Abstract
A fundamental premise of scientific research is that it should be reproducible. However, the specific requirements for reproducibility of research using electronic health record (EHR) data have not been sufficiently articulated. There is no guidance for researchers about how to assess a given project and identify provisions for reproducibility. We analyze three different clinical research initiatives that use EHR data in order to define a set of requirements to reproduce the research using the original or other datasets. We identify specific project features that drive these requirements. The resulting framework will support the much-needed discussion of strategies to ensure the reproducibility of research that uses data from EHRs.
Introduction
The widespread adoption of electronic health record (EHR) systems has created many opportunities to “re-use” the data collected as part of routine care to support various research needs. The re-use of data from EHRs has the potential to reduce the costs and time for research by decreasing the need for research-specific data collection. In addition, EHR data can be used to support observational comparative effectiveness research in real practice settings, and to explore the impact of interventions on populations that are more broadly defined than inclusion criteria for traditional clinical trials. A growing number of research studies and research networks are based upon the use of EHR data, creating a need to understand the structure and processes needed to assure or otherwise support the outcome of reproducibility.
Reproducibility has been variously defined as, “research where the study protocol, the electronic data set used for the paper, and the analysis code are all publicly available,”1 as ability to replicate a study,2–4 and as “the same materials manipulated with the same protocol produce the same result”.5 Still others answer the call by representing experimental protocols into machine-readable code.6 For the purposes of this discussion, and in keeping with an Institute of Medicine definition derived from data quality, a study is reproducible when a second study arrives at the same conclusion,7 i.e., same direction and similar magnitude. Multiple types of replication are described in the literature. The independence varies across the varieties of replication: 1) reanalysis of same data using the same or different data handling, programming, and statistics, by the same or different people, 2) replication of an entire experiment by same group, and 3) replication of an experiment by a different group using the same or different but valid design and methods.
The fundamental need to support and demonstrate the reproducibility of research is increasingly being called into attention. In 2015 alone, multiple journal retractions cited lack of ability to reproduce the research. Much attention has been given to reproducibility in pre-clinical research.8–10 Multifaceted solutions have been proposed10,11 with some facets implemented in some areas. By well-recognized accounts, reproducibility problems also exist in observational studies in high percentages.1,8, 12
While in the past, the quality of health record data has been called into question,13,14 quality was rarely assessed in studies or activities involving the re-use of health record data.15 A recent review has shown that the quality of health record data is still broadly lacking.16 Most recently researchers have called for publication of data quality assessment results with reports of research results.17 In addition, National Institutes of Health (NIH) solicitations for research expected to leverage health record data have required applicants to describe plans to assess data validity.18,19
While some of the factors described for pre-clinical research such as publication bias, publication pressure, incentives, and investigator training certainly apply to studies based on EHR data, additional factors specific to EHR-based studies complicate matters. Studies based on EHR data are uses of data for purposes other than those for which they were originally collected. As such, we expand the list articulated by Young and Miller to include the following threats to reproducibility for studies based on EHR data. As articulated in Tcheng, Nahm and Fendt (2010) the meaning of the data in the health record may not be an exact match with that desired for the study, or for data from different facilities.20 This has been called representational inadequacy. There can be significant variability in meaning, accuracy and availability of data between facilities, units in facilities and individuals providing clinical documentation.21 Information loss and degradation is often associated with mapping EHR data to a specific study or data model.22–24 Small changes in algorithmic cohort and data extraction definitions can have a substantial impact on membership in the result-set.25 The acceptability of data quality is dependent on how the data are used in a particular study. Thus, data quality assessment for EHR-based studies must be performed for each study.17 In addition to mapping, EHR-based studies usually require some manipulation of raw data to prepare them for analysis.21 These data processing operations can impact analysis results.3 Solutions in the form of data quality assessment recommendations26 and recommendations for reporting data quality assessment results27 have been proposed for EHR-based studies but to date, a framework for the broader issue of assuring reproducibility in EHR-based studies has not been articulated.
In our work supporting and operationalizing large EHR-based research initiatives including observational studies and controlled experiments we have witnessed increased expectations regarding the level of rigor in areas such as such as data management planning, data quality assessment, data sharing and research reproducibility. Requests for research data management and data sharing plans are becoming the norm for federally funded research. In our work supporting multiple initiatives using EHR data for clinical studies, it has become necessary to identify and articulate requirements for supporting reproducibility of EHR-based studies. These needs prompted efforts to bring the identified structure and process to fruition.
