Abstract
Hepatocellular carcinoma (HCC) is a deadly disease affecting the liver for which there are many available therapies. Targeting treatments towards specific patient groups necessitates defining patients by stage of disease. Criteria for such stagings include information on tumor number, size, and anatomic location, typically only found in narrative clinical text in the electronic medical record (EMR). Natural language processing (NLP) offers an automatic and scale-able means to extract this information, which can further evidence-based research. In this paper, we created a corpus of 101 radiology reports annotated for tumor information. Afterwards we applied machine learning algorithms to extract tumor information. Our inter-annotator partial match agreement scored at 0.93 and 0.90 F1 for entities and relations, respectively. Based on the annotated corpus, our sequential labeling entity extraction achieved 0.87 F1 partial match, and our maximum entropy classification relation extraction achieved scores 0.89 and 0. 74 F1 with gold and system entities, respectively.
Introduction
Hepatocellular carcinoma (HCC), the predominant form of liver cancer, is one of the leading cancer-related causes of death worldwide. Liver cancer, a fatal disease, has only a 17% 5-year survival rate across all stages.1 In the United States, incidence is expected to continue to increase. Progression develops differently across gender, lifestyle, and genetic environments.2 Furthermore, cancer-associated mortality and deteriorating liver function are separately advancing problems that exasperate patient condition.3 Thus, when prioritizing treatments, clinicians must reconcile between these competing risks. Despite the availability of new therapies, there are still no universally accepted treatment algorithms for HCC.4
To accurately determine the best treatment options given a specific set of HCC patient characteristics, patients must be quantified accurately according to their extent of disease. An important part of this requires determining tumor characteristics such as the number of tumors, size of tumors, degree of tumor spread, etc., all of which typically occur in the electronic medical record (EMR) as free text. While manual abstraction is time consuming and expensive, natural language processing (NLP) methods offer an automated means of extracting and normalizing free text, with the advantage of being scalable to volumes of historical data.
In this paper, we describe our work in annotating and extracting tumor characteristics from free text radiology reports. We focus on identifying individual radiology report anomalies and the uncertainty associated with them with regards to malignancy, with the eventual goal of handling co-reference information for summative information. Our contributions are our tumor extraction annotation scheme and our findings of using simple machine learning framework in our tumor extraction methodology.
Background
When biopsy or resection specimens are unavailable, clinicians may rely on non-invasive imaging studies to identify and characterize malignant tumors prior to planning treatment. This is often the case for HCC, as biopsies carry significant danger of bleeding and tumor spread; further, tumor features on CT or MRI are considered highly sensitive and specific.
As in other tumor diagnostic reports such as for histology and pathology, imaging reports describe crucial information related to a tumor, including location, number, size, and spread. This information is located throughout a report in a piecemeal fashion, with anaphora, i.e. references of one sentence to another, being a persistent issue. Additionally, references may involve split antecedents, i.e. multiple “first mentions,” later referred to collectively.
These phenomena are exemplified in Table 1, where diagnosis appears in the impressions section with summative information of previously mentioned lesions from the findings sections.
Table 1.
Anaphoric and split antecedent tumor references in radiology reports
| 25: Focal lesions: |
| 26: Total number: 5 |
| 27: Lesion 1: segment 8, 2.2 x 1.4cm, image 3/8, hyper enhancing with washout on delayed phase. |
| 28: Lesion 2: segment 5, 2.0 x 1.8cm, image 3/25, hyper enhancing with washout on delayed phase. |
| 29: Lesion 3: segment 4A, 1.8cm, image 3/7, hyper enhancing with washout on delayed phase. |
| 30: Lesion 4: segment 8, 1.6 x 1.1cm, image 3/15, hyper enhancing with no definite washout. |
| 31: Lesion 5: segment 6, 0.4cm, image 3/28, hyper enhancing with no definite washout. |
| ……………….. |
| 35: Impression: |
| 36: 3 focal lesions in segment 4A, 5 and 8 are hyper enhancing with washout on delayed phase, typical for HCC. |
| 37: 2 focal lesions in segment 8 and 6 are hyper enhancing with no definite washout on portal venous/ delayed phase suggestive of indeterminate nodules. |
Moreover, previous measurements may become a confounding extraction problem because radiology reports often cite past readings. For example, Table 2 shows a previous measurement mentioned.
Table 2.
Temporal tumor references
| The previously visualized mass involving segment 5 and segment 6 has increased in size (cranial caudal measuring 11 mm, previously 8.5 mm) and now extends to involve segment 4. |
In contrast to histologic or pathologic analyses which tests directly on specific corporal samples, cross-sectional imaging covers a large volume of tissue and therefore may pick up other non-cancerous entities. Further, imaging diagnostics may be prone to uncertainty related to limitations of technology. Detected anomalies in imaging reports may be related to various cancer types, but could also be benign entities such as hemangiomas (tumors made of cells that line blood vessels), cysts (abnormal membranous sac containing fluid), pseudomasses (from imaging anomalies), or anatomic scarring. Table 3 shows examples in which tumor references are determined as malignant, benign, or indeterminate.
