Abstract
Emerging issues of team-based care, precision medicine, and big data science underscore the need for health information technology (HIT) tools for integrating complex data in consistent ways to achieve the triple aims of improving patient outcomes, patient experience, and cost reductions. The purpose of this study was to demonstrate the feasibility of creating a hierarchical flowsheet ontology in i2b2 using data-derived information models and determine the underlying informatics and technical issues. This study is the first of its kind to use information models that aggregate team-based care across time, disciplines, and settings into 14 information models that were integrated into i2b2 in a hierarchical model. In the process of successfully creating a hierarchical ontology for flowsheet data in i2b2, we uncovered a variety of informatics and technical issues described in this paper.
Introduction & Background
Emerging issues of team-based care, precision medicine, and big data science underscore the need for health information technology (HIT) tools for integrating complex data in consistent ways to achieve the triple aims of improving patient outcomes, patient experience, and cost reductions. Team-based care includes two or more disciplines providing patient-centered care in any setting; it focuses on the integration of interventions from various disciplines targeted to meet patient needs and requires data that is usable across providers.1 The former Director for the National Institutes of Health (NIH), Francis Collins, states that a long-range goal of precision medicine is to use a variety of biomedical data to detect, prevent, and treat diseases by targeting treatments based on individual characteristics and circumstances.2 The initial work of Clinical, Translational, and Science Awards (CTSA) built the foundation for speeding up the translation of basic science into clinical practice and had a focus of an information infrastructure to support research. Additional NIH funding extended that focus through the development of the computational infrastructure for biomedical computing to extract, share, and analyze data from electronic health records (EHRs) and other data sources. One such tool is i2b2 (Informatics for Integrating Biology and the Bedside), which is a scalable informatics framework for integrating clinical and other health data to support big data science. Typically, patient data include encounters, demographics, diagnoses, procedures, medications, vital signs, and laboratory results. There is a need to expand the data typically represented in i2b2 with additional team-based patient assessments and interventions. These data are needed to conduct big data science that effectively targets the combinations of interventions based on individual patient characteristics and circumstances.6,7 Flowsheet data from EHRs includes time-based structured and semi-structured data is essential to represent team-based care and if included in i2b2 can support research that can be highly granular with longitudinal tracking of patient clinical states and status for achieving and measuring patient outcomes.
Unlike medical diagnoses, medications, or laboratory data, standard ontologies for flowsheet data do not yet exist. In this paper, the term “flowsheet” describes a structured method within an EHR for organizing, capturing, and displaying patient status measures over multiple points in time. Flowsheets include a variety of concepts representing patient assessments, problems, goals, interventions, and outcomes by date and time. Flowsheets are used by a variety of disciplines (nurses, therapists, dieticians, chaplains, some physicians, and others), primarily in acute care, but also in other settings (emergency department, outpatient clinics, transitional care units, and rehabilitation centers). In the University of Minnesota’s clinical data repository (CDR), flowsheets represent 34% of the entire data stored.5 Examples of flowsheet content include review of systems, response to care, psychosocial and caregiver data, and data for quality metrics such as tobacco use. Flowsheet data are useful for research, such as symptom assessments and management, patient teaching, or the effect of discharge planning and 30-day readmissions. Because of the variety and complexity of flowsheet data, unique challenges exist for incorporating this data into i2b2.
Flowsheet data challenges
Numerous challenges exist for organizing flowsheet data to track highly granular longitudinal patient clinical states, interprofessional interventions, and patient outcome status for patients. The sheer volume of the data slows the extract, transfer, and load (ETL) process. In addition to the volume of data, there is considerable redundancy and customization within and across health systems, resulting in many unique identifiers (IDs) and value sets for the same observation.4 For instance,, the same observation for pain rating 0–10 can have different formats including numeric, free text, or a choice list, resulting in duplicate flowsheet observations with unique IDs. The replication of similar flowsheet measures occurs for a variety of reason: 1) different staffs build the flowsheets, often creating new flowsheet measures (observation) and value sets, 2) software upgrades result in deprecated measures without a robust way to track changes over time and, 3) different clinical disciplines customize wording for a question or value set for the answer. As a result, one or many similar observations (such as pain rating 0–10) need to be linked to a single concept in i2b2 for researchers to have useful information for populations of interest over time that may receive care from different disciplines in multiple settings. Another challenge is that numeric values differ across observations in the units they represent, such as weight in pounds or kilograms or urine catheter balloon size (i.e. 10 mL). Abstract concept-based information models are needed to normalize flowsheet data and subsequently map the data to standardized terminologies.
