least 1 developmental GO term).
2.4 Construct Bi-Partite Networks
To visualize the results, we created bi-partite networks [18] for each SVB. BMDDs are included if they are enriched for overlapping genes with the SVB of interest. Each SVB (shown on the left side of the network) is linked to BMDDs (shown on the right side of the network) that are enriched in overlapping genes that are depicted in the middle portion of the graph. Only genes with a developmental GO process are included in the graph. We used DAVID [19,20] to annotate the genes and identify functional gene modules. Network visualization was performed using Cytoscape [21].
3. Results
3.1 Using Existing Data Repositories to Assemble Key Datasets
3.1.1 Seasonally Varying Biofactors (SVBs)
Our original search returned 3,627 articles related to seasonal variation in humans. Therefore, we included additional query terms for biological compounds such as hormones, vitamins, and immune-related cells that are thought to vary seasonally. This allowed us to identify 22 SVBs that are known to vary seasonally in humans. We also found 2 compounds (Homocysteine and Glutaric acid) that are not known to vary seasonally and one that varies seasonally in animals (Corticosterone) but with no human studies currently. We focused on the 22 SVBs that are confirmed to vary seasonally in humans by published studies indexed by PubMed. We used these as input for the enrichment algorithm. Table 4 contains the references supporting the seasonal relationship and references that refute the relationship, if any exist.
Table 4.
Biofactors With Seasonal Dependencies Extracted from the Literature
| Seasonal Relationship | |||
|---|---|---|---|
| Biofactor | Notes | Reference Supporting |
Reference Refuting |
| Hormone | |||
| Parathyroid Hormone (PTH) | PTH and vitamin D are slightly anti-correlated | [17,24] | |
| Estrogen | (modulated through Vitamin D) | [25] | |
| Estradiol | [26] | ||
| Testosterone | (modulated through Vitamin D) | [25] | |
| Progesterone | (modulated through Vitamin D) | [25] | |
| Vitamins/Minerals | |||
| Vitamin A (retinol, beta-carotine) | Bitot eye spots are a sign of vitamin a deficiency | [9,10,27,28] | |
| Vitamin B9: Folate and Folic Acid | [11,29] | ||
| Vitamin B12 | [30] | ||
| Vitamin C (Ascorbic acid) | [9,31,32] | ||
| Vitamin D | [17,24,33] | ||
| Vitamin E | [9] | ||
| Vitamin K | Vitamins K and D regulate osteocalcin | [33,34] | |
| Calcium | [17,24,33] | ||
| Phosphate | [33] | ||
| Immune Cells | |||
| Neutrophil | [35,36] | ||
| Eosinophil | [35,37,38] | ||
| Basophil | [38] | [37] | |
| Other Cells/Metabolites | |||
| Hemoglobin | [39] | ||
| Uric | Uric acid | [40] | |
| Creatine | [41] | ||
| Lactic | Lactic acid | [42] | |
3.1.2 Birth Month Dependent Diseases (BMDDs)
We combined results from our SeaWAS study with a carefully curated set of diseases related to birth month that we developed previously. This file is available with the supplement and includes the PubMed ID, publication year, disease area (high-level disease category), and a binary variable indicating whether the study found or failed to find the association. In this feasibility study of the algorithm’s framework, we used 7 randomly chosen BMDDs with one constraint: that the number of distinct disease genes differed largely. For example, 21 genes were implicated in reproductive performance while 1253 genes were implicated in asthma.
3.1.3 DisGeNET
SVBs and BMDDs were mapped to DisGeNET to extract genes associated with each SVB and BMDD. Examples of the extraction process are given in methods Tables 1 and 2 . For this proof-of-concept, we ran the DisGeNET extraction on 7 random BMDDs with different gene set sizes. We also used DisGeNET to extract the genes related to the 22 SVBs given in Table 4 (folate and folic acid are counted as separate SVBs but merged under vitamin B9 in Table 4).
3.2 Enrichment Results
Our enrichment algorithm investigates the overlap among gene sets from the BMDD and each SVB. It compares this overlap to an average across 100 randomly generated gene sets of the same size (i.e., number of genes) as the particular BMDD of interest. The average overlap score from the 100 random sets is compared against the actual number to determine significance using Fisher’s exact test. We adjusted the p-values using the Bonferroni correction method. We then ranked each significant BMDD-SVB pair by the ORs. Results are shown in Table 5 with the top three associations in bold.