Table 3.
Uncertainty related to tumor malignancy
| Example Passage | Tumor status |
|---|---|
| 1.9 x 1.8 cm hyperenhancing mass on the arterial phase with enhancing pseudocapsule, corresponding washout on portal venous phase as well as T2 hyperintensity and restricted diffusion, characteristic of HCC. | Malignant |
| There are multiple scattered hepatic hypodensities that exhibit no enhancement and likely represent cysts. | Benign |
| In segment 4a, there is a stable hypovascular lesion which is indeterminate and could represent a regenerative nodule. Would recommend MRI with Eovist specifically to further evaluate this lesion. | Indeterminate |
Although not explored here, we ultimately hope to infer overall patient information such as number and size of malignant tumors; these data are crucial for cancer staging.
Related work
We group related work under two categories: studies identifying cancer information and studies that parse radiology reports.
Cancer information extraction
The challenge of tumor extraction is not new and there is much to be learned from previous work. Rule-based systems for these tasks typically involved a dictionary look-up, context and negation checking, and heuristic algorithms to structure results. Scores range widely between systems as well as between distinct variables within a single system, depending heavily on the selection of extraction variables. Coden et al. 5 focused on finding hierarchical concepts such as anatomical site, grade value, date, primary tumor, etc., from pathology reports and organized results into structured classes, achieving F1 scores ranging for 0.65 to 1.0. Ashish et al. 6 trained and tested on pathology reports from the University of California Irvine data warehouse and looked for structured classes such as TNM stage, capsule invasion, lymph invasion, chronic inflammation, and vascular invasion, with per field F1 performances ranging from 0.78 to 1.0. Ping et al. 7 used regular expressions and structured entities extraction using heuristic algorithms for liver cancer information, with 0.92–0.996 F1 score. The machine learning equivalent of these works used statistical methods. Ou and Patrick8 took a conditional random fields (CRFs) approach to extracting cancer-related entities, such as diagnosis, metastasis, site, size, and specimen type, from processed primary cutaneous melanoma pathology reports. Afterwards, entities were populated into structured reports using rules. F1 performance for populating fields was at 0.85.
Radiology report parsing
The general task of parsing reports have been explored since the early days of biomedical informatics, with a heavy emphasis on comprehensive linguistic annotation that subsequently mapped to a separate parallel domain knowledge base. These have contributed to systems such as MedLee and others.9–13 Continuing in this tradition, Taira et al. 14 detailed their system that includes deep linguistic annotation with dependency parses fortified with a detailed radiology report domain ontology. Their strategy started with identifying concepts using custom dictionaries, then dependency parsing entities using statistical methods in their parser module. Once parsed, relations from their radiology ontology were constructed using their semantic interpreter module, which either used rule-based logic or a statistical maximum entropy classifier. Finally, their frame constructor bundled together their concepts and relations. They reported parsing performance of 87% recall and 88% precision. Meanwhile, their conversion of dependency parses into relations were evaluated at 79% and 87% recall and precision, respectively.
Our contributions
Our work is most comparable to Taira et al. 14 and Ou and Patrick.8 While Taira et al. 14 radiologic findings with a rich and complex knowledge representation, we chose a relatively simple targeted annotation approach, like Ou and Patrick,8 with the aim of achieving our specific goals (versus wholesale document medical concept encoding for future queries), which required significantly less investment. Our system is unique in our emphasized use of machine learning methods. Whereas Taira et al. 14 used rule-based methods then statistical methods for entity and relation extraction, Ou and Patrick8 used statistical and rule-based, respectively. We approached named entity recognition as a sequential labeling classification task, and modeled relation extraction as a statistical classification.
Methods
To build our system, we created, annotated, and evaluated on a clinical corpus of 101 randomly selected radiology reports originally from a cohort of 160 HCC patients from the University of Washington (UW) Medical Center system. We annotated this corpus for tumor information, using 31 reports to test inter-annotator agreement. We used the annotated corpus to train and test our extraction system in five fold cross validation.
Corpus annotation
Annotation schema
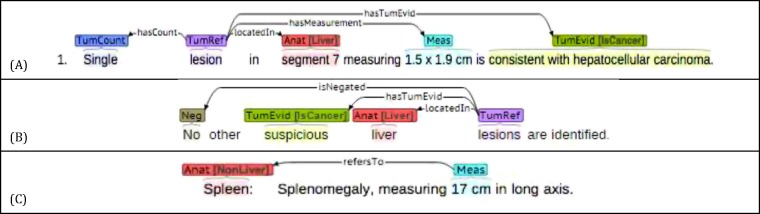
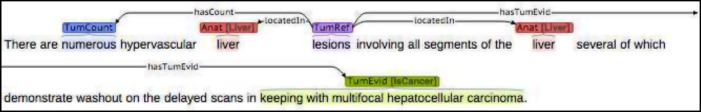
Our template schema was designed by a biomedical informatics graduate student and a medical student. Each template was represented as a composition of entities, spans of text with assigned label names, and relations, directed links between entities. In our task, entities captured anatomic entities, tumor references, sizes, number, cancer diagnosis, whereas relations ensured that the proper descriptions linked to the items they characterized. Figure 1 includes example sentences annotated with entities and relations. We used Brat,15 a web-based annotation tool, for our annotation software.