Research for creating a useful and generalizable ontology from flowsheet data for representation in i2b2 is just beginning. Waitman et al. 4 used data mining pruning and clustering analyses to reduce flowsheet data into templates (screen views), groups of similar questions displayed within screens, and individual flowsheet measures with their value sets. If a measure or choice in a value set was not used at least monthly, it was removed from the data set. A similarity score was calculated to merge groups of flowsheet observations followed by calculating a similarity score to merge templates that incorporated similar groups. This automated method of reducing similar flowsheet data resulted in a streamlined ontology. Each implementation of an EHR is unique and the generalizability of this method to other data warehouses is unknown. Warren et al. 3 used a focus group approach to identify an ontology for flowsheet data. This approach provided a logical conceptual method of organizing flowsheet data at a high level. While this is potentially a generalizable model for flowsheet data, it was not actually put into use for organizing flowsheets in i2b2. In a subsequent study, Harris et al. 8 used a data-derived method was used for creating an information model for pressure ulcers and extending data standards. The result was a model that approved by HL7 and is in testing as an e-Measure. However, no attempt was reported for issues encountered in implementing the model in i2b2 or other tools for research. Finally, a similar data-driven process for creating an information model was developed by the investigators of the current study for pressure ulcers.5 Subsequently an additional 13 information models were created and mapped to flowsheet data in a pilot study of 199, 665 patient encounters from one CTSA’s CDR.9
I2b2 architecture
CTSAs frequently use i2b2 for cohort discovery and or data delivery; it was developed at Harvard as1 of 7 tools to build a computational infrastructure for national biomedical computing.10 i2b2 is designed as an open source, web-based tool with an informatics framework for integrating EHR and other patient data within and across health care enterprises. The i2b2 tool can include additional data such as tumor registry data or other data sets to support research and quality improvement activities. The core modules include data and file repositories, identity and ontology management, workflow and project management, and natural language processing. Additional plug-ins exist to extend i2b2 functionality. The tool can help to achieve a number of research objectives; cohort discovery is one such objective. Researchers can explore and request data via a simple interface in i2b2 to determine numbers of patients meeting their research criteria to support research proposals. When configured with a common data model, i2b2 supports aggregating data requests across academic health centers and health care systems through Shrine11.
In i2b2, data are considered observations, which are facts about the patient organized in an OBSERVATION_FACT table along with patients, providers and visits organized in other tables as a star schema in the data repository cell. The observations are a collection of entity-attribute-value triples that are linked to the other components of the schema. Relationships between concepts in i2b2 are hierarchical and therefore limit the relationships in an ontology to concepts and their associated sub concepts in a strict hierarchy. The only way for a sub concept to be associated with more than one concept is to repeat that sub concept within the hierarchy under that concept. This organization is straightforward when considering diagnoses, procedures, laboratory tests, and drugs. However, it becomes more challenging to represent additional information about a concept such as the dose and dosage form of a drug. The modifier mechanism exists to support this additional information but adds complexity to the representation and retrieval schemes since modifiers are inherited by sub concepts. In particular, the representation of flowsheet measures that have one of several text phrases in a choice list as a value, becomes problematic—each value in a choice list requires a different identifier in order to be used as a unique observation type in i2b2.
A separate i2b2 ontology management cell is used to represent and navigate the ontologies that are used to organize the observations into logical collections and categories for cohort identification. The Unified Medical Language System (UMLS) provides organizations of various types of observations such as diagnoses (ICD9, ICD10), procedures (CPT, ICD9) and drugs (RxNORM). These organizations are used to construct simple hierarchical ontology structures for identifying observations of interest. Each leaf node in such a hierarchy is associated with a concept code that identifies one specific type of observation (e.g. a diagnosis or laboratory test) that is part of the observation table. The purpose of this study was to demonstrate the feasibility of creating a hierarchical flowsheet ontology in i2b2 using data-derived information models and determine the underlying informatics and technical issues.
Method
The specific aims of this study were to 1) demonstrate the feasibility of creating a hierarchical ontology of flowsheet measures using information models, 2) identify informatics issues encountered in normalizing loading and displaying these information models and their associated observations in i2b2, and 3) describe the technical issues encountered.
