Abstract
Objective
Systemic juvenile idiopathic arthritis (SJIA) is an inflammatory disease of childhood, in which cells of the monomyelocytoid lineage are felt to be key effector cells. Monocytes from SJIA patients have a distinct phenotype, with features of both M1 well as M2 alternative activation. MicroRNAs are critical regulators of monocyte polarization and function; however, cellular microRNAs in SJIA have not been systematically examined.
Methods
MicroRNA Taqman arrays were used to determine expression profiles of monocytes from children with SJIA. Expression of miR-125a-5p and its contribution to monocyte polarization was examined using in vitro polarized THP1 and primary human monocytes.
Results
110 microRNAs were differentially expressed in monocytes from patients with active SJIA, including molecules implicated in rheumatoid arthritis pathogenesis, cytokine production, and monocyte polarization. miR-125a-5p was identified as highly upregulated in active SJIA monocytes compared to those from children with clinically inactive disease or with active polyarticular JIA, and correlated with systemic features. In vitro, monocyte mir125a-5p expression was increased after polarization with M2b or M2c conditions. Interestingly, inhibition of miR-125a-5p showed that this microRNA contributed to full polarization of M2b regulatory macrophages. Conversely, miR-125a-5p overexpression enhanced M2b polarization and altered other polarized populations, including increasing production of M2 markers. Indeed, in vitro overexpression of this microRNA altered macrophage phenotype towards that observed in SJIA.
Conclusions
Children with active SJIA have profound alterations in expression of microRNAs implicated in monocyte function and polarization. Additionally, one of these, miR-125a-5p, is a regulator of immunoregulatory M2b macrophages.
Introduction
Systemic juvenile idiopathic arthritis (SJIA) is considered a subtype of juvenile idiopathic arthritis, but is unique in having chronic arthritis as well as systemic features including intermittent but long lived fever, rash, serositis and adenopathy (1). It is also associated with high risk for development of macrophage activation syndrome (MAS), an overwhelming episode of inflammation causing coagulopathy, liver dysfunction and hemodynamic instability (2). In contrast to other subtypes of JIA, SJIA has features consistent with an autoinflammatory disease, characterized by dysfunction of innate immune effector cells including neutrophils, monocytes and macrophages, and natural killer cells, and production of high levels of proinflammatory cytokines (3). Children with SJIA demonstrate increased levels of circulating monocytes with upregulation of monocyte/macrophage differentiation genes (4–8), as well as high levels of monocyte-derived proinflammatory cytokines (4,9,10).
Myeloid cells adopt several distinct polarization phenotypes based upon stimuli and their resulting activation. These include classical “M1” proinflammatory macrophages, with elaboration of inflammatory cytokines and increased microbicidal activity, as well as several distinct subtypes of “M2” or alternatively activated macrophages participating in tissue remodeling, resolution of inflammation, and scavenger functions (11,12). The best studied of these alternative phenotypes is M2a, induced by IL-4 or IL-13, leading to macrophages optimized for phagocytosis, wound repair and tissue remodeling (13). Other M2 populations appear to have potent immunoregulatory phenotypes (14). Among these are M2b macrophages, which produce high levels of IL-6 and TNFα but also the anti-inflammatory cytokine IL-10 and CCL1 (15,16), and M2c macrophages that also secrete IL-10 and are involved in phagocytosis of apoptotic cells (17–19). Proper molecular control of macrophage polarization involves the integration and regulation of numerous cellular signaling pathways (20). In this regard, microRNAs are increasingly recognized as important modulators of macrophage polarization (21). These transcriptional negative regulators serve to fine-tune gene expression programs involved in cell differentiation, metabolism and immunity, and growing evidence suggests that miRNAs contribute to the pathogenesis of inflammatory diseases, including adult rheumatoid arthritis (RA) (22). However, to date only limited studies have examined expression of selected microRNAs in childhood rheumatic diseases including JIA (23–26).
Monocytes in active SJIA have a distinct functional phenotype, with features of both classical M1 activation and several alternatively activated populations, including regulatory monocytes. SJIA patients have high serum levels of both proinflammatory monocyte-derived cytokines and IL-10, typically produced by M2-like cells (5,9,10,27,28). In contrast, gene expression profiling has largely revealed patterns associated with alternative macrophage activation (4,6,7). Monocytes also display high surface expression of both M1-associated activation markers, as well as those associated with M2a and M2c activation including CD163 (29). Thus, monocytes in SJIA patients appear to have a mixed polarization phenotype, with features of both classically and alternatively activated populations; however the molecular mechanisms that regulate this activation in SJIA are unknown Several studies have examined microRNA expression profiles during macrophage polarization and found marked differences associated with distinct polarization states (21,30,31). However, few of these microRNAs have been directly shown to mediate macrophage polarization. One recent study found that miR-125a-5p, upregulated under several alternative activating conditions, suppressed classical activation in murine macrophages whilst promoting features of M2 activation (32).
Our long term goal is to understand the immune dysregulation associated with SJIA to improve diagnosis and treatment of this serious condition. To further this aim, microRNA expression profiles were determined from freshly isolated peripheral blood monocytes of children with SJIA. We hypothesized that monocytes from children with active SJIA would have altered expression of microRNAs involved in regulation of immune responses and macrophage polarization in particular. We also hypothesized that differentially expressed microRNA play key roles in macrophage polarization.
Patients and Methods
Patients
This study was approved by the Institutional Review Board of Cincinnati Children's Hospital Medical Center, and informed consent was obtained from all patients and/or their legal guardians. SJIA was diagnosed based on the International League of Associations for Rheumatology diagnostic criteria (1), with the exception of patient 09FT6 who was diagnosed with SJIA after a prolonged period of fever, systemic inflammation, adenopathy and serositis but without frank arthritis, and an extensive negative workup for infection and malignancy. Patients with new-onset SJIA were defined as newly diagnosed patients enrolled before initiation of any treatment aside from non-steroidal anti-inflammatory drugs. Note that for several patients, samples were obtained and treatment initiated with disease duration <6 weeks, in agreement with the operational definition of SJIA as described (33). Patients were considered as having active SJIA if they had presence of any active arthritis; any systemic features including rash, fever, adenopathy or hepatosplenomegaly; or elevated ESR or CRP. Patients with polyarticular JIA were diagnosed based upon ILAR criteria and had active disease with at least 4 active joints at time of sampling. Patients were considered to have clinically inactive disease (CID) based on the Wallace criteria (34). Patients were considered to have MAS based on diagnosis assigned by the treating physician. Patients were enrolled and peripheral blood samples were collected during routine clinic visits. Laboratory information was gathered from testing done during the routine clinical care of the patients.