Figure 1.
Examples of the radiology reports annotated with entities and relations
Our entities had the following types: (1) Anatomy: anatomic locations in the human body (e.g., “segment 5” or “left lobe”) with attributes (Liver, NonLiver), (2) Measurement: quantitative size in the text (e.g., “2.2 x 2.0 cm”), (3) Negation: indicator to some negation of a tumor reference (e.g., “no”) (4) Tumor count: number of tumor references (e.g., “two” or “multiple”), (5) Tumor reference: a radiologic artifact that may reference a tumor (e.g., “lesion” or “focal density”), and (6) Tumorhood evidence: diagnostic information regarding the tumor (e.g., “characterisitcs of HCC”, “indeterminate”, “suggestive of cyst”) with attributes (isCancer, isBenign, inDeterminate).
Our relations were defined as directed links between the two entity types, often with either a tumor reference (preferred) or a measurement as the source, or the head, of the directed relation. They are described as follows: (1) hasCount: relation between a tumor reference to a tumor count, (2) isNegated: a negation cue, starting from the tumor reference to the negation entity, (3) locatedIn: marks in which anatomy a tumor reference or measurement was found, (4) hasMeasurement: present tense relation between a tumor reference to a measurement, (5) hadMeasurement: past tense relation between a tumor reference or measurement, to a measurement, (6) hasTumEvid: relates a tumor reference or measurement to a tumorhood evidence, (7) refersTo: relates a measurement to an anatomy indicating a measurement of anatomy. Templates were constructed by collating all connected entities and relations.
Annotation guidelines
Our annotation approach sought to maximize information while minimizing annotation workload. Therefore we made a few important high-level annotation decisions: (1) only the “findings” and “impressions” section of the reports were annotated, (2) we annotated either a tumor reference or a measurements (the starting point of the relation or “the head”) in all available lines, but only annotated other entities if they were related to our tumor reference or measurements or if it appeared in a line with annotations, (3) we annotated radiographic evidence of tumorhood evidence, e.g. “hypervascular with washout,” and (4) we added extra relations from a measurement to a anatomy when they referred to different locations. Relation attachments over multiple lines were allowed, though we did not mark for co-referring information and each tumor reference was treated separately.
We decided on (1) because, we found that the “findings” and “impressions” sections comprehensively harbored the radiologic information in the report. Other parts of reports had comparatively more unimportant tumor information, e.g. in the “indication” section, “please determine size and location of tumor.”
Our reason for designating tumor references and measurements as heads, in decision (2), was part of our strategy to maximize annotation simplicity. For example, we avoided a lot of excess annotation by not allowing pronouns such as “this,” “these,” “the largest” as a tumor reference. Measurements were allowed as heads because in absence of a nearby tumor reference, a size was the most reliable indicator of tumor information, e.g. “1. Segment VII: 2.6 x 2.4 cm, hyper enhancing with washout.” By only annotating entities related to these heads, we avoided lines without any information of interest. We annotated other entities within a line, not necessarily related to an event, to provide negative example cases. For example, an anatomy entity may only be near a tumor without actually having been invaded (Figure 2.A), or instead a measurement may be measuring an anatomy entity instead (Figure 2.B).
Figure 2.
(A LEFT) Example in which an anatomy entity does not necessarily refer to the tumor reference location (B RIGHT) Measurements of anatomies were marked by the refersTo relation
We decided on (3), to admit radiographic evidence as tumor evidence to avoid later needing to refer to outside lines for tumorhood evidence, as findings in a single line may not always include a diagnosis, e.g. “HCC,” as in Figure 3.
Figure 3.
Tumorhood evidence based on radiographic evidence
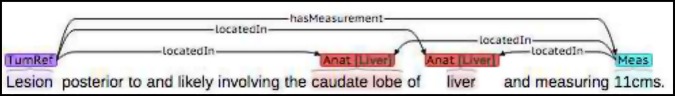
Finally, whenever there was possibility of ambiguity between what size refers to a lesion in which location, we required extra relations, decision (4), as shown in Figure 4.
Figure 4.
When multiple measurements and anatomy were present, additional disambiguating relations were added
Evaluation
Our evaluation was carried out at three levels: (a) entity, (b) relation, and (c) template levels. We used precision, recall, and F-1 measure16 as our inter-annotator agreement measure (where one annotator was held as the gold standard). These are defined according to the equations:
where TP = true positive, FP = false positive, FN = false negative, and F1 is the geometric mean of precision and recall. Two entities were considered matching if they had the same label, attribute (if appropriate), and document offset text spans. Relations were considered matching if both of its entities matched, and the relation types both matched. Two templates were considered matching if all its entities and relations matched that of the other template. Partial entity match allowed document to be counted as matching if document text spans at least overlapped and their labels matched. Similarly relation partial matching was defined on whether the two pair of entities partially matched and if the relation type was correct. Partial template match was defined by whether all entities and relations were partially matched.