Data was obtained from one CTSA funded CDR after approval by the Institutional Review Board. The entire CDR includes EHR data from one health system composed of six hospitals and 45 clinics in a Midwest state. More than 2 million patients and 4 billion rows of unique data are included were from patient encounters (inpatient and outpatient), demographics, medical diagnoses, procedures, laboratory results, medications, notes, and flowsheet data. A random subset of data for this study was extracted from the CDR and was composed of 66,660 patients with 199,665 encounters of all types documented between October 20, 2010 and December 26, 2013. The flowsheet data represented 562 templates (screen views), with 2,693 groups of observations, 14,450 unique flowsheet measures (observations), and 153,049, 704 data points or observations. Excluded were unique flowsheet data for pediatrics and obstetrics, due to the knowledge by researchers in these specialties. Additionally specialized assessments or data collected for research purposes also were excluded, such as the cardiac catheter lab.
In previous work, an interprofessional research team composed of health system, community, and academic partners collaborated to derive 14 information models used in this study. The research team included domain experts in the EHR system build and deployment, quality measures, acute care, behavioral health, computer science, and informatics. The 14 data-derived information models include behavioral health (abuse, psychiatric mental status, suicide-harm, violence); cardiovascular; falls; gastrointestinal; genitourinary; neuromusculoskeletal; pain; peripheral neurovascular (with VTE); pressure ulcers; respiratory; and, vital signs. Priority topics include high-risk clinical quality measures, physical review of systems, and behavioral health research priorities. Results of this study phase are reported elsewhere.5,9
An iterative inductive manual process was used to map flowsheet data across time, disciplines, and settings within the database to concepts relevant to researchers. The approach was both a top down and bottom up iterative process to name concepts and map flowsheet measures to those concepts. Data were extracted from the CDR, retaining the relationship of the template (screen views), group (of observations), and flowsheet measures (observations) along with their data type and value sets. Concepts related to an information model were derived from the data, research, and evidence-based guidelines. When a concepts was identified in the data, the groups and templates were searched for related concepts, such as type, indicators, onset, and duration related to pain as shown in Figure 1. Validation of information models included: one team member developing the model, a second team member evaluating the completeness of the model, and the entire research team reviewing the model for logical consistency within and across models. The reason for a manual process was to derive rules that subsequently could be incorporated into a semi-automated tool for replication of models across EHRs. Information models were constructed in Microsoft Excel using term-based search methods to identify concept names in templates, groups, and measures related to the information models of interest. We used an iterative approach of review by multiple team members, identification of additional terms, and subsequent searches. Each information model was composed of a hierarchy of increasingly granular terms. Level 1 represents the major concept i.e. pain, Level 2 addresses a term for care planning (assessment, problem, goal, intervention, or outcomes), and Level 3 up to Level 5 contains a concept related to pain such as type of pain; pain indicators, locations, or orientation; with finer levels of granularity as shown in Figure 1.
Figure 1.
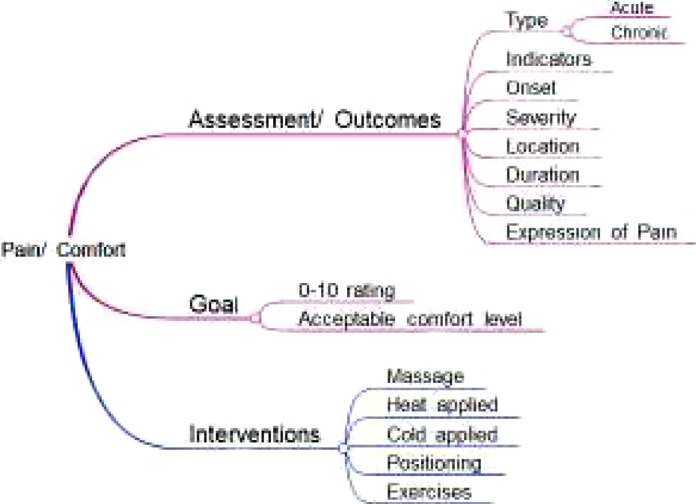
Example CDM for pain
Loading the cdms into i2b2
As the first step in the ETL process of loading EHR flowsheet data into i2b2, we created a master look-up table. For measures whose values were numeric, the lookup table entry consisted of the flowsheet measure identifier from the EHR. When the choice list was associated with a measure, we constructed separate measure-choice list combinations that each had a unique identifier based upon the EHR measure identifier, followed by a number for each choice list i.e. 12345-1, 12345-2, etc. Each of the resulting identifiers represents a separate i2b2 concept ending in a leaf node. Each information model was used to construct a hierarchical ontology of terms that could be used to identify specific flowsheet measures in i2b2.