Table 5.
BMDD-SVB Enriched Overlapping Gene Sets Sorted by OR
| BMDD Disease | Enriched SVB | OR | −log(p) * |
|---|---|---|---|
| Asthma | Eosinophil | 23.545308 | 92.3610942 |
| Neutrophil | 7.608800 | 41.7971736 | |
| Vitamin D | 7.597455 | 4.9508901 | |
| Phosphate Gene Set 1 (Phosph) | 6.787030 | 36.4156313 | |
| Hemoglobin | 6.684969 | 19.1382008 | |
| Uric | 5.672420 | 12.1008132 | |
| Calcium Gene Set 2 (Calcinosis) | 4.085383 | 5.4623019 | |
| Calcium Gene Set 1 (Calci) | 3.511367 | 9.7828493 | |
| Phosphate Gene Set 2 (Phosphate) | 5.095905 | 10.1171083 | |
| Parathyroid Hormone | 4.494405 | 23.8948578 | |
| ADHD | Eosinophil | 3.805091 | 5.2892789 |
| Parathyroid Hormone | 3.553309 | 11.9138662 | |
| Neutrophil | 3.308624 | 9.2041648 | |
| Phosphate Gene Set 1 (Phosph) | 3.302909 | 9.7728579 | |
| Calcium Gene Set 1 (Calci) | 2.679110 | 7.5585981 | |
| Calcium Gene Set 2 (Calcinosis) | 2.043983 | 7.1930726 | |
| Fibrillation | Calcium Gene Set 2 (Calcinosis) | 12.287454 | 7.1930726 |
| Phosphate Gene Set 1 (Phosph) | 6.440454 | 9.7728579 | |
| Parathyroid Hormone | 6.437664 | 11.9138662 | |
| Calcium Gene Set 1 (Calci) | 6.380333 | 7.5585981 | |
| Neutrophil | 6.252987 | 9.2041648 | |
| Eosinophil | 6.242087 | 5.2892789 | |
| Reproductive | - | - | - |
| Cardiovascular | Vitamin K | 45.187116 | 5.8861829 |
| Folic Acid | 14.548556 | 6.5377502 | |
| Vitamin D | 13.319860 | 6.1949509 | |
| Phosphate Gene Set 2 (Phosphate) | 11.560768 | 23.1079644 | |
| Phosphate Gene Set 1 (Phosph) | 9.246976 | 39.8563236 | |
| Uric | 9.174204 | 17.0939677 | |
| Calcium Gene Set 2 (Calcinosis) | 8.543921 | 13.0824611 | |
| Calcium Gene Set 1 (Calci) | 8.492730 | 28.6820297 | |
| Neutrophil | 7.892360 | 28.6839072 | |
| Eosinophil | 6.946996 | 19.5349288 | |
| Hemoglobin | 6.384460 | 11.7733926 | |
| Parathyroid Hormone | 4.149517 | 13.8335245 | |
| Cardiomyopathy | Vitamin D | 10.145258 | 3.851662 |
| Eosinophil | 8.923802 | 25.690966 | |
| Lactic | 8.159469 | 6.974233 | |
| Hemoglobin | 7.666338 | 15.743251 | |
| Calcium Gene Set 2 (Calcinosis) | 7.616024 | 10.981833 | |
| Neutrophil | 6.468582 | 21.428566 | |
| Phosphate Gene Set 1 (Phosph) | 6.284259 | 22.014083 | |
| Calcium Gene Set 1 (Calci) | 6.049040 | 16.056148 | |
| Uric | 5.784556 | 6.909913 | |
| Parathyroid Hormone | 4.781197 | 16.279232 | |
| Phosphate Gene Set 2 (Phosphate) | 4.342993 | 3.422636 | |
| Mitral Valve | Phosphate Gene Set 1 (Phosph) | 16.780523 | 5.1995428 |
| Calcium Gene Set 1 (Calci) | 13.587347 | 3.2560223 |
greater than 3.0 is significant after Bonferroni correction
The top SVBs associated with each BMDD are biologically intuitive. Cardiovascular disease is known to involve vitamin K regulation with the anti-coagulant drug warfarin targeting the well-studied vitamin K gene: VKORC1. The two top SVBs related to asthma (a known immune-related condition) are also immune related: eosinophils and neutrophils [22]. Atrial fibrillation’s top hits are calcium related and atrial fibrillation is associated with increased calcium release from the sarcoplasmic reticulum [23].