Inter-annotator agreement
After agreeing on a final annotation schema, the biomedical informatics graduate student and medical student tested inter-annotator agreement on a set of randomly selected 31 radiology documents. At the first annotator meeting, agreement was scored at 0.84, 0.73, 0.54 F1 for entities, relations, and templates, respectively. After refining guidelines further, the annotators re-annotated on the same set. The final entity, relation, and template agreements improved to 0.88, 0.78, 0.61 F1, with partial scores of 0.93, 0.90, and 0.70 F1. The full breakdown is shown in Tables 4, 5, and 6. Reported templates are broken down into categories by their constituent relations for finer- grained analysis. For example, if refersTo was a relation in the template, it is categorized as an AnatomyMeasure template; if the template has an isNegated relation, it is a Negative template. Singletons were all templates with a single entity. The remaining templates were categorized as tumor events.
Table 4.
Partial F1 agreement for entities
| Label | TP | FP | FN | P | R | F1 |
|---|---|---|---|---|---|---|
| Anatomy | 316 | 32 | 49 | 0.91 | 0.87 | 0.89 |
| Measurement | 159 | 1 | 2 | 0.99 | 0.98 | 0.99 |
| Negation | 23 | 0 | 3 | 1.00 | 0.88 | 0.94 |
| Tumor count | 65 | 2 | 5 | 0.97 | 0.93 | 0.95 |
| Tumor reference | 245 | 7 | 14 | 0.97 | 0.94 | 0.96 |
| Tumorhood evidence | 159 | 14 | 24 | 0.92 | 0.87 | 0.89 |
| ALL | 967 | 56 | 97 | 0.95 | 0.91 | 0.93 |
Table 5.
Partial F1agreement for relations
| Label | TP | FP | FN | P | R | F1 |
|---|---|---|---|---|---|---|
| hadMeasurement | 15 | 0 | 2 | 1.00 | 0.88 | 0.94 |
| hasCount | 64 | 3 | 6 | 0.96 | 0.91 | 0.93 |
| hasMeasurement | 94 | 6 | 8 | 0.94 | 0.92 | 0.93 |
| hasTumEvid | 155 | 23 | 27 | 0.87 | 0.85 | 0.86 |
| isNegated | 24 | 0 | 3 | 1.00 | 0.89 | 0.94 |
| locatedIn | 279 | 25 | 39 | 0.92 | 0.88 | 0.90 |
| refersTo | 24 | 4 | 7 | 0.86 | 0.77 | 0.81 |
| ALL | 655 | 61 | 92 | 0.92 | 0.88 | 0.90 |
Table 6.
Partial agreement for templates
| Label | TP | FP | FN | P | R | F1 |
|---|---|---|---|---|---|---|
| AnatomyMeas | 20 | 5 | 8 | 0.80 | 0.71 | 0.76 |
| Negative | 17 | 7 | 10 | 0.71 | 0.63 | 0.67 |
| Singleton | 34 | 23 | 24 | 0.60 | 0.59 | 0.59 |
| TumorEvent | 161 | 57 | 62 | 0.74 | 0.72 | 0.73 |
| ALL | 232 | 92 | 104 | 0.72 | 0.69 | 0.70 |
The medical student annotator annotated the remaining 70 reports of the corpus. The total number of entities, relations, and templates for the 101 radiology report corpus were 3211, 2283 and 1006, respectively.
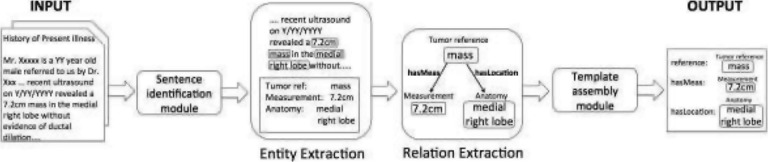
Entity and relation extraction
We used the annotated corpus to train and evaluate an extraction system, in five fold cross-validation. Figure 5 presents the overall system architecture. A sentence identification module identified sentences of interest. Entity types were extracted from the isolated sentences using regular expression for the measurement entity and CRFs on the remaining others. Relations were identified using a direct classification of enumerated pairwise entity to entity candidate relations. Afterwards, templates were assembled by traversing the graph of connected entities and relations. Evaluations were the same as those used for inter-annotator agreements.
Figure 5.
System pipeline: A report is first processed to identify sentences, before entities and relations are identified and assembled into templates.
Preprocessing
Radiology reports were processed beforehand to remove excess white spaces and blank lines using report-specific heuristics. Sentences were identified using NLTK punkt module.17 Only sentences belonging to the “findings” and “impressions” sections, as tagged by our in house section chunker,18 were kept as per our annotation guidelines.