Results
The first aim of the study was to demonstrate the feasibility of loading and displaying the 14 information models in a hierarchical ontology in i2b2. The ETL process that imported the flowsheet data from the sample of 199,665 encounters related to the 14 information models resulted in approximately 81 million new rows in the i2b2 OBSERVATION_FACT table. The 14 information models included 1,865 unique flowsheet measures. When measures that had associated choice lists were combined with each answer, the result was 10,484 unique concept identifiers in i2b2. Figure 2 portrays a portion of the flowsheet hierarchy. Each level in i2b2 provides a count of patients associated with the concept at that level. The name of the information model is displayed as Level 1 (e.g. Genitourinary [GU]). Level 2 is assessment/ intervention. Level 3 and subsequent Levels are concepts specific to GU i.e. incontinence, symptoms, etc. At the lowest level of the hierarchy or leaf node, the name includes the local flowsheet measure code in parentheses.
Figure 2.
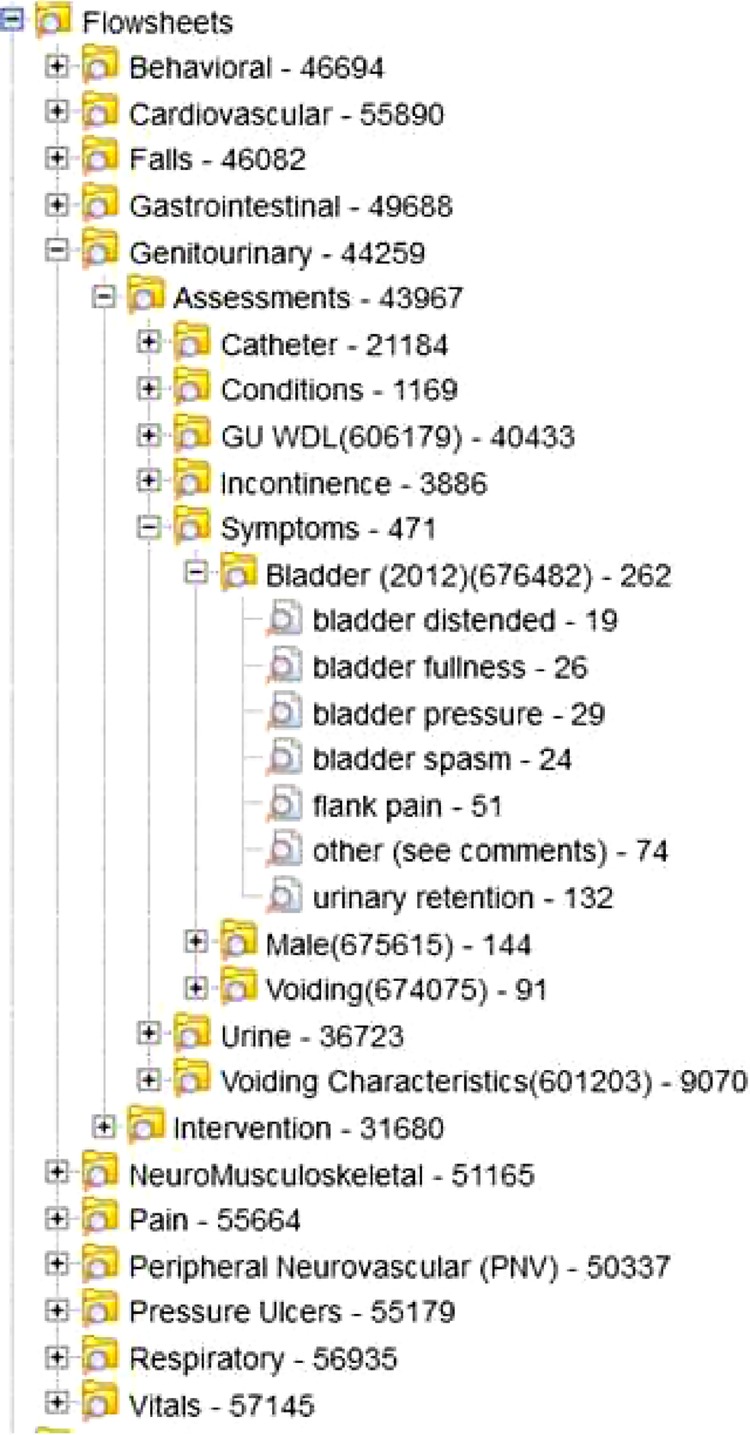
i2b2 flowsheet ontology for 14 CDMs
Aim 2 of the study was to describe the informatics issues encountered. The overarching issue was the challenge inherent in converting data from a structure optimized for data capture and visualization, to a structure optimized for research representation and retrieval. Redundancy of answers in a value set, and redundancy of flowsheets for the same concept occurred. For instance, clinicians documented “within defined limits” (WDL), when patients meet particular parameters viewable on the screen. They also can choose WDL except and then document the exception. However, there were variations in the value set for the GU WDL measure that included: Ex, Ex., No New, WDL, WDL (No new row needed), ex, no. Another issue observed was a number of instances where different flowsheet measures appear to be representing the same concepts. For example, there were 7 measures of blood pressure and 10 different measures for heart rate. Often these measures are associated with different specialty areas or settings – inpatient, outpatient, ED, ICU, respiratory therapy, etc. There also were a number of instances of measures with similar names representing different concepts as evidenced by the “answers” for the measures. An additional informatics challenge addresses flowsheet measures that represent more than once concept within a single value set. For instance, a behavioral health assessment value set of grooming-hygiene includes both the type of grooming (bath, handwashing, or shower) and the amount of assistance required (independent, prompts, with supervision, with assistance, or total care) in the choice list. Finally, the concepts represented by flowsheet measures had to be transformed to a hierarchical model of class and subclass, typically interpreted as an “is-a” relationship due to the limited relationship types in i2b2.