Methods and Materials
We use our experience from three large EHR-based research initiatives (cases 1-3) to outline the data-related requirements for research reproducibility, and distill the features of research projects that drive these requirements Case 1: the Health Care Systems Research Collaboratory (the Collaboratory). Functioning as the coordinating center for the Collaboratory,29 we have the privilege of working along side ten large pragmatic randomized clinical trials as they are implemented within delivery of routine care in health systems and using healthcare data. Case 2: collection of health record data on the MURDOCK Community Registry and Biorepository (the Registry). Described elsewhere,30,31 the Registry is an observational longitudinal cohort study that collects self reported and health record data on thirty-four medical conditions, eight procedures, hospitalizations and medications as well as biological samples. We have experienced and learned a tremendous amount acquiring and processing the health record data from 20 local facilities for the Registry. Case 3: (the Warehouse) includes development and maintenance of our institution’s clinical data warehouse and provision of warehouse data to support clinical studies.32–34 A clinical data warehouse is an important and active component of most large medical centers today and while our implementation and tools may differ, we believe that the requirements for the clinical data warehouse use case are generalizable to most healthcare organizations. The objective for our examination of these cases was to 1) identify what research reproducibility means in the context of secondary use of EHR data, i.e., the requirements for research reproducibility, and 2) identify the aspects of secondary data use projects that drive the requirements.
Young and Karr (2011) have articulated some high-level documentation needed to replicate a study, the study protocol, the electronic data, and the analysis code. These of course applied to all three of the case studies and we did not explore these categories further. Rather, we analyzed areas where the case studies differed in ways that data move, grow and change: 1) whether data change after the researcher receives them, 2) whether and in what ways the data grow throughout the study, and 3) whether and in what ways the data move throughout the study. For example, in a retrospective study using health record data, data are extracted from a clinical data warehouse or other clinical information system. Changes after the researcher receives the data are most often ignored in that no updates or additional data extracts are provided. These data do not grow or shrink in any way, and after the initial receipt are usually not sent anywhere else, except a likely archival step. The researcher will usually need to standardize or otherwise transform the data. In summary, data did not move significantly, data did not change, but data were transformed. Alternately, in a longitudinal study using health record data such as the Registry, the data change and are expected to grow in terms of newly occurring observations and potentially through addition of new data elements over time (or receipt of some data may be terminated). An investigator may also have a need to standardize or otherwise transform the data. These aspects, especially in the case of the longitudinal study require additional infrastructure to support the traceability aspects of reproducibility. We mapped these cases to these fundamental ways in which data grow, change, and move in space and time to identify necessary infrastructure needed to support the traceability aspect of reproducibility.
Results
While not dynamic, reproducibility in all three cases, in particular replication of the experiment by collecting data from different subjects, would require very specific definition for each data element. We noted this because it adds to those items listed by Young and Miller. Access to knowledge about the data is just as important as access to the data because how and where the data originate can impact their availability and meaning. For example, there will be differences in data availability of one facility performs a procedure in-house versus referring out to an independent facility. Likewise if data are provided from a data warehouse, they may have already undergone processing and such processing can impact the meaning of the data. For example combining lab values from bed-side analyzers with results from institutional and contract labs in an institutional data warehouse. How the values are interpreted may be impacted by such definition. Further, data definition can change over time as changes in the source systems are made and new systems are brought on line or as new data sources are included. These variations in the values stored in a data element affect the meaning of the data and may alter the results obtained from the data. Thus, we consider data definition and changes in data definition over time to be essential documentation for research reproducibility.
Data access.
An initial step for using EHR data that virtually all EHR-based research projects will encounter is obtaining the data or access to data. Ethics and often, additional institutional approval for data use is usually required. Approval to use data identifies which data can be used and in what way. Access to individuals who can develop, test, and execute computer programs to extract the needed data can also impact accessibility of the data.
Data transfer.
Although federated models of EHR-based research do not require actually extracting data and sending data to another institution or system, other models do rely on data transfers. If data are extracted and transferred, such transfer marks the receipt of data by the research team. In this case, the history of data receipt including the original values at the time of receipt becomes an important traceability milestone in the project. The state of data as received is necessary to reconstruct a study. Preserving data as received supports reproducibility.
Data transformation.