Control patients were recruited from healthy children undergoing routine phlebotomy at CCHMC . Patients who were acutely ill, with a prior history of autoimmune or hematologic diseases, or taking anticonvulsant or immunosuppresive medications were excluded.
Sample collection and RNA extractions
Fresh whole blood was collected in ACD solution A vacutainer (Becton Dickinson, Franklin Lakes, NJ) tubes for each participant. CD14+ monocytes were immediately separated within 120 minutes of collection using whole blood CD14 microbeads and the AutoMACSPro (Miltenyi Biotec, San Diego, CA). Total RNA was extracted immediately following cell separation using the mirVana miRNA isolation kit (Life technologies, Carlsbad, CA). RNA quantity was assessed on the Qubit Fluorometric quantitation system (Life Technologies). RNA quality was also assessed on a Bioanalyzer (Agilent, Wilmington,DE) prior to running the TaqMan ®Array Human MicroRNA Card A (Life technologies).
Microarray analysis
20-25ng of total RNA in 2.1 μl was placed in a 3.15 μl reverse transcription reaction using the Megaplex™RT primer pool A (Life technologies). An additional pre-amplification step was performed using the Megaplex ™PreAmp primer pool A (Life technologies) in a 25 μl reaction. 9 μl of PreAmp reaction diluted 1:4 with the TaqMan mastermix was loaded onto the TaqMan ®Array MicroRNA card and ran through the Viia7 Realtime PCR instrument (Life technologies) according to the manufacturer's instructions. This card contains 384 individual microRNA and control RNA.
Cell culture and monocyte polarization
THP-1 human monocytic cell line (ATCC) was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. Prior to polarization THP-1 cells were differentiated overnight with addition of 50 ng/ml PMA. Primary human monocytes were isolated as described above and maintained in RPMI 1640 supplemented with 10% FCS.
THP-1 cells or primary human monocytes isolated as above were left untreated in RPMI with 10% FCS or polarized as follows: M1 (20 ng/ml IFNγ (R&D Systems, Minneapolis, MN) and 10 ng/ml LPS (Sigma Aldrich)), M2a (20 ng/ml IL-4 (R&D Systems)), M2b (100 ng/ml LPS in IgG-coated tissue culture wells), M2c (either 0.5 ng/ml TGFβ or 50 ng/ml IL-10 (R&D Systems)). IgG-coated wells were prepared as described (21). Cells were exposed to each polarizing conditions for the times indicated in each experiment.
MicroRNA antagomir and mimic
To knock-down microRNA expression, THP-1 cells were differentiated overnight as described above and then treated with 300nM negative control or miR-125a-5p micrOFF antagoMIR (Ribobio, Guangzhou, China). Macrophages were then polarized as described above and after 24 hours RNA was extracted using TRIzol (Life Technologies). To augment microRNA expression, THP-1 cells differentiated overnight with PMA were transfected with pre-miR microRNA precursors. Briefly, negative control pre-miR or miR-125a-5p (Dharmacon, Lafayette, CO) were incubated using RNAiMAX reagent using protocol described by Gantier and colleagues (35), with a final mimic concentration of 300nM. Macrophages were then polarized and RNA extraction as described above. Efficiency of microRNA inhibition or overexpression was determined using individual Taqman assays as described below.
Gene Expression
Realtime RT-PCR was used to quantify expression of transcripts induced during macrophage polarization. Briefly, total RNA was isolated as described and purity and concentration were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher, Waltham, MA). Approximately 1 μg of RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time PCR was performed in 20 μl reactions using Viia7 Realtime PCR Instrument with gene-specific primers and SYBR Green Supermix (Life Technologies). Message copy numbers were normalized against the copy number of the housekeeping gene GAPDH as described (36). Primers used are listed in Supplementary Table 1.
For quantification of miR-125a-5p, individual Taqman assays (Life Technologies) were performed using Bio-Rad iCycler thermocycler and a Viia7 Realtime PCR Instrument. Small RNA levels were calculated using the comparative CT method and normalized to RNU48, as this RNA has previously been reported to have stable expression levels during monocyte polarization (21).
Data Analysis
MicroRNA expression was analyzed using GeneSpring GX version 13 (Agilent, Wilmington, DE). The entities were normalized to the median of the housekeeping microRNA RNU48. Differentially expressed microRNA were identified by ANOVA with Benjamini and Hochberg correction for false discovery rate, and/or a Mann Whitney test. Hierarchical clustering using Wards linkage was used to group the microRNA and samples by expression pattern.
Results
Differential expression of microRNA in patients with active SJIA
RNA was obtained from peripheral blood monocytes from 27 patients with SJIA as well as 15 healthy controls. Demographic, clinical and laboratory features of patients and controls are shown in Table 1 and Supplementary Table 2. There was no significant difference between the age of patients with active established SJIA, CID or controls, and no difference in disease duration between patients with active established disease and CID. As expected, patients with new-onset disease were significantly younger than those with active established SJIA and controls (p=0.03), and had significantly higher serum ferritin, CRP and ESR (Table 1).
Table 1. Summary of clinical and laboratory characteristics for patients in this study.
| N | F:M | Median Age, years (IQR) |
Disease duration, months (IQR) |
Active arthritis (%) |
Active joints (IQR) |
Fever (%) |
Rash (%) |
HSM (%) |
Adenopathy | Ferritin, ng/ml (IQR) |
CRP, mg/dl (IQR) |
ESR, mm/hr (IQR) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| New-onset SJIA | 7 | 4:3 | 3 (2, 10) | 3 (1, 4) | 7 (100%) | 5 (3, 19) | 7 (100%) | 5 (71%) | 5 (71%) | 3 (43%) | 6489 (261, 12961) | 13.3 (8.9, 14.7) | 65 (61, 105) |
| Active established SJIA | 9 | 9:0 | 12 (8.5, 13.5) | 33 (19.5, 93.5) | 8 (89%) | 4 (1.5, 5.5) | 0 | 4 (44%) | 2 (22%) | 1 (11%) | 50 (20, 108) | 0.2 (0.2, 1.3) | 6 (4, 26) |
| CID SJIA | 11 | 8:3 | 8 (6, 18) | 80 (48, 103) | 0 | 0 | 0 | 0 | 0 | 0 | 36 (7, 92) | 0.3 (0.2, 0.7 | 7 (6, 10) |
| Controls | 13 | 6:7 | 13 (7.5, 15.5) | NA | NA | NA | NA | NA | NA | NA | ND | ND | ND |
HSM: Hepatomegaly and/or splenomegaly. CRP: C-reactive protein. ESR: erythrocyte sedimentation rate. NA: not applicable. ND: not determined.