Aim 3 of the study was to describe technical issues encountered. A number of flowsheet measures, whose values, could not easily be represented as searchable i2b2 facts was a significant issue. This required transformation or processing of data versus direct copying into i2b2. These included flowsheet measures with free text responses. Similarly, a number of measures allowed for multiple selections from a choice list. We created a general solution (algorithm) for converting a measure with multiple associated clinical findings into multiple observations. The goal was to make these searchable, answers were separated into their constituent elements and each made a separate row in the OBSERVATION_FACT table. This work was further hindered when the EHR allowed free-text responses to flowsheet measures even when a choice list was available via a comment or “other” option. This was handled by considering such response to be an additional alternative within the choice list and assigning it a separate concept identifier. How these are to be treated was left up to the individual doing the searching related to the concept.
Flowsheet measures with associated numeric values also proved somewhat problematic as the units of measure were not directly associated with the flowsheet measure value. It was observed that often the units of measure would be part of the descriptive name (e.g. fluid volume (ml) but this was not always the case. From a clinical point of view, this may not be an issue as the units for a particular measure may be well known and out of range values easy detected, but this could prove troublesome in formulating searches for patients with a measure value in a particular range.
The i2b2 ontology design presented some unanticipated issues. For example, there was an undocumented restriction in the allowable character set with the following characters not allowed in naming concepts – “* | / \ “ < >? %”. A number of measures used these characters and transformation was required prior to uploading into i2b2. For instance, GU assessment included names such as Signs/Symptoms (Male), GU Conditions/Symptoms, or Treatment/Device/Implant. In addition, i2b2 requires that any names at a given level of the hierarchy be unique within the first 32 characters from every other name within that same level of the same hierarchy. This caused initial issues in assigning names at the lowest levels of the hierarchy. We changed the process from fully specifying the concept name, which may exceed 32 characters, to splitting the concept into two or more levels when measures were named the same. An example in the Neuromusculoskeletal information model is the concept Bilateral Lower Extremities Muscle Tone Assessment. This concept was transformed into: Level 1: Neuromusculoskeletal, Level 2: Assessment, Level 3: Muscle tone location, Level 4: Extremities, and Level 5: Bilateral. This level of granularity was necessary due to the numerous flowsheet measures assessments of muscle tone location of extremities (left, right, upper, lower, bilateral or a combination of these).