Sentence identification
To avoid classifying sentences with no annotations, we first selected sentences of interest. Based on the analysis of our corpora, we found that 90% of relations were from entities on the same line, and around 7% were from entities connected to the entities on the next line. To identify these sentences, we mimicked the annotation strategy of first finding the tumor reference or measurement before marking other entities. Sentences of interest on the first line (S1) were identified using regular expressions on measurement values, e.g. “(\\d+) cm” (a number before a “cm” word), and a word list of radiographic tumor reference terms, listed in Table 7, created from the top unigrams accounting for 90% by frequency for tumor references. S1 sentences, along with the sentence following it (S2) sentences, were passed to subsequent steps. This resulted in a sentence identification recall and precision of 94% and 69%.
Table 7.
S1 tumor reference word list
| Focal | foci | enhancing |
| hypervascular | hypodense | lesion |
| Mass | nodule | tumor |
Entity extraction
Entities were extracted from the sentences identified using one of two strategies: regular expression lookup and sequential label classification. The original regular expressions used to identify measurement values in sentence identification were taken as the measurement entities. For the remaining entities, anatomy, negation, tumor reference, tumorhood evidence entities, we used CRFs classified using CRFSuite.19
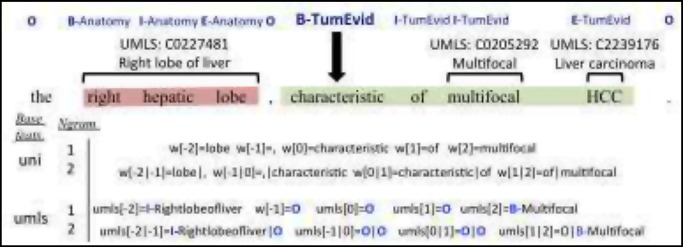
We created CRF features by identifying several base features, then generatively creating the final more complex features by tuning several variables: window-size, n-gram numbers, and tag sets (for entity features){BIOE, BIO, IOE, IO}, as illustrated in Figure 6. For example, suppose our base feature is unigrams. Then if we choose a window size of ±2, and n-grams of 1 and 2, then the final features would be all unigrams and bigrams within ±2 words of a word. For base features that may span over multiple words, such as tagged UMLS concepts, we additionally experimented with different tag sets. Table 8 gives a more detailed description of our base features. We also implemented two augmenting parameters, which replaces the UMLS feature with a more general term if a concept id is part of the specified list. These two lists were for liver anatomic parts and carcinoma concepts.
Figure 6.
Entity CRF features for a window size ±2, unigrams and UMLS concept base features, 1 - and 2-grams, BIOE label tagging, and BIO feature tagging
Table 8.
Entity extraction base features descriptions
| Feature | Feature Description |
|---|---|
| UNIGRAM | Unigram fcase-sensitive] |
| LEMMA | Lemma (lower-cased) |
| PARTS-OF-SPEECH | Parts of speech |
| LABELLED DEPENDENCY | Label of the relation from the word to its head combined with the lemma of its head word |
| DEPENDENCY HEIGHT | Height of a word in its dependency tree |
| UMLS CONCEPT | {BIOEQ-tagged normalized name |
| UMLS SEMANTIC TYPE | {BIOE}-tagged UMLS semantic type |
For example, if any concept ID part of the carcinoma list is found, instead of its specific preferred name used as a feature, it will be <CARCINOMA>. Liver concepts were identified from taking all liver anatomic subparts as specified in the Foundation Model of Human anatomy. Carcinoma concepts were generated from taking sub concepts of C3263 (Neoplasm By Site) from the National Cancer Institute thesaurus.20 S1 and S2 sentences were trained separately.
Sentences were tokenized, tagged tor parts-oi-speech, dependency parsed using ClearNLP.21 UMLS features were extracted using MetaMap,22 with word sense disambiguation turned on. During experimentation, we optimized for the feature parameters, as well as the optimal CRF tag set for the learned labels. When tag labels overlapped, we merged tags. For example, in Figure 7, “nodules” would have the tag “B- TumRef_I-TumEvid [isCancer]. ”
Figure 7.
A single sentence can have multiple tumor reference subjects, with overlapping entities
Relation extraction
Once sentences were identified with entities, they were run through a relation classifier. All possible pairwise relations between entities in S1 and corresponding S2 sentences were enumerated and classified using several machine learning algorithms. Given two entities, the direction was determined based on the entity types, e.g. the tumor reference is always the head, or the first-appearing measurement if no tumor reference is found. We experimented with a c4.5 decision tree, maximum entropy, and a support vector machine (SVM) classifier, implemented through MALLET23 and LibSVM24 with default parameters. We report the classifier with the best performances. Our features were related to the entities involved, the dependency paths between them, and the words around them. They are described in detail in Table 9. We tuned two variables in our experiments: window size for the SURRWORDS feature and the machine learning classifier.
Table 9.