For our primary analysis, all patients with active SJIA (new-onset or established disease) were compared both to patients with CID and healthy controls using multiway ANOVA. Taken together, 110 distinct microRNAs were identified with differential expression in monocytes from patients with active SJIA compared to both those with CID and control samples (Figure 1 and Supplementary Table 3). Several of these have been previously reported to be differentially expressed in PBMC and synoviocytes in adult rheumatoid arthritis (37–40). Several microRNA species with increased expression in active SJIA have also been reportedly induced in a variety of polarized macrophage phenotypes (21,30) (Table 2).
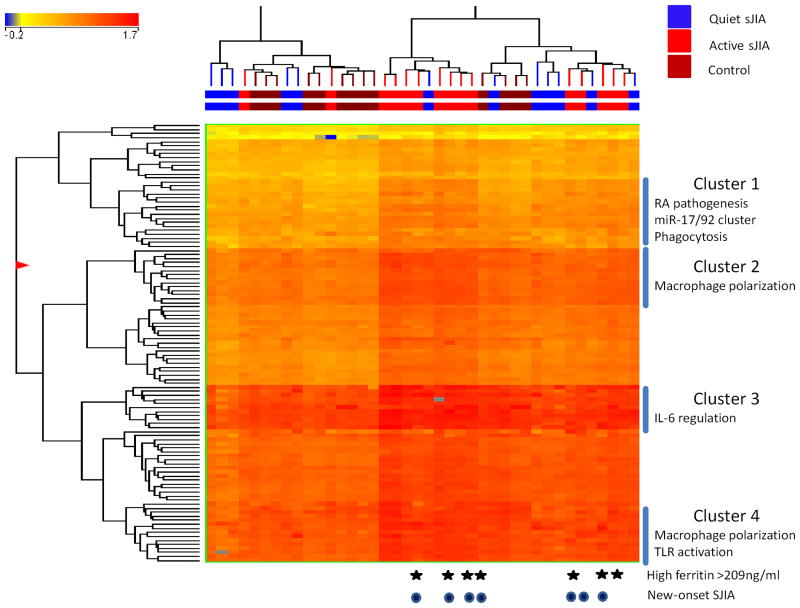
Figure 1.
Clustering analysis of differentially expressed microRNAs between patients with active SJIA and those with CID and healthy controls. The list of differentially expressed genes was generated using a three-way ANOVA comparing patients with active SJIA to those with CID and controls, with Benjamini and Hochberg corrected p<0.05. In this tree, each row represents a separate microRNA and each column represents a separate sample. The normalized expression levels for each microRNA in each sample is indicated by color. The colored line above the tree indicates patients with CID (blue), active SJIA (red) or controls (brown). Patients with high serum ferritin >209ng/ml are indicated with stars. Patients with new-onset (untreated) SJIA are indicated with dark circles.
Table 2. Specific microRNA upregulated in monocytes from children with active SJIA previously reported to be upregulated in macrophage polarization.
| microRNA ID | M1 (LPS+IFNγ) (21,30) | M2a (IL-4) (21,30) | M2b (LPS+IC) (21) | M2c (IL-10) (30) |
|---|---|---|---|---|
| miR-21 | ↑ | |||
| miR-22 | ↑ | |||
| miR-27a | ↑ | ↑ | ||
| miR-99a | ↑ | ↑ | ||
| miR-125a-5p | ↑ | ↑ | ↑ | |
| miR-146a | ↑ | |||
| miR-146b | ↑ | |||
| miR-181a | ↑ | ↑ | ||
| miR-193a | ↑ | |||
| miR-222 | ↑ | ↑ | ||
| miR-500 | ↑ | ↑ | ||
| miR-502 | ↑ |
LPS: lipopolysaccharide. IC: immune complexes
Differentially expressed microRNAs were analyzed by hierarchical clustering (Figure 1); two control samples had low RNA concentrations and >10% of microRNA not detected and were excluded from clustering analysis. Patients with active systemic JIA, both new-onset and established disease, clustered together into two distinct groups. No significant difference between these two clusters was observed with respect to disease duration, patient age, biologic treatment or high/low ferritin (see below). In contrast, patients with CID largely clustered together with control patients.
Our previous work revealed markedly different PBMC gene expression patterns between SJIA patients with elevated levels of ferritin (≥209 ng/ml) and those with normal or more modest increases in ferritin (4). Based on this, four specific microRNAs were identified with differential monocyte expression based on ferritin level: miR-16, miR-24, miR-186, and miR-342-3p (Supplementary Table 4). Finally examination of differentially expressed microRNAs between patients with new-onset disease and those with active established SJIA identified 3 microRNAs: miR-99a, miR-100 and miR-133a (Supplementary Table 5).
Several distinct clusters contributed to the heterogeneity of monocyte microRNA profiles (Figure 1). Cluster 1 included microRNAs previously shown to be elevated in RA including miR-142-3p, miR-146a/b and miR-150. It also included three members of the well-studied miR-17/92 cluster, which has recently been shown to contribute to inflammatory cytokine production by RA synovial fibroblasts (41,42). Finally it included microRNA which regulate macrophage phagocytosis (43). Cluster 2 contained several microRNAs that are increased in different polarized macrophage populations. Cluster 3 largely contains microRNAs with unknown functions in macrophages, although let-7e and miR-342-5p are involved in IL-6 regulation (44,45). Finally, cluster 4 also contained several microRNA increased in alternatively activated populations of macrophages, as well as those implicated in dampening effects of Toll-like receptor (46–48). Taken together our analysis demonstrates that monocytes from patients with active SJIA upregulate several clusters of microRNAs involved in macrophage polarization and function.
Differential expression of microRNA in patients with CID compared to controls
Although the clustering analysis shows that microRNA expression profiles from patients with CID were very similar to controls, 37 microRNAs were found with significant differences in expression levels in monocytes between these groups (Supplementary Table 6). Of these microRNAs with altered expression in patients with CID compared with controls, 31 had increased expression and 6 had decreased expression. Interestingly, of the 31 microRNAs with increased expression in children with CID, 21 were further elevated in active SJIA, suggestive of a continuum of monocyte activation. In contrast, levels of the remaining 10 microRNAs, while elevated compared to controls, were not significantly different between patients with active SJIA and CID. This includes miR-223, which has been shown to be elevated in monocytes infiltrating the synovium of patients with RA (37). This suggests that some aspects of monocyte microRNA expression seen in active SJIA persist despite patients having no clinical signs of active disease.
