Abstract
Mutations in cardiac troponin C (D75Y, E59D and G159D), a key regulatory protein of myofilament contraction, have been associated with dilated cardiomyopathy (DCM). Despite reports of altered myofilament function in these mutants, the underlying molecular alterations caused by these mutations remain elusive. Here we investigate in silico the intra-molecular mechanisms by which these mutations affect myofilament contraction. Based on the location of cardiac troponin C (cTnC) mutations, we tested the hypothesis that intra-molecular effects can explain the altered myofilament calcium sensitivity of force development for D75Y and E59D cTnC, whereas altered cardiac troponin C – troponin I (cTnC-cTnI) interaction contributes to the reported contractile effects of the G159D mutation. We employed a multi-scale approach combining molecular dynamics (MD) and Brownian dynamics (BD) simulations to estimate cTnC calcium association and hydrophobic patch opening. We then integrated these parameters into a Markov model of myofilament activation to compute the steady-state force-pCa relationship. The analysis showed that myofilament calcium sensitivity with D75Y and E59D can be explained by changes in calcium binding affinity of cTnC and the rate of hydrophobic patch opening, if a partial cTnC interhelical opening angle (110°) is sufficient for cTnI switch peptide association to cTnC. In contrast interactions between cTnC and cTnI within the cardiac troponin complex must also be accounted for to explain contractile alterations due to G159D. In conclusion, this is the first multi-scale in silico study to elucidate how direct molecular effects of genetic mutations in cTnC translate to altered myofilament contractile function.
Graphical Abstract
Introduction
Dilated cardiomyopathy (DCM), a widely prevalent acquired cardiac muscle disease, is characterized by dilatation of the heart and impaired contractile function of the ventricles1. DCM is the most common reason for heart transplantation, occurring in approximately five to eight people per 100,000 and causing approximately 10,000 deaths and 46,000 hospitalizations each year in the United States. It is estimated that 30% to 35% of DCM cases are linked to mutations in the genes encoding contractile proteins 2–3, including titin, actin, myosin, tropomyosin and all three subunits of cardiac troponin (cTn) 4. By altering key myofilament properties such as calcium sensitivity of myofilaments, mutations in cTn may directly contribute to the development of DCM. A key motivation of this study is to understand how the intramolecular dynamics underlying known DCM mutations in the calcium binding domain of cTnC translate to altered myofilament contractile response.
Cardiac Troponin (cTn), a macromolecular protein in the thin filament complex of the sarcomere in cardiomyocytes, is a key regulator of contractile mechanics. Structurally, cTn consists of three subunits: troponin C (cTnC), troponin I (cTnI) and troponin T (cTnT)5. Binding of the signaling ion Ca2+ to the N-terminal regulatory domain of cTnC results in structural and dynamic changes that trigger contraction6–7. As a consequence of calcium binding to the regulatory domain of cTnC, there is an increased exposure of hydrophobic residues on the surface of cTnC (between helices A and B). This precipitates stronger association with the switch peptide region of cTnI and reduced inhibitory peptide interaction of cTnI with actin, allowing increased tropomyosin mobility that allows strong myosin binding and contraction 6,8. Structurally, TnC is one of the best studied regulatory proteins in the sarcomere. Numerous structures of the TnC regulatory domain 9–12 as well as the full-length, two-lobed molecule exist 13–14. The regulatory domain, a highly α-helical molecule that constitutes the first 89 N-terminal residues of the troponin C protein, consists of five a-helices (N, A–D). Previous computational studies have addressed modeling many cTn-related processes on a molecular scale 15–23.
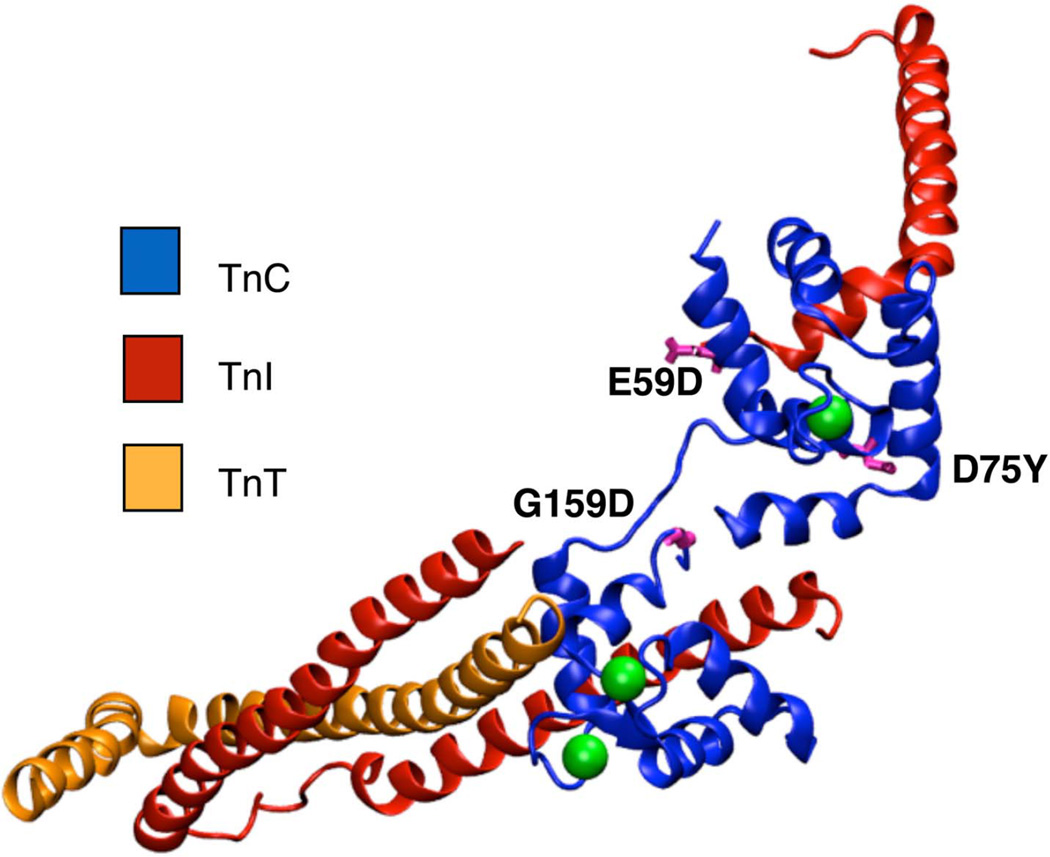
At least three clinically observed mutations in cTnC have been proposed to be associated with DCM (Figure 1). Two of these mutations, D75Y and E59D, are located in or near the Ca2+-binding site II in the regulatory domain of cTnC. Studies in skinned cardiomyocytes have reported a marked decrease in calcium sensitivity for the double mutant (D75Y/E59D) and D75Y alone, in spite of the fact that the mutations do not influence the intracellular calcium homeostasis 18,24. Interestingly the E59D mutation alone was phenotypically comparable to wildtype cTnC in its effects on myofilament function 18,24. Another cTnC DCM mutation, G159D, is located in the C-terminus of cTnC, distal to site II, in close proximity to the I-T arm interaction region of cTnI and cTnC. Experimental reports for the G159D mutation have suggested contrasting phenotypes, with increased myofilament Ca2+ sensitivity in humans 2 and unaltered contractility in rats25.
Figure 1.
Crystal structure of cTn (cTnC in blue, cTnI in red, cTnT in orange, calcium in green) complex showing the location of the DCM mutations D75Y, E59D and G159D (in pink).
The molecular mechanisms by which these cTnC mutations cause contractile alterations remain unclear. Intra-molecular dynamic changes in cTnC can cause alterations in: (a) the calcium binding affinity of cTnC; (b) the rate of calcium dissociation from cTnC; (c) the forward rate of cTnC conformation transition; (d) the reverse rate of cTnC conformation transition; and (e) the structure of cTnC such that there are differences in charge within the exposed hydrophobic patch. Additionally, cTnC mutations can affect its interactions with cTnI and downstream contractile proteins. D75Y and E59D are closer to the regulatory calcium binding site of cTnC, while G159D lies distal to the site II in the C-terminus of cTnC and is closer to the cTnC-cTnI IT arm binding site. Based on the locations of these mutations, we hypothesized that for the D75Y and E59D mutations, the intra-molecular effects within cTnC are sufficient to explain reported contractile defects, whereas altered cTnC-cTnI interaction is required to explain the effects of G159D on contraction.