Relation extraction feature descriptions
| Feature | Feature Description |
|---|---|
| CLOSESTREF | 1 if the head entity is closest left or right tumor reference, e.g. fclosestLeftRef:0, closestRightRef:1# |
| DIFFLINES | 1 if two entities are on the same line, (e.g. sameLine: 1) |
| ENTNUM | Number of each type of entity in corresponding line (e.g. num-l1[Anatomy]:2, num-l1[TumCount]:2) |
| ENTWORDS | 1 for every word inside an entity, represented by its lemma (e.g. en1-nodule:1, ent2-segment:1) |
| POSSIBLELABELS | 1 if relation label type is a possible between two entities (e.g. candidateLabel-locatedIn:1) |
| ONLYPOSSIBLEHEAD | 1 if head entity is the only tumor reference or measurement in the lines being considered fe.g. onlyHead:0# |
| SHORTESTPATH | The shortest path distance between two entities through the dependency tree (e.g. minPath:3) |
| SHORTESTPATH.HEADS | Within the shortest path, 1 if words within path have the labels of tumor reference, measurement, or the second entity label (e.g. minPath[tumorref]:1) |
| SUBTREE | Minimum distance between head entity to another second entity of the same label type in its dependency subtree (not including the second entity) (e.g. subTreeNextCand[samelabel]:1) |
| SURRWORDS | 1 for every word within the word window of the entities, (e.g. uni-ent1[-2]=multiple:1, uni-ent2[1]=with:1) |
We report our results compared to a simple baseline. The simple baseline takes the S1 and its S2 sentence and creates a template by attaching all entities to first occurring tumor reference, or measurement if tumor references are not available. If more than one relation is possible according to our annotation guidelines, we put the highest frequency relation and do not attach a relation if no relation is possible.
Results
Table 10 and 11 shows entity extraction performances for exact and partial match, respectively, consolidated by label. Our final feature configurations included a window size of ±1 word, 1-grams, and BIO tagging for both features and labels. Our higher performing entities, the measurement and tumor reference, were expected given the rule-based nature of measurement extraction and the strategy of sentence classification. Precision was high across all entities, which is perhaps a result of our tagging scheme and overlapping entities, which combines to very specific tags. Our entity overall extraction performance 0.87 F1 was lower compared to inter-annotator agreement 0.93 F1, which is often considered the upper bound for a task. Specifically, negation, tumor count, and tumorhood evidence were at considerably lower with 0.08, 0.12, and 0.19 F1 difference.
Table 10.
Entity extraction (exact)
| Label | TP | FP | FN | P | R | F |
|---|---|---|---|---|---|---|
| Anatomy | 789 | 103 | 254 | 0.88 | 0.76 | 0.82 |
| Measurement | 472 | 9 | 17 | 0.98 | 0.97 | 0.97 |
| Negation | 59 | 6 | 14 | 0.91 | 0.81 | 0.86 |
| Tumor count | 126 | 6 | 48 | 0.95 | 0.72 | 0.82 |
| Tumor reference | 678 | 64 | 124 | 0.91 | 0.85 | 0.88 |
| Tumorhood evidence | 315 | 85 | 315 | 0.79 | 0.50 | 0.61 |
| ALL | 2439 | 273 | 772 | 0.90 | 0.76 | 0.82 |
Table 11.
Entity extraction (partial)
| Label | TP | FP | FN | P | R | F |
|---|---|---|---|---|---|---|
| Anatomy | 828 | 65 | 215 | 0.93 | 0.79 | 0.86 |
| Measurement | 480 | 1 | 9 | 1.00 | 0.98 | 0.99 |
| Negation | 59 | 6 | 14 | 0.91 | 0.81 | 0.86 |
| Tumor count | 127 | 5 | 47 | 0.96 | 0.73 | 0.83 |
| Tumor reference | 714 | 25 | 88 | 0.97 | 0.89 | 0.93 |
| Tumorhood evidence | 359 | 40 | 271 | 0.90 | 0.57 | 0.70 |
| ALL | 2567 | 142 | 644 | 0.95 | 0.80 | 0.87 |
Our feature configurations for relation extraction was a window size ±3 words from each entity, using a maximum entropy classifier. The system relation and template extraction performance are shown in Table 12 and 13, with both gold and system entities. Even with gold entities, the hadMeasurement and the refersTo relations were comparatively low-performing at 0.35 and 0.67 F1 scores, respectively. Given gold entities, our system reached 0.89 F1 for relation extraction and 0.64 for tumor extraction, with 0.72 F1 for the TumorEvent template subcategory. Meanwhile, when using system entities both relation and template extraction suffered more than 10% degradation, suggesting that improvements in the entity extraction upstream task will lead to improvements in the overall system.
Table 12.
Relation extraction (partial match)
| Baseline | System | |||
|---|---|---|---|---|
| Entities | Gold | System | Gold | System |
| hadMeasurement | 0.00 | 0.00 | 0.35 | 0.36 |
| hasCount | 0.90 | 0.76 | 0.95 | 0.79 |
| hasMeasurement | 0.85 | 0.81 | 0.89 | 0.85 |
| hasTumEvid | 0.86 | 0.61 | 0.89 | 0.63 |
| isNegated | 0.97 | 0.86 | 0.98 | 0.87 |
| locatedIn | 0.82 | 0.71 | 0.89 | 0.77 |
| refersTo | 0.00 | 0.00 | 0.67 | 0.63 |
| ALL | 0.83 | 0.69 | 0.89 | 0.74 |
Table 13.