miR-125a-5p is elevated in patients with active SJIA
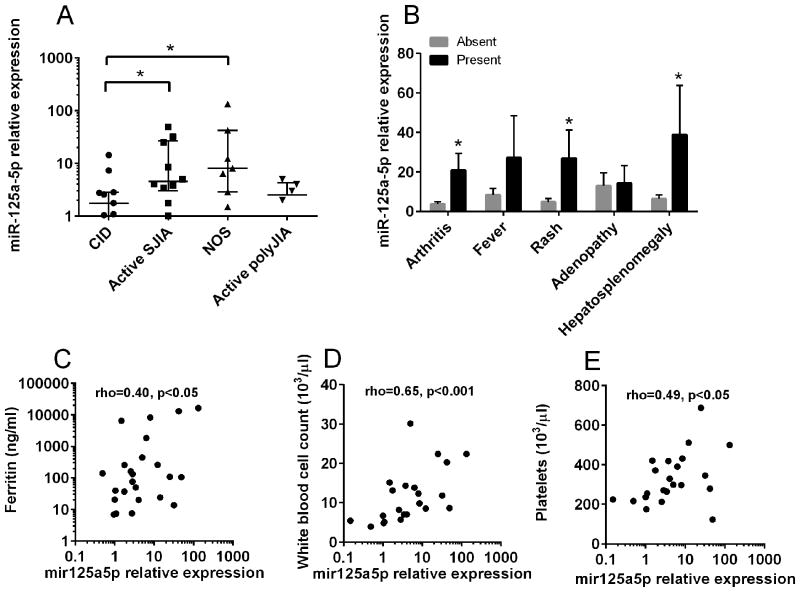
These results show that monocytes from patients with active SJIA have elevations in microRNAs that have been reported in multiple polarized populations (Table 2). One such microRNA is miR-125a-5p, which has recently been shown to play roles in alternative activation of murine macrophages (32). Here, miR-125a-5p was significantly elevated in patients with both new onset SJIA and active established SJIA compared to those with CID (Figure 2a). Increased monocyte expression of miR-125a-5p was also observed in patients with active arthritis, rash, or hepatosplenomegaly compared to patients lacking those features (Figure 2b). However miR-125a-5p expression did not correlate with the active joint count (R=0.06, p=0.75). There was significant correlation between monocyte miR-125a-5p levels and the serum ferritin level (Figure 2c), peripheral blood white cell count (Figure 2d), and platelet count (Figure 2e). No significant correlation between monocyte miR-125a-5p levels and other laboratory markers of inflammation, or with hemoglobin concentration, was observed (data not shown). To examine whether elevated monocyte miR-125a-5p levels were specific for active SJIA, as compared to other chronic childhood arthropathies, microRNA levels were also determined in monocytes from children with active polyarticular JIA. In patients with active polyarticular JIA, the levels of monocyte miR-125a-5p were significantly lower than those found in monocytes from patients with active SJIA (Figure 2A; p<0.05). In addition, miR-125a-5p levels in these patients was not significantly different from that found in clinically-inactive SJIA (Figure 2a). Taken together these data demonstrate increased monocyte expression of miR-125a-5p in patients with active SJIA with systemic features and that expression correlates with systemic inflammation, rather than degree of arthritis.
Figure 2.
Elevated levels of miR-125a-5p in monocytes from patients with active SJIA and associated with features of systemic disease. Expression levels of miR-125a-5p were determined by Taqman qRT-PCR and normalized against RNU48. A, Expression of miR-125a-5p in patients with active SJIA, new-onset disease (NOS), active polyarticular JIA, and CID. Lines indicate median and error bars indicate SEM. B, Expression of miR-125a-5p in patients with SJIA with presence (black bars) or absence (gray bars) of clinical features. *=p<0.05, Mann-Whitney U-test. C-E, Correlation of miR-125a-5p level with serum ferritin (C), white blood cell count (D), and platelet count (E) as determined by Spearman rank correlation method.
Increased expression of miR-125a-5p in M2 macrophages with regulatory phenotypes
Several studies have examined miR-125a-5p expression by polarized monocytes and macrophages, with differing results based on specific cell type and polarizing conditions (21,30,31). To explore this further, the expression of well characterized macrophage polarization markers was first verified in in vitro-stimulated cells. PMA-differentiated human THP-1 monocytic cells and primary CD14+ human monocytes were polarized as described towards M1, M2a, M2b, or M2c, after which gene expression was examined using quantitative RT-PCR. As expected, by 24 hours THP1 cells showed significant and specific upregulation of the interferon-induced chemokine MIG in M1 polarized cells, CCL13 in M2a polarized cells, CCL1 in M2b polarized cells, and CD163 in M2c-IL10 polarized cells (Supplementary Figure 1). IL-6 and TNFα were increased in both M1 and M2b conditions, and IL-10 in M1, M2b and M2c-IL10. Similar results were found in primary human monocytes from healthy donors with 42 hours of treatment (Supplementary Figure 2).
Using these conditions, in THP1 macrophages miR-125a-5p expression was significantly increased in cells polarized with LPS and immune complexes towards an M2b regulatory macrophage phenotype (Supplementary Figure 3a). There was also increased miR-125a-5p expression in M2b polarized primary monocytes, as well as when treated with TGFβ (Supplementary Figure 3b). Taken together, monocyte miR-125a-5p expression increases when cells are polarized towards M2 alternatively activated macrophages with immunoregulatory phenotypes, particularly M2b.
miR-125a-5p expression contributes to generation of M2b macrophage phenotypes
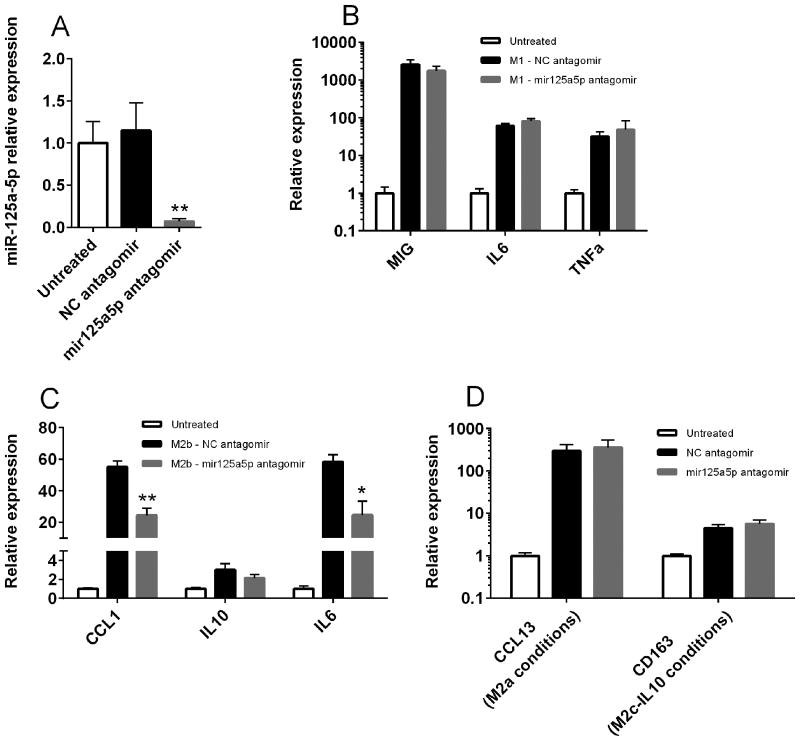
To examine whether miR-125a-5p contributed to the generation of polarized macrophage phenotypes, THP1 monocytes were treated with either negative control or miR-125a-5p antagomir, and mature microRNA levels were quantified using qRT-PCR. Treatment with specific miR-125a-5p antagomir significantly decreased cellular levels of this microRNA by up to 90% compared to negative control antagomir or untreated cells (Figure 3a). Treatment with the specific antagomir did not affect levels of the unrelated microRNA miR-155 and did not affect cell viability (data not shown).