Once data are received, they are often transformed in some way, for example, addresses may be standardized, formulary or location data may be coded or recoded, text data may be further processed, new concepts may be algorithmically computed, values may be imputed, and data may be mapped to different data models. Such transformations can be associated with information loss, thus, maintaining a history of all data changes is a best practice; maintaining a history of all data changes also enables complete traceability from original data to an analyzed dataset and back. In some cases, data are enhanced through association with data from other sources such as public birth or death registries, census data, geo-indexed environmental data, or criminal justice system data, to name a few. If enhancing data without altering the original values is not possible, enhancement should be treated like any other transform.
Record linkage.
Record linkage is an example and special case of a data transformation. If actual data are transferred, multi-facility studies where patients may have data in more than one of the facilities providing data require record linkage and in longitudinal case ongoing record linkage. Linking data from multiple facilities to a particular patient is as important as any other data transform because errors misrepresent the true state of the patient. Further, links can change over time and are subject to false positive and false negative matches from both algorithmic matches and manual assertions. The history associated with making and breaking links is necessary to reconstruct a study. For example, a report run before and after a record linkage run may have different patient counts due to new matches transitively bringing together previously non-linked records, or due to existing links being detected as incorrect and the link manually broken.
Algorithms.
Algorithms may be developed and tested for data transformation, for selecting relevant data, e.g., a cohort of patients with particular characteristics or patient outcomes. In some cases algorithms are created to de-identify or further transform data to facilitate data sharing and ultimately archival. Though different from the analysis, these algorithms impact the study either through direct data transformation or through selection of data for inclusion. Traceability requires the data before and after a change or selection as well as the algorithm used to create the change or make the selection. Reconstruction using the original data requires the state of the data at the time the algorithm was run as well as the algorithm. Thus, source code and associated execution logs are needed to support traceability.
Data quality assessment:
At some point in the study, usually the earliest possible point, the ability of the data to support the intended analysis should be assessed. Data quality assessment recommendations for secondary data use have been published by the Collaboratory. While data quality assessment results are not necessary for reproducibility of a study from the original data, they do help assess the comparability of independent datasets that might be used to replicate a study.
Each of the three cases involved one or more of these steps to using health record data. Each of the steps above, if undertaken on a particular project, pointed to requirements for traceability (Table 1). Requirements driven by dynamic aspects of data, i.e., growing, changing and moving, include necessity for approval of data use, assuring that the analyzed data can be traced back through all transformations to the original data, preservation of data and metadata such that they can be used to replicate the analysis, knowing which versions of algorithms operated over the data and when.
Table 1.
Requirements Associated With Dynamic Aspects of Data
| Requirement | Invoked by: | Met by: |
|---|---|---|
| Approval for data use (Regulatory basis HIPAA, Title 21 CFR Parts 50 and 56, Title 45 CFR part 46) | Use of data not originally collected for the investigator or data collected de novo | Institutional approval to use the data in the intended way and human subject consent and authorization where required. |
| Traceability (Regulatory basis Title 42 CFR 93,* 21 CFR 58.130 (c) and (e) and 58.35 (b)) | Data changing (any operations performed on data that change data values or create new ones) | Association of data values to transformation algorithm; association of original value to new or changed value (data processing procedures, algorithm metadata, value-level metadata) |
| Algorithm versioning, testing and association to data (Regulatory basis Title 42 CFR 93,* 21 CFR 58.130 (c) and (e) and 58.35 (b), Title 21 CFR Part 11) | Data changing (algorithms operating on or facilitating inferences from data) | Versioning and testing of algorithms. (algorithm metadata) |
| Preservation of original values and data documentation (Regulatory basis 21 CFR 58.130 (c) and (e), 21 CFR 58.3 (k)) | Data moving (transfers of data across system or organizational boundaries). | Data archival facility, including archival of data documentation. (all metadata) |
While Title 42 CFR 93 does not specifically call for traceability of all operations performed on data, all institutions receiving Public Health Service (PHS) funding must have written policies and procedures for addressing allegations of research misconduct. In misconduct investigations, traceability of data from it’s origin through all operations performed on the data to the analysis is necessary for developing the factual record.
In the case of federated data, where the queries travel to the data rather than the actual data moving, approaches are likely feasible to meet these requirements in remote fashion. We do not in any way imply that federated approaches are any less able to meet these requirements than non-federated ones.