Discussion
The purpose of this study was to demonstrate the feasibility of creating a hierarchical flowsheet ontology in i2b2 using data-derived information models and determine the underlying informatics and technical issues. I2b2 is a tool frequently used tool by CTSAs and other researchers for cohort discovery and data extraction. Our study demonstrated the feasibility of creating such a hierarchical flowsheet ontology in i2b2 that serves the needs of researchers seeking to use flowsheet data for cohort identification. The 14 information models used only 1,865 flowsheet measures to support the 14 information models, leaving an additional 87.1% of flowsheet measures unmapped to concepts in i2b2. When combined with the value sets, the result was 10,484 unique concepts. There were an additional 72.5 million facts not included in these models. Additional models are needed such as functional status, specialty assessments (therapists, obstetrics, or perioperative) and additional behavioral health. Flowsheet data represent a rich source for a researcher that is largely untouched in reported studies with secondary data from CDRs.
The process of creating information models for incorporating flowsheet data in i2b2 is novel. The goal of our information models was to create conceptual models useful for researchers, extracting data by a clinical topic that includes all related flowsheet measures regardless of time, setting, and discipline. Consider the following research question: What are the patient symptoms and interventions documented in flowsheets combined with other EHR data related to symptom management for osteosarcoma patients undergoing radiation treatment, surgery, and chemotherapy? Treatment occurs over a period of time and potentially in a variety of settings i.e. outpatient, emergency department, surgery, ICU, and medical-surgical units. The population of interest is patients of any age newly diagnosed between 10/1/10 – 12/31/13. During that time, nurses, therapists, social workers, and the dieticians document symptom management in flowsheets. Major symptoms include nausea, vomiting, fatigue, and depression. The value of using the data-derived information models is that all relevant flowsheet observations are mapped to concepts regardless of the variations in time, setting, or discipline. The 14 models represent characteristics of patients and thus are relevant to a range of clinical researchers.
Generalizability of results of our study has advantages not demonstrated in previous research. Waitman et al. 3 used a data mining approach for removing redundancy. The advantage of their work is the semi-automation of pruning and clustering flowsheet data. In comparison, our effort to manually create the information models and transform the data for ETL in i2b2 was considerable and not sustainable for additional topics of interest. The result from Waitman’s study was a reduction in the number of templates, groups, and measures that might be most useful to researchers. In addition, they implemented their method in i2b2 to make the data transparent to researchers. However, the uniqueness of the naming of templates, groups, and flowsheet measures limits the generalizability of their results to other health settings and EHR vendors. In comparison, the hierarchical ontology with an initial set of information models designed and imported into i2b2 for this study can be generalized across health systems and software vendors. The method of mapping flowsheets has been demonstrated (Westra et al.) to support comparative effectiveness research; however, the actual effort implementing such models will be unique to each setting.
Harris et al. 8 demonstrated a robust data-driven information model involving six clinical sites. They specified the concepts, definitions, and terminologies for observations and value sets related to pressure ulcers. Their study demonstrates the full range of work needed for generalizability, but results were limited to a single information model. Subsequent work is needed for our study to formalize definitions and code concepts with terminologies for reliable use of the models by others. Similar to Harris et al., the assessments need to be mapped to LOINC and problems, observations, and value sets for assessments mapped to SNOMED CT. Furthermore, our work needs validation across health settings and software vendors.
There were a number of challenges and shortcomings in i2b2 for representing the 14 information models. It is a straightforward and simple to use tool for identifying cohorts of patients who possess a particular set of characteristics. To make this feasible with large data sets and diverse types of data it imposes a set of restrictions on how that data is organized that facilitates rapid search of large datasets. Those restrictions make it quite difficult and we suspect more computationally expensive to incorporate flowsheet data. Characterizing that expense will be the subject of future study. In addition, the strict hierarchical structure and limited data representations allowed by i2b2 create considerable difficulties in fully characterizing the organization of flowsheet measures and their inter-relationships. For example, it does not allow data items with text values that can be tested for a value as it can do for laboratory tests. While workarounds are possible, they are far from ideal in creating parsimonious data representations and ontologies.