Figure 3.
miR-125a-5p is required for M2b macrophage polarization. A, THP-1 monocytic cells were differentiated with PMA and left untreated (white bar) or treated with negative control (black bar) or specific miR-125a-5p antagomir (gray bar) as described. miR-125a-5p levels were determined by individual Taqman assays and normalized to RNU48. Data are representative of three independent experiments performed in triplicate. B-D, THP-1 monocytic cells were differentiated with PMA and left untreated or treated with negative control or miR-125a-5p antagomir as described. Cells were then polarized under M1 (B), M2b (C), M2a or M2c-IL10 (D) conditions for 24 hours as described. Target gene expression was determined using qRT-PCR and normalized to GAPDH. Data are pooled from three independent experiments performed in triplicate. **=p<0.01; *=p<0.05 as determined by student's t-test.
Inhibition of miR-125a-5p using specific antagomir did not affect M1 polarization of THP-1 macrophages (Figure 3b). In contrast, when miR-125a-5p antagomir treated macrophages were exposed to M2b polarizing conditions, there was a >50% reduction in expression of two M2b-associated transcripts, IL-6 and CCL1 (Figure 3c). There was also a slight but non-significant decrease in IL-10 expression when cells were exposed to M2b polarizing conditions. Interestingly, miR-125a-5p antagomir treatment also did not alter M2a polarization as determined by CCL13 expression or M2c-IL10 polarization as determined by CD163 expression (Figure 3d). Taken together, these findings suggest that miR-125a-5p contributes to M2b polarization but is dispensable for generation of other polarized macrophage phenotypes.
Overexpression of miR-125a-5p enhances expression of genes associated with regulatory macrophages
Finally, we examined whether enforced overexpression of miR-125a-5p could alter markers of macrophage polarization. THP-1 cells were transfected with negative control microRNA mimic or miR-125a-5p mimic. Cells transfected with miR-125a-5p mimic had substantially increased levels of microRNA (Figure 4a), which remained elevated for >96 hours (data not shown). When cells were polarized towards M1 conditions, transfection of miR-125a-5p mimic significantly increased expression of the M2-associated genes CCL1 and CD163, without altering expression of IFNγ -induced chemokines such as MIG (Figure 4b). Similarly, when transfected cells were polarized towards M2b, there was significantly increased the expression of several polarization-induced genes including CCL1 and IL-6 (Figure 4c). Mimic overexpression also significantly increased levels of CD163 transcript, which was not otherwise induced by M2b conditions. Interestingly, miR-125a-5p mimic appeared to diminish the magnitude of both M2a and M2c polarization, with reduced induction of CCL13 upon IL-4 treatment and CD163 and IL-10 mRNA expression in M2c-IL10 conditions (Figure 4d). Taken together, these findings suggest that miR-125a-5p contributes to the polarization of M2 alternatively activated macrophages and specifically the balance between M2b regulatory macrophages and other forms of M2 macrophages. Indeed, upregulation of miR-125a-5p promoted a monocyte phenotype reminiscent of that seen in SJIA, with increased expression of IL-6 and several M2 markers including CD163.
Figure 4.
Overexpression of miR-125a-5p enhances expression of regulatory macrophage-associated genes. A, THP-1 monocytic cells were differentiated with PMA and transfected with negative control (black bar) or miR-125a-5p mimic (white bar). miR-125a-5p expression was determined by individual Taqman assays and normalized to RNU48. Data is representative of three independent experiments performed in triplicate. B-E, THP-1 monocytic cells were differentiated with PMA, transfected as described, and polarized under M1 (B), M2b (C), M2a (D), or M2c-IL10 (E) conditions for 24 hours. Target gene expression was determined using qRT-PCR and normalized to GAPDH. Data are pooled from three independent experiments performed in triplicate. *=p<0.01 as determined by student's t-test.
Discussion
Increasing evidence supports the notion that the early systemic phase of SJIA fits the paradigm of autoinflammatory diseases, with monomyeloid cells serving as key effector cells (3). However the precise phenotype and function of monocytes in this disease remains unclear. Here, we find that monocytes from patients with active SJIA have profound alternations in microRNA expression profiles, with upregulation of specific molecules involved in macrophage polarization, cytokine production, and regulation of TLR signaling. One of these, miR-125a-5p, was highly upregulated in monocytes from patients with active SJIA. miR-125a-5p is upregulated in M2b regulatory macrophages, and this work shows for the first time that this microRNA contributes to the generation of this macrophage phenotype. Several lines of evidence support the importance of miR-125a-5p elevation in SJIA monocytes. First, although we lack a validation cohort, miR-125a-5p was elevated in monocytes from two distinct groups of SJIA patients – those with untreated new-onset disease, and those with persistently active disease despite treatment. Second, miR-125a-5p levels were not elevated in monocytes from children with active polyarticular JIA. Although polyarticular JIA is not associated with the degree of systemic inflammation seen in SJIA, there is significant evidence for activation and polarization of circulating monocytes in this disease (16,49). Third, miR-125a-5p levels correlated with the degree of systemic inflammation as measured by serum ferritin level, white blood cell and platelet counts. Fourth, we and others have found miR-125a-5p similarly elevated in M2 monocytes with regulatory features, resembling the phenotype seen in SJIA. Fifth and most crucially, overexpression of miR-125a-5p altered monocyte polarization by enhancing features seen in active SJIA while diminishing other polarized phenotypes. Taken together, these data suggest that microRNAs contribute to monocyte activation and function in SJIA.