In the context of secondary data use, the entities described in Table 1, i.e., data transfers, data files, data elements, data values, transformation algorithms, and phenotype algorithms are related. For example data transfers include one or more files, and each transferred data file should be associated with a version of specifications. Likewise, those specifications list data elements whose values appear in the transferred files, and algorithms run over and sometimes transform those data values (Fig. 1). During a research study, these entities will incur changes. Through these relationships, each data value is associated with a data element, algorithms that changed or flagged it, and the file in which it was received. Thus, each data value is associated with prior instances of itself as well as the instances of it’s data element, transformation algorithm, and file from which it originated; traceability necessitates tracking of these changes over time. Each data value is also documented by the versions of the specifications for data files, data elements and algorithms (shaded in Fig. 1). Thus, to fully understand a data element, versions of these specifications should be tracked. This minimum tracking, 1) versioning of data element definitions and specifications for data files and algorithms, 2) new and changed instances of data values, data files, data transfers, and algorithm runs fully specifies each data change. These relationships are those that we built into a system called the historian for the Registry. While informative, they may not generalize to all other studies.
Figure 1.
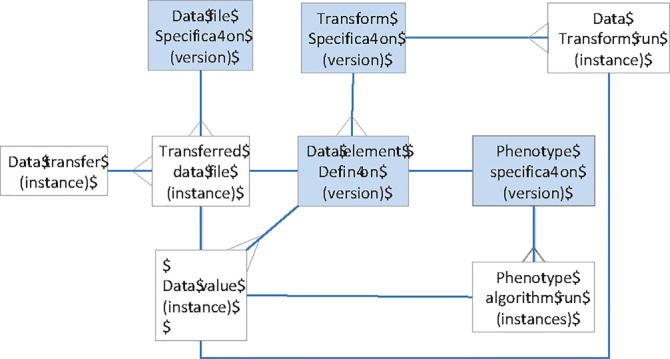
Referential Integrity Model Supporting Traceability.
In this framework, the changes to the data values and result set membership can be tracked computationally, i.e., automated. Versions of specifications require updating and tracking. The historian uses source code management software for this. This strategy requires source code to be “checked-in”. Specifications are similarly tracked. Algorithm executions are obtained from archived log files.
Discussion
Calls for research reproducibility and data sharing in the context of increasing use of EHR data to support clinical studies will force the need for explicit descriptions of the EHR data used and the limitations of those data. Our work here has identified some common requirements, features of studies that often invoke them, and provided an example of their implementation. While our work was informed by three large multi-study initiatives, unidentified requirements may exist and there may likewise be other ways of meeting them. Thus, while a substantial step forward in articulating an approach to the traceability aspect of research reproducibility for health record-based studies, this work is only a beginning.
Some of the requirements echo similar concepts in regulations for clinical studies submitted in support of marketing authorization decisions for new therapeutics, e.g., Title 21 CFR Part 11, the purpose of the Part 11 regulation is so that electronic records and electronic signatures (ERES) are as trustworthy and reliable as those on paper.35 The audit trail required in Part 11 provides for traceability. The Part 11 regulation was found by the therapeutic product industry to be onerous and two subsequent guidances were issued, the 2003 guidance on Electronic Records; Electronic Signatures (ERES) Scope and application, and the 2007 revised Guidance for Industry Computerized Systems Used in Clinical Investigations.36, 37 Reproducibility goes further then traceability, e.g., the meaning of the data are necessary as are the algorithms that operated over the data during the conduct of the study (Table 1). On the surface, reproducibility may seem too high a bar for clinical studies based on EHR data. While there is certainly an interpretation that would render this true, there is also likely a moderate approach that provides confidence in research results and is affordable within the bounds of investigator initiated or federally funded research. We provide this case synthesis and discussion to bring this dialog to the forefront.
Conclusions
Solution to traceability lies in logging, tracking and associating. Reproducibility further requires deep understanding (and documentation) of semantics, for example knowledge of how information is charted at each participating facility and how, if at all, any differences impact the meaning of the data. Both are feasible in studies based on EHR data, however, achieving traceability and more broadly reproducibility in this context likely require a level of effort beyond what has previously been applied. Open discussion will be required to strike an appropriate balance between rigor, cost and benefit.