3.3 Restrict to Genes Involved in Developmental Processes
We reduced the number of genes in our results set by restricting the gene sets to only include those involved in at least one developmental process using GO annotations. This was primarily because of our interest in genes that are involved in developmental processes and therefore may play a role in birth month associations. This drastically reduced our results set as shown in Table 6.
Table 6.
Number of Genes Involved in BMDD That Are Potentially Involved in Birth Month Contribution to Disease is Drastically Reduced After Framework Is Applied
| BMDD | No. Distinct Genes (A) | No. of Overlapping Genes from Enriched BMDD-SVB Pairs (B) | No. of Genes from B Involved in Developmental Processes (C) | % of Genes Potentially Involved in Birth Month Contribution Out of All BMDD Genes (C/A) |
|---|---|---|---|---|
| Asthma | 1253 | 439 | 140 | 0.112 |
| ADHD | 338 | 63 | 18 | 0.053 |
| Fibrillation | 318 | 105 | 45 | 0.142 |
| Reproductive | 21 | - | - | - |
| Cardiovascular | 775 | 302 | 109 | 0.141 |
| Cardiomyopathy | 717 | 250 | 89 | 0.124 |
| Mitral Valve | 82 | 24 | 15 | 0.183 |
Table 6 illustrates how the algorithm started with 1,253 genes associated with Asthma as extracted from DisGeNET. Because asthma is also known to be associated with birth month and a BMDD, we ran our algorithm to find overlapping genes between asthma and SVBs where the overlapping genes were enriched. This reduced the number of genes potentially involved in a seasonally varying process at birth down to 439 genes from 1253. We then restricted these 439 genes to only include genes known to be involved in some developmental process using GO term annotations. This further reduced the number of genes down to 140. Therefore, only 11.2% of asthma-related genes are potentially involved in developmental processes related to SVBs that could potentially lead to birth month-related effects.
3.4 Developmentally Expressed Genes Link SVBs to BMDDs in Biological Networks
Our algorithm produces output containing each BMDD, the enriched SVBs and the overlapping genes that are developmentally expressed between the BMDD and the SVB. A file containing tuples of BMDD, SVB, and gene is available in the supplement.
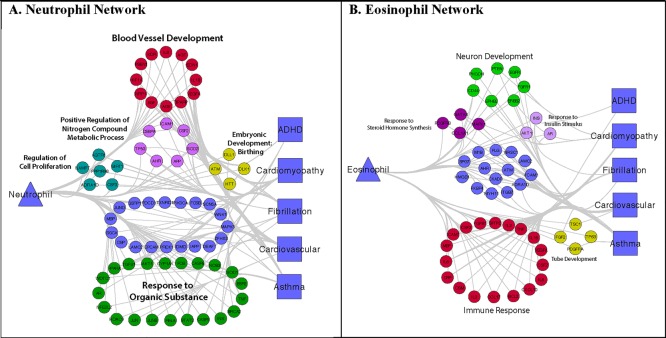
For illustrative purposes, we show three SVB networks from our feasibility study: two immune cells (eosinophil and neutrophil) and one hormone (parathyroid hormone). Full resolution images are available with supplemental information on figshare at figshare.com/s/b47610ea62d111e5b56406ec4bbcf141. The immune cells are shown in Figure 3, the SVB is represented by a triangle, the BMDD is represented by a square and circles represent the overlapping genes. Cluster annotations from DAVID are shown above or near each cluster. In the neutrophil network (Figure 3A) there are clusters involving blood vessel development, response to an organic substance, positive regulation of nitrogen compound metabolic processes, regulation of cell proliferation and embryonic development / birthing. In Figure 3B, the largest cluster includes genes related to the immune response (as expected). Other clusters are for neuron development, tube development (e.g., neural, endothelial tubes), response to sterol hormone synthesis and insulin stimulus. The same 5 BMDDs are involved in both: attention deficit hyperactivity disorder (ADHD), cardiomyopathy, atrial fibrillation, cardiovascular disease, and asthma. Eosinophils and neutrophils are in the top 3 most enriched SVBs for both asthma and ADHD (Table 5). Its possible that the genes involved in neuron development is responsible for the ADHD - Eosinophil relationship (Figure 3B).