This study represents the first systematic analysis of cellular microRNAs in any subtype of JIA. Recently a targeted analysis of serum microRNAs found elevated miR-223 in SJIA patients regardless of disease activity, which interestingly is similar to that seen with monocyte miR-223 levels in the present study (Supplementary Table 3) (24). We also observed upregulation of monocyte miR-146a, which is well studied in adult RA and increased in PBMC, synovial fibroblasts and synovial fluid itself from RA patients (37–40). However, several other microRNAs that are implicated in RA pathogenesis did not appear to be significantly elevated in monocytes from patients with SJIA. Most notably this includes miR-155, which is critical for production of adaptive immune responses after vaccination (50). This finding may highlight important pathogenic differences between diseases such as SJIA and autoimmune disorders such as RA.
This study also identified several intriguing microRNAs which may contribute to the pathogenesis of SJIA. Several members of the miR-17/92 cluster were upregulated in monocytes from patients with active SJIA. The role of this cluster in inflammation remains unclear, but it appears to be induced by TNFα in RA synoviocytes, and potentiates the NF-kB pathway leading to increased matrix-degrading enzymes and inflammatory cytokines (51). SJIA monocytes also demonstrated upregulation of several members of the let-7 family, high abundance microRNAs involved in cellular differentiation (52). Several members of the let-7 family are regulated by STAT3, and let-7a directly targets IL-6 mRNA (44). Among the microRNAs with increased expression in patients with high levels of ferritin is miR-16, which has recently been shown to promote the TLR-mediated induction of proinflammatory cytokines including IL-6 (53). Similarly miR-133a, significantly elevated in patients with new-onset SJIA, may potentiate activation of the NLRP3 inflammasome (54). Thus, considering the pivotal role for IL-1 in SJIA, dysregulation of this microRNA could contribute to the hyperinflammation seen early in the disease.
Interestingly, this study found 37 microRNA with significant differences in expression between control monocytes and those from SJIA patients with CID. This suggests that there is persistent monocyte activation and dysfunction in patients with CID, despite no outward signs of systemic inflammation. Indeed, we have recently shown that patients treated with IL-1 blockade remain at risk for MAS, even when the underlying disease appeared well controlled (55). To this end, it is interesting to note that nearly one-third (9/31) of microRNA with increased expression in CID monocytes were among the most highly elevated during active hemophagocytic lymphohistiocytosis (56).
We also identified numerous microRNAs reported to be upregulated during macrophage polarization (Table 2). Interestingly, there was not a clear expression profile consistent with a single polarized phenotype, but rather a mixture of microRNAs upregulated in multiple different conditions. This is consistent with cytokine measurements, gene expression and flow cytometry studies suggesting that monocytes in SJIA represent a mixed polarization population (4,5,7,29). This may also reflect the fact that polarized phenotypes are largely an in vitro construct, and how well they represent the phenotype of activated monocytes in inflammatory diseases is unclear.
Several reports have highlighted dysregulation of miR-125a in chronic inflammatory diseases (57,58). Notably, recent works has demonstrated that miR-125a contributes to autoimmune diseases by stabilizing T regulatory cell homeostasis (59). In this study, monocytes from patients with active SJIA had increased expression of miR-125a-5p compared to monocytes from patients with CID. The highest levels of miR-125a-5p were in patients with new-onset SJIA, who had high levels of inflammation and systemic features associated with this disease. In contrast, while there was a higher level of miR-125a-5p in SJIA patients with active arthritis compared to those without arthritis, there was no correlation with the active joint count and no elevation in active polyarticular JIA. This suggests that miR-125a-5p expression may be more strongly associated with the systemic phase of SJIA, and could support the biphasic models of SJIA pathogenesis (60).
Mir-125a-5p expression appears to be induced by signaling through TLR4 and inhibited by IFNγ and IL-4 (32,61). As such, we and others (21,30) have found the highest levels of miR-125a-5p in M2b macrophages conditions, with some induction in M2c macrophages induced by both TGFβ and IL-10 as well. Notably these are all populations of regulatory macrophages with potent immunomodulatory properties. Conflicting results regarding the expression of miR-125a-5p (and other microRNA) in prior studies are likely due to differences in the species, source (primary vs immortalized) and specific activating conditions used. Interestingly, we also find that expression of miR-125a-5p contributes to full generation of M2b macrophages. Knockdown of miR-125a-5p using a specific microRNA antagomir decreased expression of IL-6 and the M2b-specific chemokine CCL1, and miR-125a-5p overexpression enhanced expression of CCL1, IL-6, as well as CD163. Overexpression of miR-125a-5p also diminished M2a and M2c polarization, suggesting this microRNA may influence macrophage phenotype within the spectrum of M2 macrophages. Banerjee and colleagues have previously shown that miR-125a-5p may impact polarization in murine macrophages treated with either GM-CSF towards an M1 phenotype or M-CSF towards an M2 phenotype (32). The functional consequence of miR-125a-5p-mediated regulatory monocyte and macrophage polarization in humans is currently unknown and requires further study. In particular, whether miR-125a-5p-mediated monocyte activation drives disease pathology in SJIA or drives counter-regulatory functions is a critical question that remains unknown.
It is particularly noteworthy that miR-125a-5p is involved in the generation of M2b macrophages. They are a prototypical example of the class of IL-10 producing regulatory macrophages, and distinct from M1 classically activated macrophages, as well as M2a “alternatively activated” macrophages induced by IL-4 or IL-13 (14). Indeed, our data shows that miR-125a-5p is dispensable for other macrophages phenotypes, but mediates full production of IL-6 and CCL1, a chemokine specifically produced by M2b regulatory macrophages (16). IL-6 levels are highly elevated in patients with active SJIA (9), and IL-6 blockade is highly effective at treatment of this condition (62). CCL1 expression has not been specifically examined in monocytes from SJIA; our previous gene expression studies of PBMC found a small but non-specific increase in CCL1 expression (4). Overexpression of miR-125a-5p also appears to affect the balance between IL-6 and CCL1-producing M2b macrophages and other forms of M2 macrophages, including CD163-expressing M2c macrophages. While both of these populations represent regulatory macrophages able to produce IL-10, M2c macrophages are highly efficient at engulfment of apoptotic cells (17), and CD163 is a hallmark of the emergence of MAS clinical features (63,64). One could speculate that miR-125a-5p expression is involved in the generation of hemophagocytes and the pathogenesis of MAS.
Several putative targets for miR-125a-5p have been proposed which could mediate its effects on macrophage polarization. Banerjee and colleagues demonstrated that the effects of miR-125a-5p on murine macrophage phenotypes were mediated at least in part by the transcription factor Kruppel-like factor 13 (32). MiR-125b, which is a homolog of miR-125a, has been shown to regulate interferon regulatory factor 4 (IRF4) in macrophages (65). Interestingly, IRF4 is induced in M2a polarized macrophages, and regulates a subset of IL-4 induced genes in this phenotype (66).