Template extraction (partial match)
| Baseline | System | |||
|---|---|---|---|---|
| Entities | Gold | System | Gold | System |
| AnatomyMeas | 0.00 | 0.00 | 0.49 | 0.26 |
| Negative | 0.84 | 0.57 | 0.82 | 0.57 |
| Singleton | 0.25 | 0.25 | 0.35 | 0.32 |
| TumorEvent | 0.69 | 0.42 | 0.72 | 0.44 |
| ALL | 0.60 | 0.38 | 0.64 | 0.42 |
Discussion
A significant hurdle for our entity extraction task was that, different from traditional entity extraction tasks (e.g. the i2b2 2010 challenge), our entities were not always noun phrases or even well-contained chunks of information. For example, we annotated “hepatic” such as in “hepatic lesions” to be an anatomy liver entity.” Ou and Patrick8 reported similar experiences in their extraction. Our tumorhood evidence experience entity extraction was particularly interesting in this respect. Particularly, tumorhood evidence based on radiographic evidence, such as if “hypervascularity” or “enhancement” in addition to “washout” cues were present, the contained text would be considered positive for cancer. These mentions of “enhancement” could occur as adjectives to other entities, e.g. “enhancing lesion” or “hypervascular lesion,” and the “washout” may be mentioned very far from the “enhancement,” resulting in long spans of identified text with spurious words. On the other hand, if both positive mentions were not met, then cues were not highlighted, e.g. “enhancing with no definite washout.”
Other issues included medical abbreviations. This occurred for anatomy terms, e.g. “SMV,” (short for superior mesenteric vein) as well as tumorhood evidence terms, e.g. LR3 (short for LI-RADS, a coding system for tumor malignancy). Overtraining on context was another problem. For example negation and tumor counts worked better in short sentences, or around words they were most often found near. Tumorhood evidence isBenign evidence were difficult to differentiate since a non cancerous entity could be a number of things, e.g. “nonocclusive chronic thrombi,” “cysts,” “likely related to old trauma/fracture,” “differential includes infection.”
Our partial match performance entity extraction performance was comparable to Ou and Patrick8 who achieved an overall 0.84 F1 score for a variety of entities on their training set. That said, we have many opportunities with which simple adjustments can make large improvements. For example, we may condition tumorhood evidence instead as a classification on our tumor reference or measurements (e.g. “Is lesion cancerous?”). In so doing, we can also conveniently re-introduce outside sentence information (e.g. previous sentence unigrams), incorporate our already extracted evidence, and address the abbreviations issue for LI-RADS. Having found the tumor references, we can do additional checks to ascertain for missed tumor counts or negation indicators.
Relation classification had the most trouble in several situations: (a) when a previous measurement was linked to a previous line, (b) relations to anatomy entities, and (c) when there were multiple tumor references or measurements in a single sentence. Because of our definitions of S1 and S2 lines, any line with a measurement is considered its own S1 line, therefore a sentence such as “This compared to a prior measurement of approximately 6.4 x 6.0 cm” may not be linked to a prior tumor reference or measurement as was in annotation. Some prior measurements were also associated with past dates, which we did not handle in our system. As with some of our entities, we may reframe hadMeasurement relation classification as a classification on the measurement entity for past vs. current. Relations to an anatomy can have a locatedIn and refersTo label or no label at all. In our evaluation, we found cases of each being correctly identified, however, there were some misclassifications. The consequence would be missing relations and mislabels between referTo and locatedIn. Though our refersTo relation performance was quite low, we only added this as a negative case for our locatedIn relation. In the last situation, (c), either too many relations were enumerated or too little. This was a product of both our annotation method, in which measurements may be attached to more specific anatomy entities, as well as our classification method, where each relation is classified individually. We may correct these by piping classification results that consider relations in a sentence jointly.
Our relation extraction performed similarly to other statistical methods, such as Taira et al.’s 79% and 87% recall and precision. Not directly comparable to our relation extraction or our template evaluation definitions, but of interest to compare, we report performances of similar information extraction subcategories from related works. Coden et al. 5 reported evaluations of 0.82, 0.65, and 0.93 F1 for primary tumor, metastatic tumor, and lymph nodes class structures. Ou et al. 8 achieved 0.84, 0.92, 0.29, 0.92, 0.33, 0.29, 0.84, 0.93, 0.90 F1 for clinical diagnosis, diagnosis, distant metastasis, lymphovascular invasion, microsatellites, other lesions, site and laterality, size of specimen, and tumor thickness fields.
Our template extraction performances were low, in large part because of our punishing metric. Our templates were in fact graphs that required all relations to be exact even if it could provide equivalent information, as in Figure 8. Moreover our annotations were fairly detailed, so that there was often repeats of the same information regarding the same information within a template. As an example, Figure 9 is considered incorrect because “hypervascular […] demonstrate washout” should have been highlighted as tumorhood evidence isCancer. Although constructing a more flexible evaluation would require arbitration for cases such as in Figure 4, it would provide a more lenient measure.
Figure 8.
Incorrect extra relations to the measurement makes the entire template incorrect
Figure 9.