Figure 3.
Immune Cell Bi-Partite Networks Connecting SVBs to BMDDs Via Overlapping Genes Involved in Developmental Processes. Full resolution images are available on figshare.
Figure 4 shows the network for parathyroid hormone (PTH). There are three main genetic processes involved: positive regulation of the development process, receptor linked signal transduction, and response to hormone stimulus. Not only is parathyroid hormone a hormone, but it also is involved in regulation or other hormones. Figure 2 illustrates the somewhat anti-correlated relationship PTH has with vitamin D that has been described by others [17]. Both PTH and vitamin D are hormones that regulate each other through complex mechanisms. Positive regulation of the development process to also be enriched in this network connecting PTH and BMDD this fits with the involvement of this SVB with a contribution to disease risk that is due to birth month.
Figure 4.
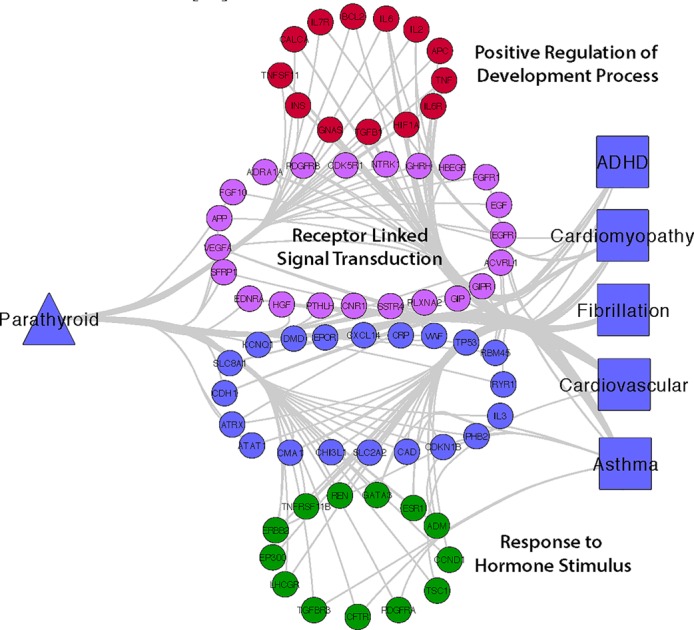
Network Connecting Parathyroid Hormone with BMDDs Via Overlapping Genes Involved in Developmental Processes. Full resolution images are available on figshare.
4. Discussion
4.1 Value of a High-throughput Birth Month-Disease Dependency Genetic Algorithm
The relationship between genes, environment and disease has been discussed by medical researchers since the early days of genetics [43]. Because the relationship between genes and the environment is complex, researchers originally investigated single environmental exposures and how those exposures influenced disease risk via genetic changes [44]. An improvement on this original concept was the development of the Environment-Wide Association Study (EWAS) that explored a large variety of environmental exposures (not just one as was done previously) and then explored how those exposures effected one single disease: Type 2 Diabetes [45]. While an improvement over the work that was conducted previously, EWAS was still limited to exploring one disease at a time.
Our SeaWAS study revealed multiple birth month-disease dependencies (called BMDDs). Additionally, 92 other articles revealed additional information on BMDDs. None of these epidemiological studies sheds light on the genetic underpinnings of BMDDs as they focus primarily on observational data. Therefore, a method was required that could investigate diverse environmental triggers across a plethora of diseases and disease types (e.g., reproductive, mental, immune, and respiratory diseases). To address this gap, we developed an algorithmic framework to uncover enriched SVBs related to BMDDs.
In addition to finding SVBs enriched in BMDDs, we also explore the overlapping genes implicated in both the SVB and the BMDD. We limit our investigation to only those genes that are known to be involved in developmental processes to hone in on those genes that are potentially responsible for birth month disease dependencies. We describe in this paper our exploration of 7 BMDDs and we highlight 3 biological networks related to key SVBs and the BMDDs they potentially modulate.