To understand how molecular level changes precipitated by these mutations manifest at the level of myofilament contraction we employed multi-scale computational models, combining molecular dynamics, Brownian dynamics and Markov models of myofilament activation. To address our hypotheses, we used a modified, previously described model26 that includes Markov states representing the molecular events of calcium binding to cTnC and conformational changes in cTnC. We derived state transition rates from BD and MD simulations and compared computed steady-state force-pCa curves with experimental measurements from protein exchange experiments in rat cardiac myofilaments. This novel approach of coupling molecular-level effects to models of myofilament contraction can elucidate whether the observed molecular processes are sufficient to explain experimentally measured myofilament properties.
Material and Methods
System preparation
Four different human N-terminal or full length cardiac troponin C ((N)cTnC) Ca2+-bound systems were investigated: wildtype cTnC, G159D cTnC (both based on PDBID 1J1E), D75Y NcTnC and E59D NcTnC (both based on PDBID 1AP4). The initial preparation of the systems, including introduction of mutations, is described in 15–16. The Ca2+-bound cTnC and cTnC-G159D systems contained three bound Ca2+ ions (one in N-terminal lobe and two in C-terminal lobe). The Ca2+-bound NcTnC-D75Y and NcTnC-E59D systems contained one bound Ca2+ ion (in N-terminal lobe). No free unbound Ca2+ ions were present. Well-equilibrated structures after 150 ns of NAMD MD were used as input for the Anton system preparation. The scripts convertNAMDtoMaestro.2.5.py, viparr.py and build_constraints.py were used to generate cms system files. An in-house perl script was used to fix the atomic number of potassium ions to 19 before running viparr. The script mae2dms converted the cms files into dms format. Subsequently guess_chem, refinesigma and subboxer were used to prepare all the input files for the actual simulations.
Anton MD simulations
All Anton simulations were performed under the NPT ensemble at 300 K using a Berendsen thermostat and barostat. Bonds involving hydrogen atoms were constrained using the SHAKE algorithm 27, allowing for a time step of 2 fs. Structures were saved every 100.002 ps. Production runs were carried out on the 512 node Anton machine, running 75 jobsteps for wildtype Ca2+-bound cTnC, 40 jobsteps for Ca2+-bound cTnC-G159D, 40 jobsteps for Ca2+-bound NcTnC-D75Y, and 40 jobsteps for Ca2+-bound NcTnC-E59D. This corresponds to a total simulation time of 8.76 µs for wildtype Ca2+-bound cTnC, 4.67 µs for Ca2+-bound cTnC-G159D, 7.16 µs for Ca2+-bound NcTnC-D75Y, and 7.16 µs for Ca2+-bound NcTnC-E59D.
Interhelical angle analysis and estimation of free energy cost of opening the hydrophobic patch
The degree of opening of the hydrophobic patch can best be described by the interhelical angle between helices A and B 28. For the analysis of interhelical angles, angles were calculated using interhlx (K. Yap, University of Toronto). Using interhelical angle analysis, the degree of openness for every frame of the trajectory was determined. The analysis was performed equivalent to what was described in 17. Defining a cutoff angle below which a state will be characterized as open and above which a state will be characterized as closed, the Boltzmann distribution of states was used to derive the free energy difference ΔG from the occupancies of the open and closed states. , where kB is the Boltzmann constant, T is the temperature of the system and Nclosed and Nopen are the number of systems found in the closed and open states, respectively during the simulation. An A/B interhelical angle of 90° is generally considered the most accurate criterion for defining a structure to be open, since experimentally determined cTnI-bound structures of cTnC exhibit interhelical angles of around 90°. However, in the absence of cTnI, both apo and Ca2+-bound cTnC are found in the closed conformation, with an interhelical A–B angle of ~ 135°. For the analysis, several different cutoff angles were used. Assuming that for example structures with interhelical angles below equally-spaced values between 90° and 130° are considered open, free energy differences for the system transitioning into these “semi-open” conformations can be computed. This allowed extrapolation of the free energy even if the trajectory did not contain any opening event to 90°. We assumed a linear extrapolation to be valid since previously a linear ΔG vs. cutoff angle behavior was observed for a NcTnC mutant that opened all the way in the simulations 17 and there was nothing obvious about the systems used in this analysis that should prevent them from opening the entire way.
Clustering
For clustering, frames every 8 ps were extracted from the MD trajectories. Alignment was based on all C and D helix Cα atoms (cTnC residues 54–87) within 7 Å of the site II Ca2+. Subsequent clustering was performed by RMSD using GROMOS++ conformational clustering 29. A RMSD cutoff of 0.4 Å (wildtype cTnC), 0.5 Å (cTnC-G159D), 0.5 Å (NcTnC-D75Y), and 0.4 Å (NcTnC-E59D) was chosen, respectively. These cutoffs resulted in 6 (wildtype cTnC), 6 (cTnC-G159D), 6 (NcTnC-D75Y), and 4 (NcTnC-E59D) clusters that represented at least 90% of the respective trajectories. The central members of each of these clusters were chosen to represent the calcium binding loop conformations within the cluster and thereby the conformations sampled by the trajectory.
Brownian dynamics simulation
Similar to studies presented in 16, Brownian dynamics simulations were performed with BrownDye 30 to estimate calcium association rates. PQR files for representative protein structures determined by a cluster analysis were generated using pdb2pqr 31. There were 6 (wildtype cTnC), 6 (cTnC-G159D), 6 (NcTnC-D75Y), and 4 (NcTnC-E59D) representative structures which were used in the BrownDye simulations. Since RMSD cutoffs between 0.4 and 0.5 Å have been chosen, these structures are similar. The calcium pqr file was generated using a charge of +2 and an ionic radius of 1.14 Å. APBS 32 was used to generate the electrostatic fields for the protein and the calcium ion in openDX format. Bd_top was used to generate all necessary input files for the BrownDye runs. A phantom atom of zero charge and negative radius (−1.14 Å) was introduced after the first execution of bd_top. The phantom atom was placed at the position of the calcium ion from the trajectory frame. It has no influence on the association rate constant calculation and serves solely to be able to define a reaction criterion that is spherically symmetric around the expected binding position of the calcium. The reaction criterion was chosen to be 1.2 Å within the calcium binding site for all structures in the analysis. 200,000 single trajectory simulations were performed on 8 parallel processors using nam_simulation. The reaction rate constants were calculated using compute_rate_constant from the BrownDye package30. A weighted average of the rate constants of each of the representative cluster centers yielded an estimate of the overall rate constant for the system. The weight for each representative conformation was determined by the number of structures in the cluster.
Markov state model of myofilament activation
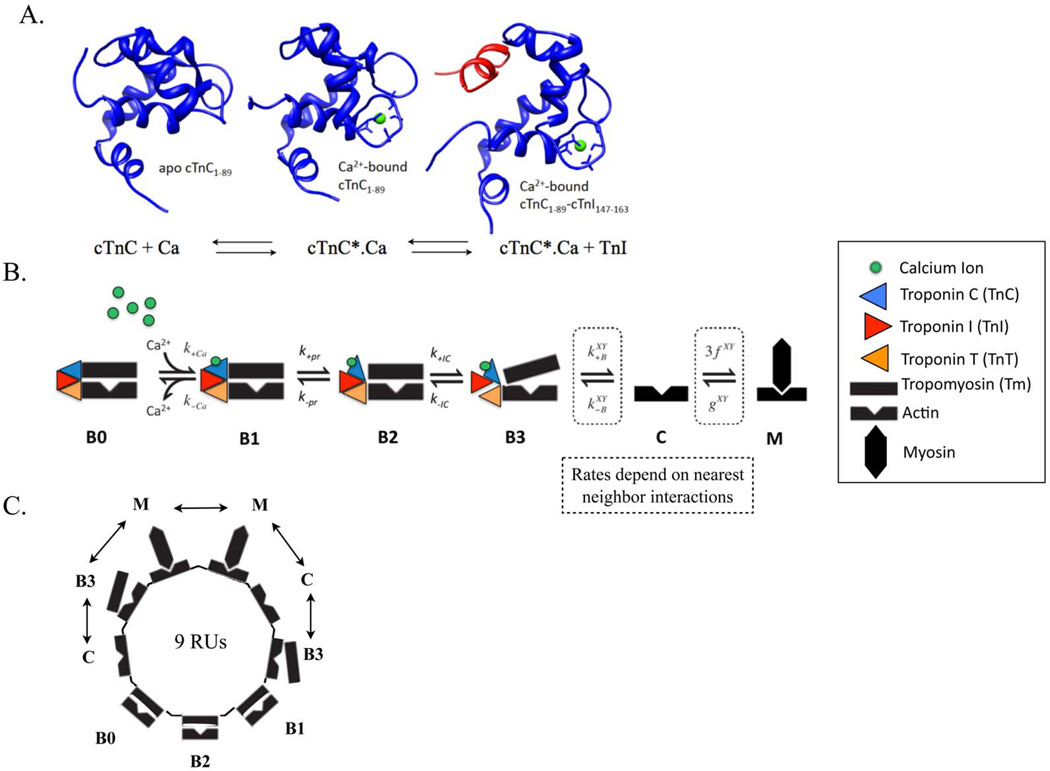
Calcium binding to cTnC triggers a conformational change in cTnC (Figure 2A). This structural change exposes a hydrophobic patch within cTnC that facilitates the binding of cTnC to cTnI and triggers downstream intermolecular events leading to contraction.