Acknowledgements
The MURDOCK Study is based in Kannapolis, North Carolina and is made possible with a generous gift from Mr. David H. Murdock to Duke University. This publication was also made possible by the National Institutes of Health (NIH) Common Fund, through a cooperative agreement (U54 AT007748) from the Office of Strategic Coordination within the Office of the NIH Director. Our institutional infrastructure supporting clinical studies, including the maintenance of our clinical data warehouse, is in part supported by the Clinical and Translational Science Award to Duke University grant number (UL1TR001117) and the associated institutional commitment. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the U.S. Department of Health and Human Services or any of its agencies.
References
- 1.Young S, Karr A. Deming, data and observational studies: A process out of control and needing fixing. Significance. September 2011:116–120. [Google Scholar]
- 2.Bissell M. The risks of the replication drive. NATURE. 21 November 2013;503:333–4. doi: 10.1038/503333a. [DOI] [PubMed] [Google Scholar]
- 3.Young SS, Miller HI. Are Medical Articles True on Health, Disease? Sadly, Not as Often as You Might Think. Genetic Engineering & Biotechnology News. May 1, 2014;34(9) [Google Scholar]
- 4.Freedman LP, Cockburn IM. Simcoe TS (2015) The Economics of Reproducibility in Preclinical Research. PLoS Biol. 2015;13(6):e1002165. doi: 10.1371/journal.pbio.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plant A, Parker GC. Translating Stem Cell Research from the Bench to the Clinic: A Need for Better Quality Data. Stem cells and development. 2013;22(18) doi: 10.1089/scd.2013.0188. [DOI] [PubMed] [Google Scholar]
- 6.Hayden EC. The automated lab. NATURE. 2014;516:131–2. doi: 10.1038/516131a. [DOI] [PubMed] [Google Scholar]
- 7.Division of Health Sciences Policy, Institute of Medicine. Washington, DC: National Academy Press; 1999. Assuring Data Quality and Validity in Clinical Trials for Regulatory Decision Making: Workshop Report: Roundtable on Research and Development of Drugs, Biologics, and Medical Devices. [PubMed] [Google Scholar]
- 8.Ioannidis JPA. Contradicted and Initially Stronger Effects in Highly Cited Clinical Research. JAMA. July 13, 2005;294(2) doi: 10.1001/jama.294.2.218. [DOI] [PubMed] [Google Scholar]
- 9.Baker M. Irreproducible biology research costs put at $28 billion per year. NATURE News. 9 June 2015.
- 10.Collins FS, Tabak LA. NIH plans to enhance reproducibility. NATURE. 2014;505:612–3. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Journals unite for reproducibility. NATURE. 2014;515 doi: 10.1038/515007a. [DOI] [PubMed] [Google Scholar]
- 12.Young SS, Bang H, Oktay K. Cereal-induced gender selection? Most likely a multiple testing false positive. Comment in Proc. R. Soc. B. 2009;276:1211–1212. doi: 10.1098/rspb.2008.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnum JF. The misinformation era: the fall of the medical record. Ann Intern Med. Mar 15 1989;110(6):482–484. doi: 10.7326/0003-4819-110-6-482. [DOI] [PubMed] [Google Scholar]
- 14.van der Lei J. Use and Abuse of Computer- Stored Medical Records. Meth. Inform. Med. 1991;30(2) [PubMed] [Google Scholar]
- 15.Meads S, Cooney JP. The medical record as a data source: use and abuse. Top Health Rec Manage. 1982;2:23–32. [PubMed] [Google Scholar]
- 16.Chan KS, Fowles JB, Weiner JP. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev. Oct 2010;67(5):503–527. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 17.Kahn MG, Brown JS, Chun AT, Davidson BN, Holve E, Lopez MH, Meeker D, Ryan PB, Schilling LM, Weiskopf NG, Williams AE, Zozus MN. Recommendations for transparent reporting of data quality assessment Results for observational healthcare data. eGEMS Journal. 2015 doi: 10.13063/2327-9214.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. RFA-RM-13-012, NIH Health Care Systems ResearchCollaboratory - Demonstration Projects for Pragmatic Clinical Trials Focusing on Multiple Chronic Conditions (UH2/UH3). Posted on August 7, 2013.
- 19.Zozus MN, Hammond WE, Green BB, Kahn MG, Richesson RL, Rusincovitch SA, Simon GE, Smerek MM. Assessing data quality for healthcare systems data used in clinical research. Available from www.nihcollaboratory.org.