4.2 Highlighting One Well-Studied Disease: Asthma
The top SVBs enriched for asthma were eosinophil (OR=23.545), neutrophil (OR=7.601) and vitamin D (OR=7.597) (Table 5). The relationship between eosinophils (key cells in the immune response) and asthma is well known and studied [46]. At the same time asthma exacerbations and underlying gene expression changes are also known to vary seasonally [15]. As revealed by our framework, there is also literature support for a relationship between eosinophil changes and season [35,37,38]. Our framework revealed 42 genes in common between asthma and eosinophils that are also involved in developmental processes (the entire eosinophil network is shown in Figure 3B). The immune response was the key functional process involved in this network along with neuron development (which could help to explain the interesting relationship between eosinophils and ADHD in our network).
Table 5 reveals an interesting relationship between asthma and ADHD: they both share eosinophils and neutrophils among their top 3 SVB enrichments (asthma also has vitamin D and ADHD has parathyroid hormone—which are also related to each other). When we investigated the biological network (Figure 3B), the enrichment in neuron development genes is revealed. Importantly, asthma patients are known to be at increased risk for developing ADHD and this increased risk was observed even after adjusting for urbanization and comorbid allergic diseases suggesting an underlying etiology behind the two diseases [47]. Others have also studied the relationship between asthma and ADHD [48] without uncovering a clear genetic/biological mechanism for the relationship.
Studies show that fetal outcomes following an environmental exposure can vary based on the trimester of exposure. Specifically there have been studies related to famine [49] and air pollution [50]. Our SeaWAS study found both ADHD and asthma to be associated with birth month, but the relative risk curves were different (ADHD risk peaked in Nov. while asthma risk peaked in Sept.) [6]. These differences in birth month risk suggest the possibility of a trimester effect (if the exposure is constant for both diseases). Gelardi et al. found that eosinophil cell counts increased almost four-fold in March when compared to any other month (data from Southern Europe) [35]. Hence, babies born in November would be experiencing their first trimester during March while babies born in September would be in their second trimester and a trimester-exposure effect of eosinophils on development could be partially responsible. Importantly, our framework enables researchers to construct biological networks that connect complex associations between SVBs and BMDDs through their shared underlying genetic pathways. This enables researchers to formulate and test hypotheses behind disease etiology and progression.
4.3 Limitations and Future Work
Our study is limited by the information contained and available on PubMed regarding biofactors that vary seasonally (SVBs) and BMDDs. Therefore, neither of these lists is fully complete as there may be other studies not reported in PubMed, studies not translated into English and so forth that would prevent us from using their findings in our framework. Our initial query to PubMed returned 3,627 articles related to seasonal variation in humans. Because we were primarily interested in biological compounds that vary seasonally (such as hormones, vitamins, immune cells), we added additional query terms (as specified in the methods section of the paper). Ideally we would manually review all 3,627 articles but this was not feasible, therefore we may be missing some lesser-known SVBs. Future work includes expanding this work beyond the proof-of-concept presented here to all BMDDs and SVBs. This includes developing a master list of biofactors. Additionally, the fetal-maternal barrier warrants further investigation as the placenta is known to be susceptible to environmental effects [51]. Incorporating knowledge on the epigenetics of the placenta could help with understanding the underlying disease mechanism [51].
5. Conclusion
We present a framework that combines existing data repositories (PubMed, GO, and DisGeNET) to uncover biological mechanisms underlying birth month - disease dependencies (BMDDs) using known Seasonally Varying Biofactors (SVBs). Our framework allows us to link 1) epidemiological data on birth month-disease relationships and 2) genetic data on gene-disease associations recorded in existing public data repositories. Our algorithm finds enriched BMDD-SVB pairs using the genes involved in both the disease and the SVB. We then investigate the overlapping genes in these enrichments and trim away genes not known to be involved in developmental processes using GO annotations. Our framework produces a bipartite network that connects enriched SVBs with BMDDs through their overlapping developmental gene sets. Thus allowing us to form biological hypotheses around the genetic mechanisms underlying birth month-disease dependencies. As a proof-of-concept, we present results from 7 BMDDs across all identified known SVBs. We show biological networks from 3 SVBs while highlighting asthma.
Acknowledgments
MRB supported by NLM Training grant T15 LM00707 and R01 GM107145 (NPT). Authors report no conflicts of interest. Thanks to Joseph Romano for comments on an earlier version of the manuscript.