In summary, we report the first comprehensive examination of cellular microRNA expression in children with SJIA. One hundred ten specific microRNAs were identified with increased expression in monocytes from children with active SJIA, including both microRNAs with known roles in innate immune responses as well as those without known functions in monocytes. There was also significant microRNA dysregulation persisting in monocytes from patients with no overt signs of active disease. Monocyte miR-125a-5p is significantly elevated in patients with active SJIA, associated with presence of systemic features and correlates with laboratory markers of inflammation. We also show that miR-125a-5p contributes to regulatory macrophage polarization. Indeed, in vitro overexpression of miR-125a-5p appeared to drive macrophages towards a polarization phenotype reminiscent of that seen in active SJIA, with expression of both inflammatory cytokines and M2-associated markers. This works suggests that microRNAs could serve as potential biomarkers for both the diagnosis and response to therapy in SJIA. In addition, microRNAs could be attractive targets for therapies aimed at modulating the function of monocytes and macrophages to control the unremitting inflammation associated with this disease.
Supplementary Material
Acknowledgments
We would like to thank all the patients and families who agreed to participate in this study. We also acknowledge our clinical research coordinators Allen Watts, Krista Solomon and Anne Johnson for patient recruitment. We thank the Cell Processing and Manipulation Core in the Translational Cores, and the physicians and nurses at CCHMC for obtaining and processing peripheral blood samples used for monocyte polarization. We also thank the CCHMC Translational Research Trials Office for providing the regulatory and administrative support for this endeavor. Dr. Schulert is supported by K12 HL119986 from the National Institutes of Health and a Scientist Development Award from the Rheumatology Research Foundation. Dr. Grom is supported by RO1-AR059049 and Drs. Lovell and Grom by PO1-AR048929 from the National Institutes of Health.
Footnotes
COI statement: Dr. Grom has received consulting fees and has research collaborations with Novartis and NovImmune. Daniel J. Lovell has received consulting fees from AbbVie Inc., AstraZeneca, Centocor, Bristol-Myers Squibb, Pfizer, Regeneron, Hoffman La-Roche, Novartis, UBC, Xoma, and Genentech, served on speaker bureaus for Wyeth Pharmaceuticals, and served on data and safety monitoring boards for Amgen and Forest Research. All other authors declare no conflicts of interest.
References
- 1.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 2.Ravelli A, Grom AA, Behrens EM, Cron RQ. Macrophage activation syndrome as part of systemic juvenile idiopathic arthritis: diagnosis, genetics, pathophysiology and treatment. Genes Immun. 2012;13:289–298. doi: 10.1038/gene.2012.3. [DOI] [PubMed] [Google Scholar]
- 3.Mellins ED, Macaubas C, Grom AA. Pathogenesis of systemic juvenile idiopathic arthritis: some answers, more questions. Nat Rev Rheumatol. 2011;7:416–426. doi: 10.1038/nrrheum.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–3804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 5.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogilvie EM, Khan A, Hubank M, Kellam P, Woo P. Specific gene expression profiles in systemic juvenile idiopathic arthritis. Arthritis Rheum. 2007;56:1954–1965. doi: 10.1002/art.22644. [DOI] [PubMed] [Google Scholar]
- 8.Macaubas C, Nguyen K, Deshpande C, Phillips C, Peck A, Lee T, et al. Distribution of circulating cells in systemic juvenile idiopathic arthritis across disease activity states. Clin Immunol. 2010;134:206–216. doi: 10.1016/j.clim.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benedetti F de, Massa M, Robbioni P, Ravelli A, Burgio GR, Martini A. Correlation of serum interleukin-6 levels with joint involvement and thrombocytosis in systemic juvenile rheumatoid arthritis. Arthritis Rheum. 1991;34:1158–1163. doi: 10.1002/art.1780340912. [DOI] [PubMed] [Google Scholar]
- 10.Ling XB, Park JL, Carroll T, Nguyen KD, Lau K, Macaubas C, et al. Plasma profiles in active systemic juvenile idiopathic arthritis: Biomarkers and biological implications. Proteomics. 2010;10:4415–4430. doi: 10.1002/pmic.201000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 13.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 14.Fleming BD, Mosser DM. Regulatory macrophages: setting the threshold for therapy. Eur J Immunol. 2011;41:2498–502. doi: 10.1002/eji.201141717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:1298–307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sironi M. Differential regulation of chemokine production by Fc receptor engagement in human monocytes: association of CCL1 with a distinct form of M2 monocyte activation (M2b, Type 2) J Leukoc Biol. 2006;80:342–349. doi: 10.1189/jlb.1005586. [DOI] [PubMed] [Google Scholar]
- 17.Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol. 2012;189:3508–20. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–63. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 19.Williams L, Jarai G, Smith A, Finan P. IL-10 expression profiling in human monocytes. J Leukoc Biol. 2002;72:800–9. [PubMed] [Google Scholar]
- 20.Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–7. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo X, Ranade K, Talker R, Jallal B, Shen N, Yao Y. microRNA-mediated regulation of innate immune response in rheumatic diseases. Arthritis Res Ther. 2013;15:210. doi: 10.1186/ar4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucherini OM, Obici L, Ferracin M, Fulci V, McDermott MF, Merlini G, et al. First report of circulating microRNAs in tumour necrosis factor receptor-associated periodic syndrome (TRAPS) PLoS One. 2013;8:e73443. doi: 10.1371/journal.pone.0073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamiya Y, Kawada J-I, Kawano Y, Torii Y, Kawabe S, Iwata N, et al. Serum microRNAs as Potential Biomarkers of Juvenile Idiopathic Arthritis. Clin Rheumatol. 2015 doi: 10.1007/s10067-015-2922-1. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Wu F, Xin L, Su G, He F, Yang Y, et al. Differential plasma microRNAs expression in juvenile idiopathic arthritis. Mod Rheumatol. 2015:1–34. doi: 10.3109/14397595.2015.1060663. [DOI] [PubMed] [Google Scholar]
- 26.Jiang K, Hu Z, Chen Y, Jarvis JN. A176: deep sequencing reveals differential small RNA expression in serum exosomes of children with juvenile idiopathic arthritis. Arthritis Rheumatol. 2014;66(Suppl 1):S230. [Google Scholar]
- 27.Shimizu M, Yachie A. Compensated inflammation in systemic juvenile idiopathic arthritis: role of alternatively activated macrophages. Cytokine. 2012;60:226–232. doi: 10.1016/j.cyto.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Maeno N, Takei S, Imanaka H, Yamamoto K, Kuriwaki K, Kawano Y, et al. Increased interleukin-18 expression in bone marrow of a patient with systemic juvenile idiopathic arthritis and unrecognized macrophage-activation syndrome. Arthritis Rheum. 2004;50:1935–1938. doi: 10.1002/art.20268. [DOI] [PubMed] [Google Scholar]