Creation of data-derived information models could be enhanced by a tighter integration of academic informaticians with clinical informaticians, information technology analysts, and EHR builders. Our research team includes members of both the health system and the academic setting. Results are invaluable for demonstrating the downstream challenges of a big bang implementation, continued allowance of a non-standard build, and lack of a dedicated data architect role to assure data continuity between versions of the EHR and between clinical disciplines. The results were shared with the health system leadership to influence standardization of flowsheet build across settings and disciplines, plan for optimization phases, and evaluate upgrades that include flowsheet measure and value set changes. Standardizing build in the current system can positively affect documentation efficiencies for clinical staff, improve reporting of data both within and outside of the organization, and streamline research queries. Each of these improvements can positively affect our patients through improved real time use of data.
The results of our study are also proving useful for research. There are studies in process that now include flowsheet data mapped to our data-derived information models. One of the investigators (JIP) is conducting a data mining study to discover factors that might provide new insights into catheter-associated urinary tract infection (CAUTI). She is including flowsheet measures from the GU ontology in order to include all possible measures that may have influence on CAUTI occurrence. Another investigator is using the vital sign information model to understand factors during anesthesia that predict unanticipated ICU admission. In the current i2b2 implementation, vital signs are mapped to a single, most frequently used flowsheet ID for each vital sign. However, prior to and during anesthesia, vital signs might be linked to devices such as an O2 Saturation monitor or an arterial line. If a single measure of heart rate, blood pressure, or respiration were used, critical information might be missing to address the research question of interest.
There are a number of limitations to our study. The 14 information models were derived from a subset of the data and not the all flowsheets in the CDR. In addition, only the simplest forms of validation (review by a peer expert) were undertaken to validate these models. Further work is needed to assure that the information models include all relevant flowsheets and are valid and complete representations of the associated topics. One limitation is the manual process and use of Excel spreadsheets. Prototype software is under development to semi-automate mapping of flowsheet data to information models, but further work is needed.
Conclusion
The purpose of this study was to demonstrate the feasibility of creating a hierarchical flowsheet ontology in i2b2 using data-derived information models and determine the underlying informatics and technical issues. This study is the first of its kind to use information models that aggregate team-based care across time, disciplines, and settings into 14 information models that were integrated into i2b2 in a hierarchical model. In the process of successfully creating a hierarchical ontology for flowsheet data in i2b2, we uncovered a variety of informatics and technical issues described in this paper.
References
- 1.Mitchell P, Wynia M, Golden R, et al. Core principles & values of effective team-based health care. Washington, DC: Discussion Paper, Institute of Medicine; 2012. [Google Scholar]
- 2.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren JJ, Manos EL, Connolly DW, Waitman LR. Ambient findability: Developing a flowsheet ontology for i2B2. Nurs Inform. 2012;2012:432. [PMC free article] [PubMed] [Google Scholar]
- 4.Waitman LR, Warren JJ, Manos EL, Connolly DW. Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data repository to support research and quality improvement; AMIA Annu Symp Proc.; 2011. pp. 1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SG, Byrne MD, Christie B, et al. Modeling flowsheet data for clinical research. AMIA Jt Summits Transl Sci Proc. 2015;2015:77–81. [PMC free article] [PubMed] [Google Scholar]
- 6.Pechacek J, Shanedling J, Lutfiyya MN, Brandt B, Cerra F, Delaney C. The national center data repository: core essential interprofessional practice & education data enabling triple aim analytics. JIC. 2015;(Special Issue) doi: 10.3109/13561820.2015.1075474. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pechacek J, Cerra F, Brandt B, Lutfiyya MN, Delaney C. Creating the evidence through comparative effectiveness research for interprofessional education and collaborative practice by deploying a national intervention network and a national data repository. Healthcare. 2015;3(1):146–161. doi: 10.3390/healthcare3010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris MR, Langford LH, Miller H, Hook M, Dykes PC, Matney SA. Harmonizing and extending standards from a domain-specific and bottom-up approach: An example from development through use in clinical applications. J Am Med Inform Assoc. 2015;22(3):545–552. doi: 10.1093/jamia/ocu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra BL, Christie B, Johnson SG, et al. Modeling flowsheet data for quality improvement and research. Applied Clinical Informatics. In Process. [Google Scholar]
- 10.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber GM, Murphy SN, McMurry AJ, et al. The shared health research information network (SHRINE): A prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16(5):624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]