References
- 1.Hippocrates Galen. In: Hippocratic Writings and On The Natural Faculties. Hutchins RM, editor. Encyclopaedia Britannica; 1952. p. 215. [Google Scholar]
- 2.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland MR, Hripcsak G, Albers DJ, Wei Y, Wilcox AB, Wei J, et al. Discovering medical conditions associated with periodontitis using linked electronic health records. Journal of Clinical Periodontology. 2013;40(5):474–82. doi: 10.1111/jcpe.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland MR, Tatonetti NP. Are All Vaccines Created Equal? Using Electronic Health Records to Discover Vaccines Associated With Clinician-Coded Adverse Events; AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science; 2015. pp. 196–200. [PMC free article] [PubMed] [Google Scholar]
- 5.Shah NH. Mining the ultimate phenome repository. Nat Biotech. 2013;31(12):1095–7. doi: 10.1038/nbt.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boland MR, Shahn Z, Madigan D, Hripcsak G, Tatonetti NP. Birth Month Affects Lifetime Disease Risk: A Phenome-Wide Method. Journal of the American Medical Informatics Association. 2015 doi: 10.1093/jamia/ocv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dopico XC, Evangelou M, Ferreira RC, Guo H, Pekalski ML, Smyth DJ, et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat Commun. 2015:6. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzybowska E, Hemminki K, Szeliga J, Chorazy M. Seasonal variation of aromatic DNA adducts in human lymphocytes and granulocytes. Carcinogenesis. 1993;14(12):2523–6. doi: 10.1093/carcin/14.12.2523. [DOI] [PubMed] [Google Scholar]
- 9.Woodhouse P, Khaw K-T. Seasonal variation of risk factors for cardiovascular disease and diet in older adults. International journal of circumpolar health. 2000;59(3–4):204–9. [PubMed] [Google Scholar]
- 10.Basu T, Donald E, Hargreaves J, Thompson G, Chao E, Peterson R. Seasonal variation of vitamin A (retinol) status in older men and women. Journal of the American College of Nutrition. 1994;13(6):641–5. doi: 10.1080/07315724.1994.10718460. [DOI] [PubMed] [Google Scholar]
- 11.Hao L, Ma J, Stampfer MJ, Ren A, Tian Y, Tang Y, et al. Geographical, Seasonal and Gender Differences in Folate Status among Chinese Adults. The Journal of Nutrition. 2003;133(11):3630–5. doi: 10.1093/jn/133.11.3630. [DOI] [PubMed] [Google Scholar]
- 12.Korsgaard J, Dahl R. Sensitivity to house dust mite and grass pollen in adults. Clinical & Experimental Allergy. 1983;13(6):529–36. doi: 10.1111/j.1365-2222.1983.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 13.Cohen HA, Blau H, Hoshen M, Batat E, Balicer RD. Seasonality of asthma: a retrospective population study. Pediatrics. 2014;133(4):e923–e32. doi: 10.1542/peds.2013-2022. [DOI] [PubMed] [Google Scholar]
- 14.Randolph C. Seasonality of Asthma: A Retrospective Population Study. Pediatrics. 2014;134(Supplement 3):S165–S6. doi: 10.1542/peds.2014-1817DDD. [DOI] [PubMed] [Google Scholar]
- 15.Bjornsdottir US, Holgate ST, Reddy PS, Hill AA, McKee CM, Csimma CI, et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS One. 2011;6(7):e21902. doi: 10.1371/journal.pone.0021902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinero J, Queralt-Rosinach N, Bravo Á, Deu-Pons J, Bauer-Mehren A, Baron M, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015;2015 doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier C, Woitge HW, Witte K, Lemmer B, Seibel MJ. Supplementation With Oral Vitamin D3 and Calcium During Winter Prevents Seasonal Bone Loss: A Randomized Controlled Open-Label Prospective Trial. Journal of Bone and Mineral Research. 2004;19(8):1221–30. doi: 10.1359/JBMR.040511. [DOI] [PubMed] [Google Scholar]