- 20.Tcheng JE, Nahm M, Fendt K. Data quality issues and the electronic health record. 6. Vol. 2. DIA Global Forum; 2010. [Google Scholar]
- 21.Zozus MN, Richesson RL, Hammond WE. Acquiring, Processing and Using Electronic Health Record Data, in The Living Textbook for the Health Care Systems Research Collaboratory. available at https://www.nihcollaboratory.org.
- 22.Kahn MG, Batson D, Schilling LM. Data Model Considerations for Clinical Effectiveness Researchers. Med Care. 2012;50(0) doi: 10.1097/MLR.0b013e318259bff4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan JR, Britell S, Delcambre LML, Kapoor V, Buckmaster JG. Representing Multi-Database Study Schemas for Reusability. Summit on Translat Bioinforma. 2010;2010:21–25. [PMC free article] [PubMed] [Google Scholar]
- 24.Cimino JJ, Ayres EJ, Remennik L, Rath S, Freedman R, Beri A, Chen Y, Huser V. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): Design, contents, functionality and experience to date. J Biomed Inform. 2014 December;52:11–27. doi: 10.1016/j.jbi.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richesson RL, Rusincovitch SA, Wixted D, Batch BC, Feinglos MN, Miranda ML, Hammond WE, Califf RM, Spratt SE. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zozus MN, Hammond WE, Green BB, Kahn MG, Richesson RL, Rusincovitch SA, Simon GE, Smerek MM. Assessing data quality for healthcare systems data used in clinical research. Available from www.nihcollaboratory.org.
- 27.Kahn MG, Brown JS, Chun AT, Davidson BN, Holve E, Lopez MH, Meeker D, Ryan PB, Schilling LM, Weiskopf NG. Williams AE, Zozus MN, Recommendations for Transparent Reporting of Data Quality Assessment Results for Observational Healthcare Data. eGEMS Journal. 2015 doi: 10.13063/2327-9214.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zozus MN, Hammond WE, Green BB, Kahn MG, Richesson RL, Rusincovitch SA, Simon GE, Smerek MM. Assessing data quality for healthcare systems data used in clinical research. Available from www.nihcollaboratory.org.
- 29.Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, Bauck AE, Cifelli D, Smerek MM, Dickerson J, Laws RL, Madigan RA, Rusincovitch SA, Kluchar C, Califf RM. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013 Dec;20(e2):e226–31. doi: 10.1136/amiajnl-2013-001926. PMID: 23956018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenenbaum JD, Christian V, Cornish M, Dolor RJ, Dunham AA, Ginsburg GS, Kraus VB, McCarthy JM, McHutchison JG, Nahm ML, Newby KL, Svetkey LP, Udayakumar K, Califf RM. The MURDOCK study: A long-term initiative for disease reclassification through advanced biomarker discovery and integration with electronic health records. Am J Transl Res. 2012;4(3):291–301. PMCID: PMC3426390. [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Dunham AA, Cornish MA, Christian VA, Ginsburg GS, Tenenbaum JD, Nahm ML, Miranda ML, Califf RM, Dolor RJ, Newby KL. The measurement to understand reclassification of disease of cabarrus/Kannapolis (MURDOCK) study community registry and biorepository. Am J Transl Res. 2012;4(4):458–470. PMCID: PMC3493022. [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE guided query tool: providing simplified access to clinical data for research and quality improvement. J Biomed Inform. 2011 Apr;44(2):266–76. doi: 10.1016/j.jbi.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath MM, Rusincovitch SA, Brinson S, Shang HC, Evans S, Ferranti JM. Modular design, application architecture, and usage of a self-service model for enterprise data delivery: the Duke Enterprise Data Unified Content Explorer (DEDUCE) J Biomed Inform. 2014 Dec;52:231–42. doi: 10.1016/j.jbi.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth C, Rusincovitch SA, Horvath MM, Brinson S, Evans S, Shang HC, Ferranti JM. DEDUCE clinical text: an ontology-based module to support self-service clinical notes exploration and cohort development. AMIA Jt Summits Transl Sci Proc. 2013 Mar 18;2013:227. [PubMed] [Google Scholar]
- 35.United States Food and Drug Administration (FDA) Title 21 Code of Federal Regulations Part 11. 1997.
- 36.United States Food and Drug Administration (FDA) FDA guidance for industry part 11, electronic records: electronic signatures - scope and application. 2003.
- 37.United States Food and Drug Administration (FDA) FDA guidance for industry computerized systems used in clinical investigations. May 2007.