Figure 2.
(A) Molecular state of cTnC (in blue) in apo state (BO in Markov model), calcium (in green) bound state (B1 in Markov model) and calcium-bound state with exposed hydrophobic patch allowing for cTnI (in red) to bind to cTnC (B2 in Markov model). (B) Schematic of one regulatory unit (RU) sequence of the six-state (BO, B1, B2, B3, C, M) Markov model of cooperative myofilament activation with nearest neighbor interactions. (C) Schematic of ring structure mathematical formulation used to solve the Markov model that incorporates looping of short segments of RUs to allow elimination of redundant states due to symmetry, resulting from the tight co-operative coupling of 9 adjacent RUs, linked by tropomyosins, depicted in one possible configuration of nearest neighbor interactions (the double-headed arrow) (see Methods and26 for more details). The nearest neighbor interactions of one RU with two adjacent RUs, X and Y – which could be in either Markov state B3, C or M (as in 26), determine the degree of co-operativity of myofilament activation in the Markov model.
To test the ability of the BD and MD simulations to predict behavior at larger scales, we modified a Markov model of thin filament regulation26. The four defined states in the original computational model are: B0, blocked myofilament state where myosin SI head binding to actin are blocked by Tm with no calcium bound to TnC; B, blocked state with calcium bound to cTnC; C, closed state where conformational changes in Tn are observed and Tm moves allowing weak binding of SI; and M, open state where strongly bound cross-bridges cycle. This model was expanded to a six-state model to accommodate myofilament states corresponding to intra-molecular cTnC states probed by BD and MD simulations (Figure 2A). Thus, the current model incorporated the following additional states: B2, blocked calcium-bound state with conformational change of TnC exposing hydrophobic patch thereby allowing binding to Tnl and B3, blocked state with TnC bound to TnI (Figure 2B). By adding states B2 and B3 to the model, it was possible to use rates derived from BD/MD simulation in the Markov model.
All simulations were done in terms of relative steady-state force as computed from the fraction of cross-bridges in state M. The model is a system of coupled differential equations guided by the following rate equations:
| (1) |
| (2) |
| (3) |
| (4) |
As in the earlier model26, actin regulatory units (RUs), are comprised of seven actin monomers, TnC, TnI (the inhibitory subunit of Tn), and Tm, together with the SI region of myosin. Each regulatory unit can bind to one myosin; and there is cooperativity in activation of RUs based on nearest neighbor interactions in state B3, C or M (Figure 2C). We solved for 9 coupled RUs in the current model. Our model requires an updated calculation of the φ, a number reflecting the probability of blocked RUs where cTnI does not inhibit movement into the Closed (C) state. The quantity must now reflect the addition of states B1 and B2:
Steady-state simulations were performed to generate force-pCa curves as determined experimentally. Therefore, all rates with respect to time reduced to zero and the system of equations was simplified considerably into a linear problem that is solved using eigen values.
State transitions and model parameterization: For each state transition we needed to parameterize the forward and reverse rates of transition. For the transition BO to Bl, the calcium binding kCa+ on-rate and kca− off-rate were obtained from literature values for rat myofilaments at 27°C 33–34. For state transition B1 and B2, kpr+ values were plugged in directly from MD simulation results and kpr− was fitted to a standardized wild-type curve at 27°C in rat cardiac myofilaments. The forward and reverse rates (kIC+ and kIC−; kB+ and kB−) for state transitions, B2 to B3 and B3 to M respectively were optimized to fit the wild type steady-state force-pCa relationship in rat myofilaments at 27°C. The apparent forward (fxy) and reverse (gxy) rates of cross -bridge cycling for state transition C to M, were calculated based on Ktr (Ktr = fxy + gxy) and ATPase Q10 values from35 with the formula,
where R1 and R2 are reaction rates at temperatures T1 and T2. The wild-type steady-state force-pCa curve was standardized to 27°C for rat myofilaments by calculating the Tmax., Ec50 and Hill coefficient nH to 27°C based on the Q10 values from the literature35 and fitted using the Hill equation,
where P(Ca2+) is the calcium-dependent force, Pmax is the maximum myofilament force, Ec50 is the calcium concentration at 50% force and nH is the Hill coefficient. In order to optimize the WT steady state force-pCa curve simulation, parameters kIC+, kIC−, µM and µB were varied in combination to minimize RMSD between simulated and experimental WT data. For all mutations, kCa+ and kpr+ rates were altered based on MD data but all other rates and co-operativity coefficients in the model were assumed to remain constant in mutated state. The matlab code for the Markov model has been provided as part of the supplemental information.
cTnC alignment - BLAST tool was used to perform protein sequence alignment corresponding to cTnC FASTA sequence for rat (Rattus Novergicus, gi 66969456) and human (Homo Sapiens; gi 2460249).
Results & Discussion
Full atom, explicit solvent simulations of full length cTnC show straightening of linker region
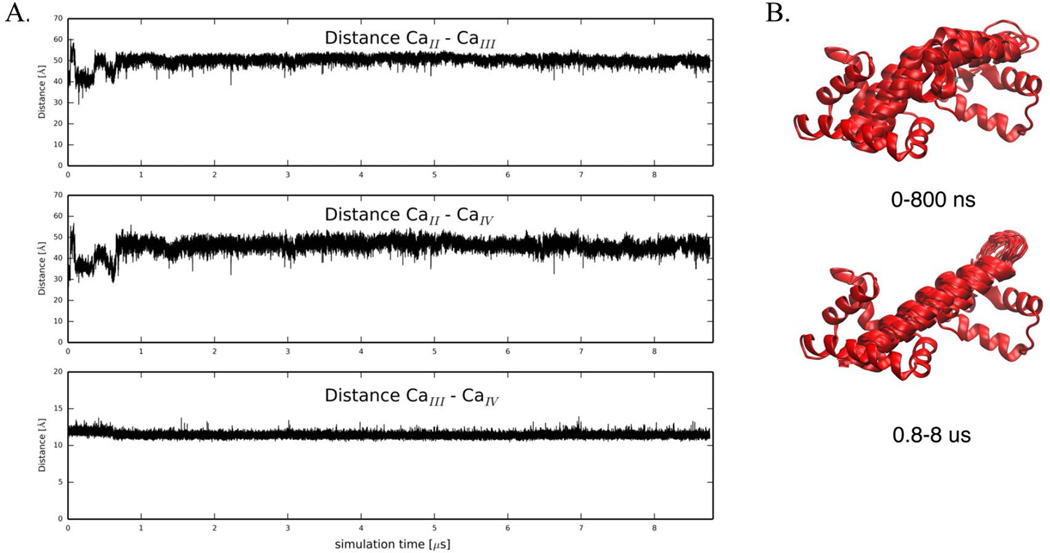
First, we investigated the molecular behavior of wildtype Ca2+-bound cTnC. The N-terminal cTnC EF-hands (helices A through D), which are known to be metal-binding sites, are labeled site I and II36. The C-terminal domain is linked to the regulatory domain by a short flexible linker (approximately residues 89–93). Four more helices (E through H) comprise two more EF-hands with calcium binding sites III and IV. Site n, the low-affinity, Ca2+-specific Ca2+-binding site is generally considered the only site directly involved in calcium regulation of cardiac muscle contraction 37. Previous simulations focused only on NcTnC and not the relative dynamics of NcTnC and CcTnC. To account for changes in the interdomain arrangement of cTnC, we monitored the pairwise distances between calcium binding sites II, III and IV as a measure of the relative distances between the domains. Pairwise distances between the three calcium ions in the 1J1E crystal structure are 41 Å (Ca2+II - Ca2+III), 32 Å (Ca2+II - Ca2+IV), and 12 Å (Ca2+III - Ca2+IV) respectively. The distance between site III and IV Ca2+ remained around its crystallographic value throughout the entire ~9 µs simulation (Figure 3A). This distance confirms structural integrity within CcTnC over the course of the simulation, but is not a measure of interdomain arrangement. The distances between site II Ca2+ and site III/IV Ca2+, respectively, were sensitive to the NcTnC-CcTnC interdomain arrangement. Over the first ~800 ns they remained around their crystallographic values (with the exception of a short-lived extension within the first 100 ns). After about 800 ns of simulation both distances increased by about 10 Å and remained in that state stably for the rest of the simulation as shown in Figure 3A. This increase in inter-calcium distance was caused by a straightening of the central cTnC helix, leading to an increased distance between NcTnC and CcTnC (see Figure 3B). Experimentally, it has been demonstrated that isolated cTnC takes on a more extended form than cTnC anchored in the cTn complex38. Similar trends are observed for the homologous calmodulin protein, for which the Ca2+-bound state assumes an elongated conformation that maximally-separates the N- and C-terminal globular domains in absence of a target protein, but subsequently collapses when a target protein is presented 39. These simulation results are therefore consistent with experimental observations.