- 18.Lorberbaum T, Nasir M, Keiser MJ, Vilar S, Hripcsak G, Tatonetti NP. Systems Pharmacology Augments Drug Safety Surveillance. Clinical Pharmacology & Therapeutics. 2015;97(2):151–8. doi: 10.1002/cpt.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd BA, Wroblewska A, Boland MR, Agudo J, Merad M, Tatonetti NP, et al. Mapping the effects of drugs on the immune system. Nat Biotech. 2015 doi: 10.1038/nbt.3367. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hove-Madsen L, Llach A, Bayes-Genís A, Roura S, Font ER, Arís A, et al. Atrial Fibrillation Is Associated With Increased Spontaneous Calcium Release From the Sarcoplasmic Reticulum in Human Atrial Myocytes. Circulation. 2004;110(11):1358–63. doi: 10.1161/01.CIR.0000141296.59876.87. [DOI] [PubMed] [Google Scholar]
- 24.Steingrimsdottir L, Gunnarsson O, Indridason OS, Franzson L, Sigurdsson G. Relationship between serum parathyroid hormone levels, vitamin d sufficiency, and calcium intake. JAMA. 2005;294(18):2336–41. doi: 10.1001/jama.294.18.2336. [DOI] [PubMed] [Google Scholar]
- 25.Lee DM, Tajar A, Pye SR, Boonen S, Vanderschueren D, Bouillon R, et al. Association of hypogonadism with vitamin D status: the European Male Ageing Study. European Journal of Endocrinology. 2012;166(1):77–85. doi: 10.1530/EJE-11-0743. [DOI] [PubMed] [Google Scholar]
- 26.Bjørnerem As, Straume B, Øian Pl, Berntsen GK. Seasonal variation of estradiol, follicle stimulating hormone, and dehydroepiandrosterone sulfate in women and men. The Journal of Clinical Endocrinology & Metabolism. 2006;91(10):3798–802. doi: 10.1210/jc.2006-0866. [DOI] [PubMed] [Google Scholar]
- 27.Xiang J, Nagaya T, Huang X-E, Kuriki K, Imaeda N, Tokudome Y, et al. Sex and seasonal variations of plasma retinol, alpha-tocopherol, and carotenoid concentrations in Japanese dietitians. Asian Pac J Cancer Prev. 2008;9(3):413–6. [PubMed] [Google Scholar]
- 28.Khan MA, Khan MD. Classification of 154 clinical cases of vitamin A deficiency in children (0–15 years) in a tertiary hospital in North West Frontier Province Pakistan. J Pak Med Assoc. 2005;55(2):77–8. [PubMed] [Google Scholar]
- 29.McKinley MC, Strain JJ, McPartlin J, Scott JM, McNulty H. Plasma Homocysteine Is Not Subject to Seasonal Variation. Clinical Chemistry. 2001;47(8):1430–6. [PubMed] [Google Scholar]
- 30.Palva I, Salokannel S. Seasonal variation in megaloblastic anaemia. British Journal of Nutrition. 1972;27(03):593–5. doi: 10.1079/bjn19720129. [DOI] [PubMed] [Google Scholar]
- 31.Paalanen L, Prattala R, Alfthan G, Salminen I, Laatikainen T. Seasonal variation in plasma vitamin C concentration in Pitkaranta, Northwestern Russia. Eur J Clin Nutr. 2013;67(10):1115. doi: 10.1038/ejcn.2012.185. [DOI] [PubMed] [Google Scholar]
- 32.Hallmann E, Lipowski J, Marszałek K, Rembiałkowska E. The Seasonal Variation in Bioactive Compounds Content in Juice from Organic and Non-organic Tomatoes. Plant Foods Hum Nutr. 2013;68(2):171–6. doi: 10.1007/s11130-013-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas AS. Seasonality of Hip Fracture and Haemorrhagic Disease of the Newborn. Scottish Medical Journal. 1993;38(2):37–40. doi: 10.1177/003693309303800202. [DOI] [PubMed] [Google Scholar]
- 34.Anai T, Matsu T, Oga M, Yoshimatsu J, Miyakawa I. Seasonal incidence of subclinical vitamin K deficiency during early newborn period. Nihon Sanka Fujinka Gakkai zasshi. 1991;43(3):342–6. [PubMed] [Google Scholar]