Missing extra radiographic tumorhod evidence cues causes entire template to be incorrect
Conclusion
In this work, we developed a sparse annotation method for tumor information extraction and present a machine learning based system for entity and relation extraction for these characteristics. Considering the complexity of our annotation and the simplicity of our features, our performances are very promising. We have characterized the errors in our system, which may be augmented by further processing. In future work we will expand our system to handle the reference solution problem, and move towards granular classification of patients into grades or stages, facilitating automated methods for quantifying patient statistics, clinical trial eligibility, and cohort identification.
Acknowledgements
This project was partially funded by the National Institutes of Health, National Center for Advancing Translational Sciences (KL2 TR000421) and the UW Institute of Translational Health Sciences (UL1TR000423).
References
- 1.American Cancer Society. Cancer Facts & Figures 2014. Atlanta: 2014. [Google Scholar]
- 2.McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15(2):223–243. vii–x. doi: 10.1016/j.cld.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JD, Roberts LR. Epidemiology and management of hepatocellular carcinoma. Infect Dis Clin North Am. 2010;24(4):899–919. viii. doi: 10.1016/j.idc.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han K-H, Kudo M, Ye S-L, et al. Asian consensus workshop report: expert consensus guideline for the management of intermediate and advanced hepatocellular carcinoma in Asia. Oncology. 2011;81(Suppl 1):158–164. doi: 10.1159/000333280. [DOI] [PubMed] [Google Scholar]
- 5.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform. 2009;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Ashish N, Dahm L, Boicey C. University of California, Irvine-Pathology Extraction Pipeline: The pathology extraction pipeline for information extraction from pathology reports. Health Informatics J. August 2014 doi: 10.1177/1460458213494032. [DOI] [PubMed] [Google Scholar]
- 7.Ping X-O, Tseng Y-J, Chung Y, et al. Information extraction for tracking liver cancer patients’ statuses: from mixture of clinical narrative report types. Telemed J E-Health Off J Am Telemed Assoc. 2013;19(9):704–710. doi: 10.1089/tmj.2012.0241. [DOI] [PubMed] [Google Scholar]
- 8.Ou Y, Patrick J. Automatic Population of Structured Reports from Narrative Pathology Reports; Proceedings of the Seventh Australasian Workshop on Health Informatics and Knowledge Management - Volume 153. HIKM ‘14; Darlinghurst, Australia, Australia: Australian Computer Society, Inc.; 2014. pp. 41–50. [Google Scholar]
- 9.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc JAMIA. 1994;1(2):142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman C. A broad-coverage natural language processing system; Proc AMIA Symp.; 2000. pp. 270–274. [PMC free article] [PubMed] [Google Scholar]
- 11.Rassinoux AM, Wagner JC, Lovis C, Baud RH, Rector A, Scherrer JR. Analysis of medical texts based on a sound medical model; Proc Annu Symp Comput ApplMed Care; 1995. pp. 27–31. [PMC free article] [PubMed] [Google Scholar]
- 12.Zweigenbaum P, Bachimont B, Bouaud J, Charlet J, Boisvieux JF. A multi-lingual architecture for building a normalised conceptual representation from medical language; Proc Annu Symp Comput Appl Sic Med Care Symp Comput Appl Med Care; 1995. pp. 357–361. [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DA, Brownlow ND, Hersh WR, Campbell EM. Automating concept identification in the electronic medical record: an experiment in extracting dosage information; Proc Conf Am Med Inform Assoc AMIA Annu Fall Symp AMIA Fall Symp; 1996. pp. 388–392. [PMC free article] [PubMed] [Google Scholar]
- 14.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiogr Rev Publ Radiol Soc NAm Inc. 2001;21(1):237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 15.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. EACL ‘12. Stroudsburg, PA, USA: Association for Computational Linguistics; 2012. BRAT: A Web-based Tool for NLP-assisted Text Annotation; pp. 102–107. [Google Scholar]
- 16.Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. J Am Med Inform Assoc. 2005;12(3):296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loper E, Bird S. NLTK: The Natural Language Toolkit; Proceedings of the ACL-02 Workshop on Effective Tools and Methodologies for Teaching Natural Language Processing and Computational Linguistics - Volume 1. ETMTNLP ‘02; Stroudsburg, PA, USA: Association for Computational Linguistics; 2002. pp. 63–70. [Google Scholar]
- 18.Tepper M, Capurro D, Xia F, Vanderwende L, Yetisgen-Yildiz M. Statistical Section Segmentation in FreeText Clinical Records. LREC. 2012.
- 19.Okazaki N. CRFsuite: a fast implementation of Conditional Random Fields (CRFs) 2007. http://www.chokkan.org/software/crfsuite/. Accessed March 8, 2014.
- 20.U.S. National Institutes of Health. National Cancer Institute: NCIthesaurus. http://ncit.nci.nih.gov/
- 21.ClearNLP. code.google.com/p/clearnlp/
- 22.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proc AMIA Annu Symp AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 23.McCallum AK. MALLET: A Machine Learning for Language Toolkit. http://mallet.cs.umass.edu.
- 24.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(3):27:1–27:27. [Google Scholar]