Figure 3.
(A) Pairwise distances between Ca2+ in binding sites II, III and IV, respectively. Distance between Ca2+II− Ca2+III, Ca2+II− CaIV, and Ca2+III− Ca2+IV over the course of a 8.76 µs MD simulations for wildtype Ca2+-bound cTnC. (B) The simulation starts with a bent central helix and remains the kink for about 800 ns. After about 800 ns, the central helix in cTnC straightens and remains straight until the end of the simulation (0.8 – 8.8 µs).
Effects of cTnC PCM mutations on calcium binding affinity of cTnC
Brownian dynamics (BD) simulations of the effects of the mutations on calcium association gave estimated association rate constants for calcium binding to the wild-type cTnC, NcTnC-D75Y, cTnC-E59D and cTnC-G159D systems (Table 1). The average calcium association rate constant of wildtype cTnC was determined from Brownian dynamics simulations on representative structures from the MD simulations of wildtype Ca2+-bound cTnC to be 1.5 • 108 1/(M•s) (Table 1). This is in good agreement with the association rate determined for wildtype NcTnC (3.1 • 108 1/(M•s), 16). Additionally these rates are in excellent agreement with experimentally determined values of 2–4 • 108 1/(M•s) for wildtype isolated cTnC 33,40–41, while Ogawa reported a diffusion limited association (> 4 • 107 1/(M•s))42. The idea behind our approach was to adjust the BD parameters so that we can reproduce values for the wildtype systems and then make predictions for mutant systems for which no published data is available. The average calcium association rate constant of NcTnC-D75Y was determined to be 5.0 • 106 1/(M•s), a 50-fold reduction from its wildtype value. This is in agreement with experimental data suggesting a decrease in calcium affinity in NcTnC-D75Y 18. The direct effect of D75Y causing decreased Ca2+ binding affinity is not surprising given the proximity of the mutation to one of the Ca2+ coordination sites (E76) in the regulatory calcium-binding site. BD simulations for E59D, performed on a set of structures representing the conformations of the calcium-binding site most commonly observed during the 7.16 µs MD simulation predicted an average calcium association rate constant of NcTnC-E59D of 1.2 • 107 1/(M•s), a 10-fold reduction from the wild type value. Since there is no effect in myofilaments, this suggests that the cTnC-E59D calcium dissociation rate may also differ in counteraction to the reduced association rate 18,40. The BD simulations for G159D predicted an average calcium association rate constant of cTnC-G159D of 1.0 • 106 1/(M•s), about a 100-fold reduction from its wild type value. Interestingly, these results suggest the mutation G159D may have direct allosteric effects on the N-terminus of cTnC affecting calcium binding to cTnC. The finding of a lower calcium association rate was unexpected since cTnC-G159D has been associated with an unaltered or increased Ca2+ sensitivity in skinned myofilaments from rat and human respectively2,25. Thus, we speculated that cTnC-cTnI interaction effects will be required to explain the steady-state contractile effects of G159D.
Table 1.
Summary of calcium association rates (from BD simulations), computed free energy differences between open and closed states (from MD simulations) and average simulation time between opening events (from MD simulations) for a cutoff angle of 110°.
| system | kon [1/(M•s)] | ΔG [kcal/mol] | Δt |
|---|---|---|---|
| wild-type Ca2+-bound NcTnC |
3.1 • 108 | 4.0 | 130 ns |
| NcTnC-D75Y | 5.0 • 106 | 3.3 | 30 ns |
| NcTnC-E59D | 1.2 • 107 | 2.7 | 40 ns |
| cTnC-G159D | 1.0• 106 | 5.2 | ~700 ns |
Effects of cTnC PCM mutations on rates of cTnC conformational change on calcium binding
Exposure of a hydrophobic patch between cTnC helices A and B is facilitated by calcium binding at site II and can be significantly modulated by mutations 17. It is crucial for binding of the cTnI switch peptide in the context of regulating muscle contraction. Since the hydrophobic patch opening frequency is in the high nanosecond to low microsecond regime, long time-scale simulations have the potential to quantitatively investigate the kinetics of the opening and closing of the cTnC hydrophobic patch. Here we elucidated the hydrophobic patch opening behavior of all three systems under investigation using microsecond Anton43 simulations. The degree of opening of the hydrophobic patch can best be described by the interhelical angle between helices A and B 28. Both apo and Ca2+-bound cTnC are found in the closed conformation 9,44, with an interhelical A–B angle of ~135° 9. The cTnI switch peptide must be present to stabilize the open conformation of the Ca2+-bound regulatory domain of cTnC (interhelical A–B angle of ~90°) 10,45 since the open conformation is only a transient state that is sampled by cTnC after Ca2+-binding 17,46. Kinetics and the free energy difference of the opening-closing process were extracted from the simulations, shedding light on the different ways that the mutations affect the process of contraction. The intramolecular conformational changes induced by Ca2+ binding and the subsequent association of the cTnI switch peptide have been measured experimentally to be significantly slower than the actual Ca2+ binding33,41. Unfortunately there is no direct experimental observable that can confirm the simulations in terms of frequency and degree of hydrophobic patch opening. However, simulated data for gain-of-function and loss-of-function mutations agreed with expected opening behavior 17. Additionally, by simulating the force-pCa curves from the BD and MD results, we obtain indirect experimental verification.
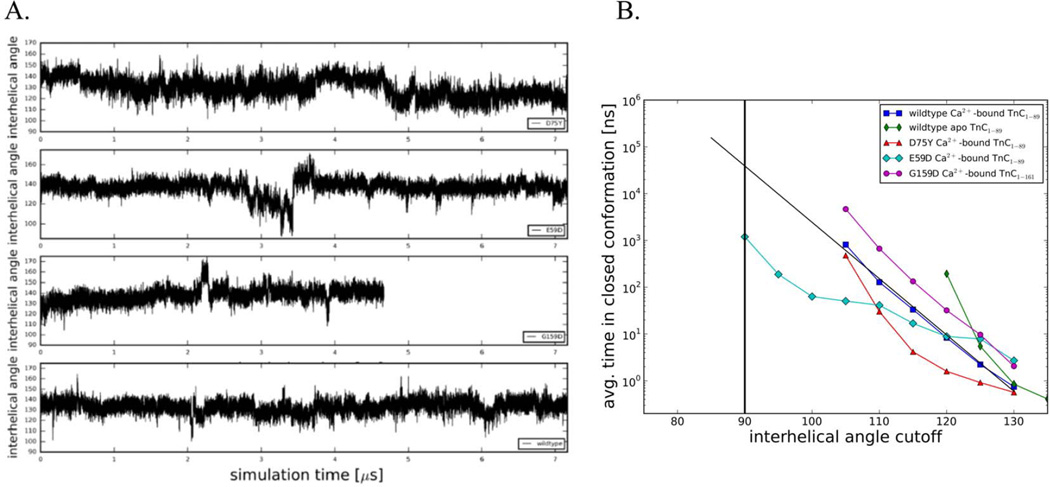
The A/B interhelical angle as a function of simulation time is a measure of the average degree of openness of cTnC. Qualitatively, NcTnC-D75Y (Figure 4A) had a similar behavior to wildtype NcTnC (see 17), in that it remained closed most of the time but occasionally ventured to transiently open conformations (~105°). A more quantitative analysis of the opening of the hydrophobic patch in cTnC can be gleaned using a set of incremental cutoff angles dividing structures into respective open and closed conformations as shown in Figure 4B. While the experimentally determined cTnI-bound structures of cTnC all exhibit A/B interhelical angles of around 90° (which will be considered the most stringent criterion of defining a structure to be open), we chose angles between 135° and 90° as cutoff angles. For each of these cutoff angles, the Boltzmann distribution of states was used to derive the free energy difference ΔG based on the occupancies of the open (≤ cutoff angle) and closed (> cutoff angle) states, . In a related analysis we also evaluated the average time between open conformations as a function of the same cutoff angles (Figure 4B). Compared with wild type NcTnC, NcTnC-D75Y had a slightly opened equilibrium A/B interhelical angle of ~130°. This causes opening to A/B interhelical angles between 110° and 130° to be related with a slightly lower free energy cost than the wildtype system. The frequency of the most opened sampled conformations (A/B interhelical angle ~05°) was identical in NcTnC-D75Y and NcTnC. Sampling of even further opened conformations would require simulations beyond the low microsecond timescale and was not feasible. Nonetheless, we are estimating that the free energy difference between a fully closed and fully open state is slightly larger for NcTnC-D75Y compared to wildtype NcTnC, even though it is hard to make accurate predictions since the shape of the NcTnC-D75Y ΔG-curve deviates somewhat from a linear relationship. We also evaluated the average time between open conformations as a function of the same cutoff angles (Figure 4B). The opening frequency of fully opened conformations (A/B interhelical angle of 90°) was extrapolated to be in the 100 µs to 1 ms range. This would correspond to a 2–20 fold decrease in the opening frequency compared to wildtype. We found that a semi-open configuration (A/B interhelical angle of 110°) was sufficient to model changes to myofilament mechanics (see below). Table 1 summarizes those values for a cutoff angle of 110°.