- 29.Macaubas C, Nguyen KD, Peck A, Buckingham J, Deshpande C, Wong E, et al. Alternative activation in systemic juvenile idiopathic arthritis monocytes. Clin Immunol. 2012;142:362–372. doi: 10.1016/j.clim.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobos Jiménez V, Bradley EJ, Willemsen AM, Kampen AHC van, Baas F, Kootstra NA. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol Genomics. 2014;46:91–103. doi: 10.1152/physiolgenomics.00140.2013. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang M, Zhong M, Suo Q, Lv K. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med. 2013;31:797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. 2013;288:35428–35436. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeWitt EM, Kimura Y, Beukelman T, Nigrovic PA, Onel K, Prahalad S, et al. Consensus treatment plans for new-onset systemic juvenile idiopathic arthritis. Arthritis Care Res. 2012;64:1001–1010. doi: 10.1002/acr.21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace CA, Giannini EH, Huang B, Itert L, Ruperto N. American College of Rheumatology provisional criteria for defining clinical inactive disease in select categories of juvenile idiopathic arthritis. Arthritis Care Res (Hoboken) 2011;63:929–36. doi: 10.1002/acr.20497. [DOI] [PubMed] [Google Scholar]
- 35.Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J Immunol. 2008;180:2117–24. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 36.Sikora KA, Fall N, Thornton S, Grom AA. The limited role of interferon-gamma in systemic juvenile idiopathic arthritis cannot be explained by cellular hyporesponsiveness. Arthritis Rheum. 2012;64:3799–3808. doi: 10.1002/art.34604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanczyk J, Pedrioli DML, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–9. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 39.Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord. 2010;11:209. doi: 10.1186/1471-2474-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippe L, Alsaleh G, Pichot A, Ostermann E, Zuber G, Frisch B, et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann Rheum Dis. 2013;72:1071–9. doi: 10.1136/annrheumdis-2012-201654. [DOI] [PubMed] [Google Scholar]
- 42.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naqvi AR, Fordham JB, Nares S. miR-24, miR-30b, and miR-142-3p Regulate Phagocytosis in Myeloid Inflammatory Cells. J Immunol. 2015;194:1916–1927. doi: 10.4049/jimmunol.1401893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalán-Campos J, et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–19. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 46.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–7. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation. Innate Immun. 2012;18:846–55. doi: 10.1177/1753425912443903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Chen Q, Song Y, Lai L, Wang J, Yu H, et al. MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 2011;585:1963–8. doi: 10.1016/j.febslet.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 49.Prokopec KE, Berntson L, Öman A, Kleinau S. Up regulated complement and fc receptors in juvenile idiopathic arthritis and correlation with disease phenotype. J Clin Immunol. 2012;32:540–50. doi: 10.1007/s10875-012-9657-4. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trenkmann M, Brock M, Gay RE, Michel BA, Gay S, Huber LC. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 2013;65:916–27. doi: 10.1002/art.37834. [DOI] [PubMed] [Google Scholar]
- 52.Kolenda T, Przybyła W, Teresiak A, Mackiewicz A, Lamperska KM. The mystery of let-7d - a small RNA with great power. Contemp Oncol (Poznań, Poland) 2014;18:293–301. doi: 10.5114/wo.2014.44467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou R, Li X, Hu G, Gong A-Y, Drescher KM, Chen X-M. miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One. 2012;7:e30772. doi: 10.1371/journal.pone.0030772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bandyopadhyay S, Lane T, Venugopal R, Parthasarathy PT, Cho Y, Galam L, et al. MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2. Biochem Biophys Res Commun. 2013;439:407–12. doi: 10.1016/j.bbrc.2013.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grom AA, Ilowite NT, Pascual V, Brunner HI, Martini A, Lovell D, et al. Rate and Clinical Presentation of Macrophage Activation Syndrome in Patients With Systemic Juvenile Idiopathic Arthritis Treated With Canakinumab. Arthritis Rheumatol. 2016;68:218–228. doi: 10.1002/art.39407. [DOI] [PubMed] [Google Scholar]
- 56.Sumegi J, Nestheide S, Aronow B, Fletcher D, Keddache M, Villanueva J, et al. MicroRNA activation signature in patients with hemophagocytic lymphohistiocytosis and reversibility with disease-specific therapy. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 58.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8:e69118. doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan W, Zhu S, Dai D, Liu Z, Li D, Li B, et al. MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat Commun. 2015;6:7096. doi: 10.1038/ncomms8096. [DOI] [PubMed] [Google Scholar]
- 60.Nigrovic PA. Review: is there a window of opportunity for treatment of systemic juvenile idiopathic arthritis? Arthritis Rheumatol (Hoboken, NJ) 2014;66:1405–13. doi: 10.1002/art.38615. [DOI] [PubMed] [Google Scholar]
- 61.Eigsti RL, Sudan B, Wilson ME, Graff JW. Regulation of activation-associated microRNA accumulation rates during monocyte-to-macrophage differentiation. J Biol Chem. 2014;289:28433–47. doi: 10.1074/jbc.M114.599316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benedetti De F, Brunner HI, Ruperto N, Kenwright A, Wright S, Calvo I, et al. Randomized trial of tocilizumab in systemic juvenile idiopathic arthritis. N Engl J Med. 2012;367:2385–2395. doi: 10.1056/NEJMoa1112802. [DOI] [PubMed] [Google Scholar]
- 63.Bleesing J, Prada A, Siegel DM, Villanueva J, Olson J, Ilowite NT, et al. The diagnostic significance of soluble CD163 and soluble interleukin-2 receptor alpha-chain in macrophage activation syndrome and untreated new-onset systemic juvenile idiopathic arthritis. Arthritis Rheum. 2007;56:965–971. doi: 10.1002/art.22416. [DOI] [PubMed] [Google Scholar]
- 64.Behrens EM, Beukelman T, Paessler M, Cron RQ. Occult macrophage activation syndrome in patients with systemic juvenile idiopathic arthritis. J Rheumatol. 2007;34:1133–1138. [PubMed] [Google Scholar]
- 65.Chaudhuri AA, So AY-L, Sinha N, Gibson WSJ, Taganov KD, O'Connell RM, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–8. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chartouni El C, Schwarzfischer L, Rehli M. Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology. 215:821–5. doi: 10.1016/j.imbio.2010.05.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.