- 35.Gelardi M, Peroni DG, Incorvaia C, Quaranta N, De Luca C, Barberi S, et al. Seasonal changes in nasal cytology in mite-allergic patients. Journal of inflammation research. 2014;7:39. doi: 10.2147/JIR.S54581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klink M, Bednarska K, Blus E, Kielbik M, Sulowska Z. Seasonal changes in activities of human neutrophils in vitro. Inflammation Research. 2012;61(1):11–6. doi: 10.1007/s00011-011-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henriksen JM. Exercise-induced bronchoconstriction. Seasonal variation in children with asthma and in those with rhinitis. Allergy. 1986;41(7):499–506. doi: 10.1111/j.1398-9995.1986.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu CM. Seasonal variation of nasal surface basophilic cells and eosinophils in Japanese cedar pollinosis. Rhinology. 1988;26(3):167–73. [PubMed] [Google Scholar]
- 39.Lee CJ, Lawler GS, Panemangalore M. Nutritional status of middle-aged and elderly females in Kentucky in two seasons: Part 2. Hematological parameters. Journal of the American College of Nutrition. 1987;6(3):217–22. doi: 10.1080/07315724.1987.10720184. [DOI] [PubMed] [Google Scholar]
- 40.Parks JH, Barsky R, Coe FL. Gender differences in seasonal variation of urine stone risk factors. The Journal of urology. 2003;170(2):384–8. doi: 10.1097/01.ju.0000071721.91229.27. [DOI] [PubMed] [Google Scholar]
- 41.Percy ME, Andrews DF, Thompson MW. Serum creatine kinase in the detection of Duchenne muscular dystrophy carriers: effects of season and multiple testing. Muscle & nerve. 1982;5(1):58–64. doi: 10.1002/mus.880050111. [DOI] [PubMed] [Google Scholar]
- 42.Svedenhag J, Sjödin B. Physiological characteristics of elite male runners in and off-season. Canadian journal of applied sport sciences Journal canadien des sciences appliquees au sport. 1985;10(3):127–33. [PubMed] [Google Scholar]
- 43.Boland MR, Hripcsak G, Shen Y, Chung WK, Weng C. Defining a comprehensive verotype using electronic health records for personalized medicine. Journal of the American Medical Informatics Association. 2013;20(e2):e232–e8. doi: 10.1136/amiajnl-2013-001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei S, Wang L-E, McHugh MK, Han Y, Xiong M, Amos CI, et al. Genome-wide gene-environment interaction analysis for asbestos exposure in lung cancer susceptibility. Carcinogenesis. 2012;33(8):1531–7. doi: 10.1093/carcin/bgs188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on Type 2 Diabetes Mellitus. PLoS ONE. 2010;5(5):e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busse W, Sedgwick J. Eosinophils in asthma. Annals of allergy. 1992;68(3):286–90. [PubMed] [Google Scholar]
- 47.Chen M-H, Su T-P, Chen Y-S, Hsu J-W, Huang K-L, Chang W-H, et al. Asthma and attention-deficit/hyperactivity disorder: a nationwide population-based prospective cohort study. Journal of Child Psychology and Psychiatry. 2013;54(11):1208–14. doi: 10.1111/jcpp.12087. [DOI] [PubMed] [Google Scholar]
- 48.Secnik K, Swensen A, Lage M. Comorbidities and costs of adult patients diagnosed with attention-deficit hyperactivity disorder. Pharmacoeconomics. 2005;23(1):93–102. doi: 10.2165/00019053-200523010-00008. [DOI] [PubMed] [Google Scholar]
- 49.Roseboom TJ, van der Meulen JHP, Ravelli ACJ, Osmond C, Barker DJP, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Molecular and Cellular Endocrinology. 2001;185(1–2):93–8. doi: 10.1016/s0303-7207(01)00721-3. [DOI] [PubMed] [Google Scholar]
- 50.Lee BE, Ha EH, Park HS, Kim YJ, Hong YC, Kim H, et al. Exposure to air pollution during different gestational phases contributes to risks of low birth weight. Human Reproduction. 2003;18(3):638–43. doi: 10.1093/humrep/deg102. [DOI] [PubMed] [Google Scholar]
- 51.Nelissen ECM, van Montfoort APA, Dumoulin JCM, Evers JLH. Epigenetics and the placenta. Human Reproduction Update. 2011;17(3):397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]