Figure 4.
(A) Interhelical angles over the course of the simulations for Ca2+-bound wildtype NcTnC, Ca2+-bound NcTnC-D75Y, Ca2+-bound NcTnC-E59D, and Ca2+-bound cTnC-G159D. Interhelical angles are a measure of the openness of the hydrophobic patch between cTnC helices A and B. An interhelical angle of ~135° corresponds to a closed conformation, while an angle of ~90° corresponds to a fully open conformation. (B) Average simulation time between opening events in dependence on the cutoff angle. Values for the Anton wild-type Ca2+-bound TnC simulation, the Anton wild-type apo TnC simulation (both from 17), and the three Ca2+-bound DMC mutation (NcTnC-D75Y, NcTnC-E59D, cTnC-G159D) simulations are shown. The widely accepted open-close cutoff criterion of 90° is marked by a vertical black line.
The cTnC-E59D mutation is interesting since experimentally it has been found to reduce calcium affinity and sensitivity of myofilaments in a cTnC-D75Y/E59D system. cTnC-E59D however was functionally benign and did not impact calcium sensitivity of myofilaments in experiments 18. Here we investigated the molecular effects of cTnC-E59D on hydrophobic patch opening using microsecond MD simulations, as shown in Figure 4A (A/B interhelical angle as a function of simulation time for NcTnC-E59D). While still being in the closed conformation on average, for about 500 ns the system was in a semi-open conformation opening up all the way to 87.4°. This is reflected in a free energy difference between the open and closed conformations of 6 kcal/mol compared to ~8 kcal/mol for wildtype NcTnC (Figure 4, Table 1). Similarly the time between full opening events (A/B interhelical angle of 90°) drops to 1 µs compared to ~50 µs for wildtype NcTnC (Figure 4). Again, Table 1 summarizes those values for a cutoff angle of 110°.
For cTnC-G159D we also computationally determined the molecular effects on hydrophobic patch opening. Experimentally this mutation has been associated with an increased 2 or unchanged 25 Ca2+ sensitivity. The hydrophobic patch remains closed through all of the 4.67 µs of MD simulation and never opens beyond 107.3°. This is reflected in a free energy difference between the open and closed conformations of ~9.5 kcal/mol compared to ~8 kcal/mol for wildtype NcTnC (Figure 4). Similarly the time between full opening events (A/B interhelical angle of 90°) increases to 800 µs compared to ~50 µs for wildtype NcTnC (Figure 4). Table 1 summarizes those values for a cutoff angle of 110°.
In-silico translation of molecular level changes to myofilament mechanics
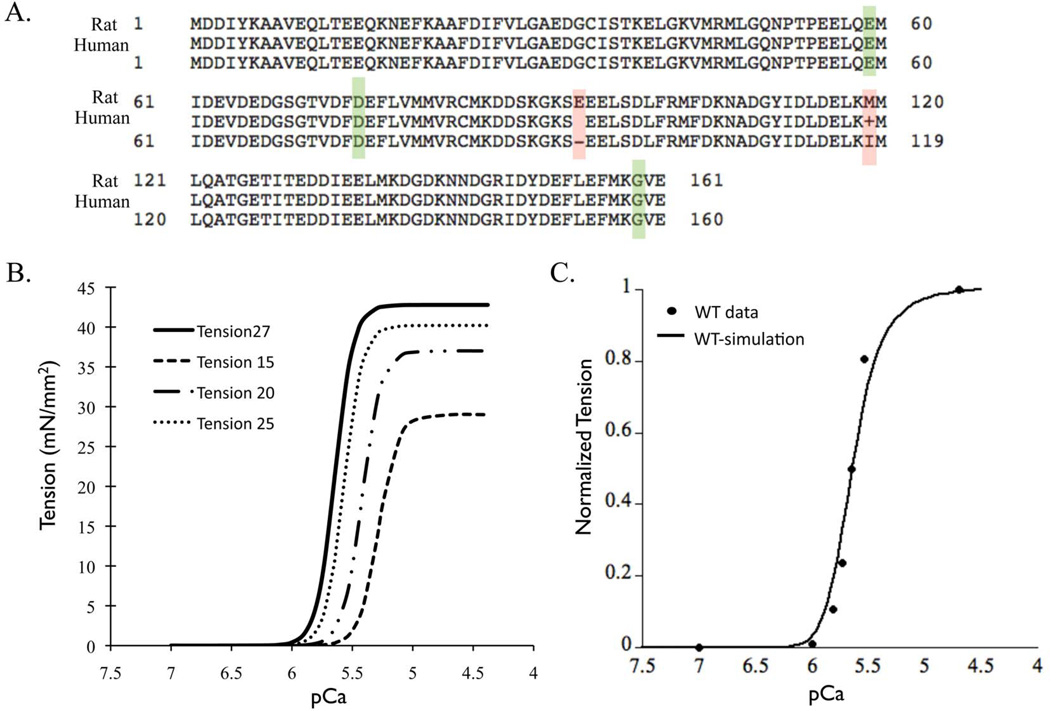
The primary sequence of cTnC is identical between rat and human with the exception of one amino acid in the linker region and one in C-terminus domain of cTnC (Figure 5A). Also there are no other known isoforms of TnC in the heart. Thus the MD and BD simulation results for human cTnC can be applied to a Markov model standardized to rat parameters.
Figure 5.
(A) Sequence alignment of cTnC for protein sequences from rat and human, highlighting the conserved DCM mutations (green box) and two residue difference (red box) in N-terminus lobe of cTnC. (B) Standardized steady-state force-pCa relationship at 27°C, 25°C, 20°C and 15°C (data extracted from35). (C) Steady-state force-pCa curve simulation (solid black trace) fit to standardized force-pCa relationship (solid black dots) for rat myofilaments at 27°C (300K).
The current six-state Markov model describes cTnC activation by calcium and co-operative myofilament activation (Figure 2B, 2C). Thin filament regulation is similar to the myofilaments in that the calcium binding to cTnC (Markov state Bl) triggers a conformational change in cTnC (Markov state B2) which leads to conformational changes in cTnI and cTnT (lumped Markov state B3) such that Tm moves allowing for weak binding of cross-bridges to bind to actin (Markov state C). The weakly bound cross-bridges then cycle to load-bearing strongly bound cross-bridges that generate force (Markov state M). Thus, the Markov model explicitly defines the physiological states of calcium binding to cTnC and the conformational changes that result in cTnC as a consequence. Also the downstream conformational changes that occur in myofilament activation are implicitly defined in the lumped Markov states B3 and C. Lastly, in the model only one cross-bridge binds per RU, however physiologically at least seven cross-bridges can bind per RU. The model computes the co-operative nature of thin filament activation of myofilaments by nearest neighbor interactions. By adding states B2 and B3 to the model, it was possible to use rates derived from BD/MD simulation in the Markov model.
In order to maintain uniformity of parametric conditions (temperature and species) when scaling up longitudinally from molecular state to myofilament state we computed the wild type myofilament properties for rat myocardium at 300K. The resultant steady-state force-pCa relationship at 300K (Figure 5B) had a pCa value of 5.64, a Hill coefficient of 5.7 and maximal tension of 42.78 mN/mm2, computed using Q10 values that were experimentally derived for steady-state force-pCa in skinned rat myofilaments 35. The simulation result from the Markov model fit to the steady-state wild type force-pCa curve for rat myofilaments at 300K within less than 5% error range (Figure 5C). It is known that kinetic rates of a given state transition vary in isolated molecular states of cTnC and integrated myofilament states 34. This was very well elucidated in a previous study 34 where the off-rates of calcium binding were studied in depth from isolated cTnC molecule to a structurally integrated myofilament preparation. Thus here we describe the choice of transitional kinetic rate parameters for each Markov state transition. The rate of calcium binding to cTnC has been measured experimentally for rat to be 1.4 • 108 (M•s)−1 33. This rate is not reported to change in isolated cTnC in solution vs. in integrated myofilaments. However, the off-rate of calcium binding to cTnC is highly sensitive to its corresponding system state (isolated or integrated). Accordingly, we chose the off-rate of calcium as measured in rigor state of myofilaments at 25°C 34. For the conformational state transition of cTnC in myofilaments we assumed that these kinetic rates are largely insensitive to system state. Thus we directly input the values from the MD simulations. Further, the transition Markov states B2 to B3 and B3 to C are lumped states as they implicitly integrate intermolecular associations between TnC-Tnl-TnT-Tm-Actin and thin filament co-operativity. Thus the parameters for these state transitions were scaled to fit the standardized wild-type steady-state force-pCa relationship. Lastly, for the state transition C to M, the apparent forward (fapp) and reverse (gapp) rate of cross-bridge cycling were derived from reported Q10 of Ktr and tension cost35. Input parameters for each state transition are summarized in Table 2 (Figure 2).
Table 2.
Best-fit parameters in six-state Markov model of myofilament activation for WT rat cardiac myofilaments at 300K steady-state force-pCa relationship.
| Paramater | WT |
|---|---|
| kca+ [1/(µM * ms)] | 0.14 |
| kCa− (1/ms) | 0.25 |
| kpr+ (1/ms) | 5699.432425 |
| kpr− (1/ms) | 19500 |
| kIC+ [1/(µM * ms)] | 0.01 |
| kIC− (1/ms) | 0.08 |
| kB+ (1/ms) | 18 |
| kB− (1/ms) | 0.04 |
| fXY(1/ms) | 0.041837669 |
| gXY(1/ms) | 0.01891771 |
| γB | 80 |
| γM | 75 |
| µM | 46 |
Myofilament effects of PCM cTnC mutations
The parameters for the optimized best-fit wild type steady-state force-pCa curve were determined (Figure 5C, Table 2). For each cTnC mutation, all parameters in the Markov model were kept constant, except for the intra-molecular values for calcium binding to cTnC and forward rate of conformational change of cTnC such that the hydrophobic patch is exposed within cTnC (See Table 3). The values of these intra-molecular cTnC state transitions were input directly as reported by MD and BD simulations for all three DCM mutations.
Table 3.
Parameters in six-state Markov model of myofilament activation for WT, D75Y–cTnC, E9D–cTnC and G159D–cTnC rat cardiac myofilaments at 300K steady-state force-pCa relationship for a cutoff angle of 110°.
| Parameter | WT | D75Y | E59D | G159D | G159D (cTnC-cTnI interaction) |
|---|---|---|---|---|---|
| kCa+ [1/(µM * ms)] |
0.14 | 0.005 | 0.012 | 0.001 | 0.001 |
| kCa− (1/ms) | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| kpr+ (1/ms) | 5699.432425 | 26435.49272 | 27055.76642 | 1337.311851 | 1337.311851 |
| kpr− (1/ms) | 19500 | 19500 | 19500 | 19500 | 19500 |
| kIC+ [1/(µM * ms)] |
0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| kIC− (1/ms) | 0.08 | 0.08 | 0.08 | 0.08 | 3.87E-04 |
| kB+ (1/ms) | 18 | 18 | 18 | 18 | 18 |
| kB− (1/ms) | 0.04 | 0.04 | 0.04 | 0.04 | 0.04 |
| fXY(1/ms) | 0.041837669 | 0.041837669 | 0.041837669 | 0.041837669 | 0.041837669 |
| gXY(1/ms) | 0.01891771 | 0.01891771 | 0.01891771 | 0.01891771 | 0.01891771 |
| γB | 80 | 80 | 80 | 80 | 80 |
| γM | 75 | 75 | 75 | 75 | 75 |
| µM | 46 | 46 | 46 | 46 | 46 |
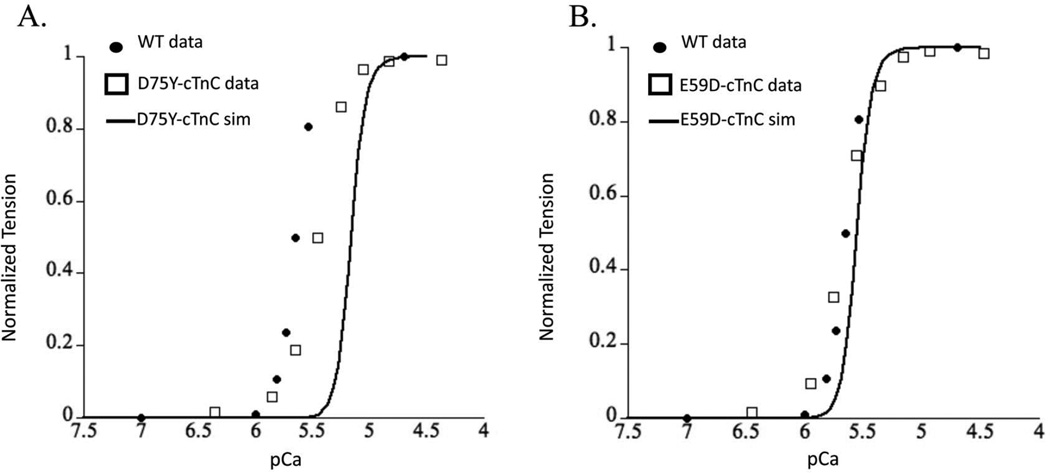
The Markov model simulation for D75Y–cTnC system resulted in a decrease in calcium sensitivity of myofilaments as seen by a rightward shifted steady state force-pCa curve (Figure 6A). This decrease in calcium sensitivity is consistent in direction with experimental recordings in skinned cTnC-D75Y mutant myofilaments 18,24. However, the magnitude of the decrease in calcium sensitivity of myofilaments was significantly exaggerated by the Markov model prediction compared to experimental data. This could be because of either the a) likely changes in the rate of hydrophobic patch closing which we assume to not change in mutations, or b) rather unlikely allosteric effects of cTnC-D75Y mutation in the CcTnC domain that may change the rate of calcium binding to cTnC and rate of hydrophobic patch exposure. This result suggests that the decrease in calcium binding affinity of cTnC coupled with the decrease in ΔG can explain the directional shift in the steady-state force-pCa relationship showing a decrease in calcium sensitivity of tension in skinned myofilaments for cTnC-D75Y.
Figure 6.
Steady-state force-pCa simulation (solid black trace) prediction for (A) cTnC-D75Y mutation and (B), cTnC-D75Y mutation plotted with experimental measures of the same for WT (solid black dots), and DCM cTnC mutations (open squares) for (A) cTnC-D75Y and (B) cTnC-D75Y from skinned myofilaments.
The Markov model of E59D–cTnC did not shift the force-pCa relationship relative to the wild-type steady state force-pCa curve (Figure 6B), consistent with experimental observations in skinned myofilaments 18,24. This was interesting as the calcium binding affinity of E59D–cTnC was decreased, but the ΔG for the E59D–cTnC conformational change was increased. While this did imply that the intramolecular alterations could offset the phenotype such that it may appear benign as observed experimentally, it is remarkable that the effects in and of themselves are sufficient to replicate the experimentally reported results when cTnI switch peptide can bind to cTnC in the partially open conformation state. The experimentally determined cTnI-bound structures of cTnC exhibit A/B interhelical angles of around 90° (which will be considered the most stringent criterion of defining a structure to be open), it is probable that cTnI is able to associate with cTnC before this fully open conformation is achieved. We explored this possibility with the Markov model. The simulation results for cTnC-E59D showed that a partially opened conformation state of cTnC at an opening angle of 110° could explain the experimental observations. Interestingly, simulation results for a 90° opening angle for E59D–cTnC showed a significant leftward shift in force-pCa relationship unlike a modest leftward shift for D75Y–cTnC. This was explained by the smaller change in AG, with an increase in opening angle, for D75Y than E59D based on MD simulation results. These results would implicate that the switch peptide of cTnI must bind to cTnC in its partial open conformation state. To validate the prediction that cTnI can bind to a partially open conformation of cTnC would require more sophisticated MD simulations with whole cTn structure and more explicitly defined Markov model state for cTnC-cTnI interaction with experimentally derived parameters. Although these are beyond the scope of the current study we can conclude that E59D–cTnC mutation intra-molecular interactions could explain experimental observations if a more relaxed opening angle for cTnI binding is sufficient.
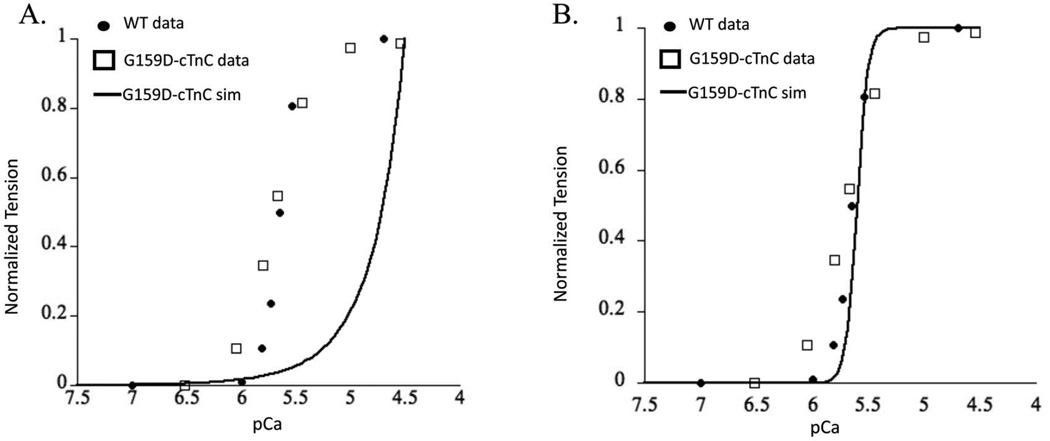
The Markov model simulation for G159D–cTnC system predicted an extremely right shifted steady state force-pCa curve (Figure 7A). This is not meaningful as the heart will likely fail under such extreme conditions and neither were such values reported from experimental observations. In fact the tightly controlled experimental results in rats suggest that G159D is a benign mutation and has no apparent effect on myofilament contractile function 25,47. The calcium affinity values for cTnC from BD simulations and delta G values from MD simulations suggest an offsetting intra-molecular interaction but clearly these effects are not sufficient to capture the experimentally reported benign phenotype. This would suggest that changes in cTnC-cTnI interactions potentially offset the resultant intra-molecular effects. Such a scenario would not be surprising given that the G159D residue is in the cTnC-cTnI interaction region. In fact when we hold all other parameters constant except for lumped state transition B2 to B3, simulation results can predict experimentally observed results (Figure 7B). It is important to note here that this fit was possible only when the KD (equilibrium constant) of the state transition between B2 to B3 was significantly lowered. Assuming that the change in this lumped state transition is only because of a change in cTnC-cTnI interaction, then this would suggest that the mutant G159D–cTnC leads to structural change in cTnC that increases its affinity for cTnI. The opening angle change to 110° was not enough to predict experimentally reported myofilament effects. These observations do suggest that cTnC-cTnI interaction is perturbed by G159D, but are certainly not conclusive. Clearly, other factors within cTnC (such as calcium dissociation rate) and beyond (such as cTnC-cTnI affinity or PKA phosphorylation of cTnI) also have profound impact on myofilament Ca2+ sensitivity. These factors are still beyond the reach of our molecular-scale and myofilament scale computational analysis. The current six-state model (Figure 2B) does not explicitly define the cTnC-cTnI interaction as a Markov state in itself but lumps it in the B3 state where cTnC-cTnI-cTnT-Tm-actin interactions take place. This is likely the reason that the Markov simulation results in an extremely right shifted contractile response. Incidentally, the cTnC-G159D mutation has been shown to interfere with the modulation of Ca2+ sensitivity through cTnI PKA phosphorylation at cTnI sites Ser 23 and Ser 24 25,48. We speculate that this effect outweighs the reversed effects on hydrophobic patch opening and calcium association. Nonetheless, the multi-scale in-silico modeling approach described here is a robust way to study as to how molecular effects can longitudinally translate to myofilament contractile response.
Figure 7.
Steady-state force-pCa simulation (solid black trace) prediction for (A) cTnC-G159D mutation and (B), cTnC-G159D mutation fitted when accounting for cTnC-cTnI interaction effects (see text for details) plotted with experimental measures of the same for WT (solid black dots), and cTnC-G159D mutant (open squares) from skinned myofilaments.
Summary and conclusions
We investigated alterations in calcium association rates to mutant cTnC using Brownian dynamics (BD) simulations and the opening frequency of the hydrophobic patch using us-timescale MD simulations. All three mutations decreased cTnC calcium association rate by 10 to 100-fold, but while D75Y and G159D caused a 10-fold decrease in hydrophobic patch opening frequency, the E59D mutation increased opening frequency by 50-fold compared with WT.
Using these findings, we scaled the molecular state results from BD and MD simulations to myofilament state by solving a six-state Markov model of myofilament contraction. This multi-scale approach enabled us to test and understand the intra-molecular effects of cTnC DCM mutations on contractile function. Results from the myofilament simulations suggest that the intra-molecular changes resulting from the cTnC mutation D75Y and E59D can explain experimentally observed directional changes in calcium sensitivity of myofilaments, provided the cTnI switch peptide can associate with the N-terminus of cTnC in a partially open state of cTnC. However, intra-molecular changes observed in G159D genotype of cTnC are not sufficient to explain the experimentally observed contractile effects. Importantly, they shed light as to how the interaction of cTnC with cTnI can offset the intra-molecular effects of G159D cTnC mutation observed in DCM.
To summarize, we report the intra-molecular changes and potential cTnC-cTnI interactions in cTnC DCM mutants that underlie experimentally observed contractile defects. This is important from a drug development standpoint as it highlights how intramolecular cTnC changes and/or cTnC-cTnI interactions can be interactive and offsetting, to the extent that a known DCM mutation may appear to be benign functionally. To our knowledge, this is the first study to integrate and leverage in silico modeling approaches from molecular level (MD simulations) to myofilament level (Markov models of contractility). This is a promising step towards demonstrating the feasibility and applicability of multi-scale modeling to gain mechanistic insights into role of known genetic DCM mutations in contractile proteins.
Acknowledgments
The authors would like to extend a special thanks to Andy McCammon for his continuing support and advice, as well as for helpful discussions regarding this work. We would also like to thank members of his research group for useful discussions. We would like to thank Peter Kekenes-Huskey for interesting discussions concerning the opening transition of the hydrophobic patch. We would also like to thank Dr. Stuart Campbell for interesting discussions concerning Markov modeling of myofilaments. This work was supported by the National Institutes of Health, the National Science Foundation, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, and the NSF Supercomputer Centers. Computational resources were supported, in part, by the National Science Foundation grant PHY-0822283, the Center for Theoretical Biological Physics. S. L. was supported by the American Heart Association and the Center for Theoretical Biological Physics. Anton computer time was provided by the National Center for Multiscale Modeling of Biological Systems (MMBioS) through Grant P41GM103712-S1 from the National Institutes of Health and the Pittsburgh Supercomputing Center (PSC). The Anton machine at PSC was generously made available by D.E. Shaw Research. This research was also supported by the National Biomedical Computation Resource (P41 GM103426) and NIH grants P50 GM094503, HL105242, and HL105242 (to ADM), and HL111197 (MR).
Footnotes
Supporting Information Available
The matlab code for the Markov model has been provided as part of the supplemental information.
References
- 1.Karkkainen S, Peuhkurinen K. Genetics of dilated cardiomyopathy. Ann. Med. 2007;39:91–107. doi: 10.1080/07853890601145821. [DOI] [PubMed] [Google Scholar]
- 2.Dyer EC, Jacques AM, Hoskins AC, Ward DG, Gallon CE, Messer AE, Kaski JP, Burch M, Kentish JC, Marston SB. Functional analysis of a unique troponin c mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circulation. Heart failure. 2009;2:456–464. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 3.Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, Burke M, Elliott PM, McKenna WJ. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 4.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 5.Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. Faseb J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 6.Li MX, Wang X, Sykes BD. Structural based insights into the role of troponin in cardiac muscle pathophysiology. Journal of Muscle Research and Cell Motility. 2004;25:559–579. doi: 10.1007/s10974-004-5879-2. [DOI] [PubMed] [Google Scholar]
- 7.Davis JP, Tikunova SB. Ca2+ exchange with troponin C and cardiac muscle dynamics. Cardiovasc. Res. 2008;77:619–626. doi: 10.1093/cvr/cvm098. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 9.Spyracopoulos L, Li MX, Sia SK, Gagne SM, Chandra M, Solaro RJ, Sykes BD. Calcium-induced structural transition in the regulatory domain of human cardiac troponin C. Biochemistry. 1997;36:12138–12146. doi: 10.1021/bi971223d. [DOI] [PubMed] [Google Scholar]
- 10.Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-1147-163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 11.Robertson IM, Sun YB, Li MX, Sykes BD. A structural and functional perspective into the mechanism of Ca(2+)-sensitizers that target the cardiac troponin complex. J. Mol. Cell. Cardiol. 2010;49:1031–1041. doi: 10.1016/j.yjmcc.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagne SM, Li MX, McKay RT, Sykes BD. The NMR angle on troponin C. Biochem Cell Biol. 1998;76:302–312. doi: 10.1139/bcb-76-2-3-302. [DOI] [PubMed] [Google Scholar]
- 13.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Mercier P, Letoumeau PJ, Sykes BD. Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies. Protein Sci. 2005;14:2447–2460. doi: 10.1110/ps.051595805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kekenes-Huskey PM, Lindert S, McCammon JA. Molecular basis of calcium-sensitizing and desensitizing mutations of the human cardiac troponin C regulatory domain: a multi-scale simulation study. PLoS Comput. Biol. 2012;8:el002777. doi: 10.1371/journal.pcbi.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindert S, Kekenes-Huskey PM, Huber G, Pierce L, McCammon JA. Dynamics and calcium association to the N-terminal regulatory domain of human cardiac troponin C: A multiscale computational study. J. Phys. Chem. B. 2012;116:8449–8459. doi: 10.1021/jp212173f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindert S, Kekenes-Huskey PM, McCammon JA. Long-timescale molecular dynamics simulations elucidate the dynamics and kinetics of exposure of the hydrophobic patch in troponin C. Biophys. J. 2012;103:1784–1789. doi: 10.1016/j.bpj.2012.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim CC, Yang H, Yang M, Wang CK, Shi J, Berg EA, Pimentel DR, Gwathmey JK, Hajjar RJ, Helmes M, et al. A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophysical journal. 2008;94:3577–3589. doi: 10.1529/biophysj.107.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varguhese JF, Li Y. Molecular dynamics and docking studies on cardiac troponin C. J. Biomol. Struct. Dyn. 2011;29:123–135. doi: 10.1080/07391102.2011.10507378. [DOI] [PubMed] [Google Scholar]
- 20.Varughese JF, Baxley T, Chalovich JM, Li Y. A computational and experimental approach to investigate bepridil binding with cardiac troponin. J. Phys. Chem. B. 2011;115:2392–2400. doi: 10.1021/jp1094504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varughese JF, Chalovich JM, Li Y. Molecular dynamics studies on troponin (Tnl-TnT-TnC) complexes: insight into the regulation of muscle contraction. J. Biomol. Struct. Dyn. 2010;28:159–174. doi: 10.1080/07391102.2010.10507350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Robertson IM, Li MX, McCully ME, Crane ML, Luo Z, Tu AY, Daggett V, Sykes BD, Regnier M. Structural and functional consequences of the cardiac troponin C L48QCa(2+)-sensitizing mutation. Biochemistry. 2012;51:4473–4487. doi: 10.1021/bi3003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayasundar JJ, Xing J, Robinson JM, Cheung HC, Dong WJ. Molecular dynamics simulations of the cardiac troponin complex performed with FRET distances as restraints. PLoS One. 2014;9:e87135. doi: 10.1371/journal.pone.0087135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dweck D, Reynaldo DP, Pinto JR, Potter JD. A dilated cardiomyopathy troponin C mutation lowers contractile force by reducing strong myosin-actin binding. J. Biol. Chem. 2010;285:17371–17379. doi: 10.1074/jbc.M109.064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biesiadecki BJ, Kobayashi T, Walker JS, Solaro RJ, de Tombe PP. The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ. Res. 2007;100:1486–1493. doi: 10.1161/01.RES.0000267744.92677.7f. [DOI] [PubMed] [Google Scholar]
- 26.Campbell SG, Lionetti FV, Campbell KS, McCulloch AD. Coupling of adjacent tropomyosins enhances cross-bridge-mediated cooperative activation in a Markov model of the cardiac thin filament. Biophysical journal. 2010;98:2254–2264. doi: 10.1016/j.bpj.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 28.Wang X, Li MX, Sykes BD. Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin 1147-163 and bepridil. J Biol Chem. 2002;277:31124–31133. doi: 10.1074/jbc.M203896200. [DOI] [PubMed] [Google Scholar]
- 29.Christen M, Hunenberger PH, Bakowies D, Baron R, Burgi R, Geerke DP, Heinz TN, Kastenholz MA, Krautler V, Oostenbrink C, et al. The GROMOS software for biomolecular simulation: GROMOS05. Journal of computational chemistry. 2005;26:1719–1751. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 30.Huber GA, McCammon JA. Browndye: A software package for Brownian dynamics. Comput. Phys. Commun. 2010;181:1896–1905. doi: 10.1016/j.cpc.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker NA, Sept D, Joseph S, Hoist MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazard AL, Kohout SC, Stricker NL, Putkey JA, Falke JJ. The kinetic cycle of cardiac troponin C: calcium binding and dissociation at site II trigger slow conformational rearrangements. Protein Sci. 1998;7:2451–2459. doi: 10.1002/pro.5560071123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little SC, Biesiadecki BJ, Kilic A, Higgins RS, Janssen PM, Davis JP. The rates of Ca2+ dissociation and cross-bridge detachment from ventricular myofibrils as reported by a fluorescent cardiac troponin C. J. Biol. Chem. 2012;287:27930–27940. doi: 10.1074/jbc.M111.337295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Tombe PP, Stienen GJ. Impact of temperature on cross-bridge cycling kinetics in rat myocardium. J. Physiol. 2007;584:591–600. doi: 10.1113/jphysiol.2007.138693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu. Rev. Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi T, Jin L, de Tombe PP. Cardiac thin filament regulation. Pflug. Arch. Eur. J. Phy. 2008;457:37–46. doi: 10.1007/s00424-008-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordina NM, Liew CK, Potluri PR, Curmi PM, Fajer PG, Logan TM, Mackay JP, Brown LJ. Ca2+-induced PRE-NMR changes in the troponin complex reveal the possessive nature of the cardiac isoform for its regulatory switch. PLoS One. 2014;9:ell2976. doi: 10.1371/journal.pone.0112976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annual review of biophysics and biomolecular structure. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 40.Tikunova SB, Davis JP. Designing calcium-sensitizing mutations in the regulatory domain of cardiac troponin C. J. Biol. Chem. 2004;279:35341–35352. doi: 10.1074/jbc.M405413200. [DOI] [PubMed] [Google Scholar]
- 41.Dong W, Rosenfeld SS, Wang CK, Gordon AM, Cheung HC. Kinetic studies of calcium binding to the regulatory site of troponin C from cardiac muscle. J. Biol. Chem. 1996;271:688–694. doi: 10.1074/jbc.271.2.688. [DOI] [PubMed] [Google Scholar]
- 42.Ogawa Y. Calcium binding to troponin C and troponin: Effects of Mg2+, ionic strength and pH. J. Biochem. 1985;97:1011–1023. doi: 10.1093/oxfordjournals.jbchem.a135143. [DOI] [PubMed] [Google Scholar]
- 43.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. Acm. 2008;51:91–97. [Google Scholar]
- 44.Sia SK, Li MX, Spyracopoulos L, Gagne SM, Liu W, Putkey JA, Sykes BD. Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J. Biol. Chem. 1997;272:18216–18221. doi: 10.1074/jbc.272.29.18216. [DOI] [PubMed] [Google Scholar]
- 45.Dong WJ, Xing J, Villain M, Hellinger M, Robinson JM, Chandra M, Solaro RJ, Umeda PK, Cheung HC. Conformation of the regulatory domain of cardiac muscle troponin C in its complex with cardiac troponin I. J. Biol. Chem. 1999;274:31382–31390. doi: 10.1074/jbc.274.44.31382. [DOI] [PubMed] [Google Scholar]
- 46.Lindert S, Kekenes-Huskey PM, Huber G, Pierce L, McCammon JA. Dynamics and calcium association to the N-terminal regulatory domain of human cardiac troponin C: A multiscale computational study. J. Phys. Chem. B. 2012;116:8449–8459. doi: 10.1021/jp212173f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gollapudi SK, Chandra M. Cardiomyopathy-related mutations in cardiac troponin C, L29Q and G159D, have divergent effects on rat cardiac myofiber contractile dynamics. Biochemistry research international. 2012;2012:824068. doi: 10.1155/2012/824068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong WJ, Xing J, Ouyang Y, An J, Cheung HC. Structural kinetics of cardiac troponin C mutants linked to familial hypertrophic and dilated cardiomyopathy in troponin complexes. J. Biol. Chem. 2008;283:3424–3432. doi: 10.1074/jbc.M703822200. [DOI] [PubMed] [Google Scholar]