Abstract
Background
The molecular mechanisms behind cerebral aneurysm formation and rupture remain poorly understood. In the past decade, microRNAs (miRNAs) have been shown to be key regulators in a host of biological processes. They are non-coding RNA molecules, approximately 21 nucleotides long, which post-transcriptionally inhibit mRNAs by attenuating protein translation and promoting mRNA degradation. We attempted to profile the miRNA and mRNA interactions and expression levels in cerebral aneurysm tissue from human subjects.
Methods
We performed a prospective case-control study in human subjects in order to characterize the differential expression of mRNA and miRNA in unruptured cerebral aneurysms compared to control tissue [healthy superficial temporal arteries (STA)]. Ion Torrent was used for deep RNA sequencing. Affymetrix miRNA microarrays were used to analyze miRNA expression, whereas Nanostring nCounter technology was used for validation of identified targets.
Results
Overall, 7 unruptured cerebral aneurysm and 10 STA specimens were collected. We identified several differentially expressed genes in aneurysm tissue with MMP-13 (Fold change 7.21) and various collagen genes (COL1A1, COL5A1, COL5A2) being among the most upregulated. In addition, multiple miRNAs were significantly differentially expressed, with miR-21 (Fold change 16.97) being the most upregulated, and miR-143-5p (Fold change −11.14) being the most downregulated. From these, miR-21, miR-143, and miR-145 had several significantly anti-correlated target genes in our cohort, associated with smooth muscle cell function, extracellular matrix remodeling, inflammation signaling, and lipid accumulation. All these processes are crucial in the pathophysiology of cerebral aneurysms.
Conclusion
Our analysis identified differentially expressed genes and miRNAs in unruptured human cerebral aneurysms, suggesting the possibility of a role for miRNAs in aneurysm formation. Further investigation for their importance as therapeutic targets is needed.
Keywords: cerebral aneurysms, gene expression, microRNA expression, molecular mechanisms of disease, deep sequencing
INTRODUCTION
Cerebral aneurysm rupture is a devastating event, with extremely high morbidity and mortality.32 The molecular mechanisms behind cerebral aneurysm formation and rupture remain poorly understood. Some investigators have attempted to identify the genetic basis of their development. These studies have focused mainly on linkage analyses14,18 in cases of familial aneurysms, or association analyses40 of single nucleotide polymorphisms (SNPs). Most recently, limited gene expression studies25,27,30,33-35 have identified genes (mainly related to inflammation, and extracellular matrix remodeling) with differential expression in aneurysm tissue. These groups utilized microarray and polymerase chain reaction (PCR) techniques. Their results are not readily comparable due to the use of different microarray platforms, gene nomenclatures, and control tissues.34 While genome-wide analysis of gene expression using deep sequencing has not been performed yet, such data allows the quantification of the expression of both annotated and unannotated genes.
In the past decade, microRNAs (miRNAs) have been shown to be key regulators in a host of biological processes.10 They are non-coding RNA molecules, approximately 21 nucleotides long, which post-transcriptionally inhibit mRNAs by attenuating protein translation and promoting mRNA degradation.10 Experiments with vascular smooth muscle cell (VSMC)-specific Dicer depletion emphasized the importance of miRNAs for VSMC homeostasis, and is therefore likely that miRNAs also play a prominent role in aneurysm formation.2 It has been shown2,12,17,26,28,29,31 that miR-29, miR-21, miR-26, miR-143, and miR-145 play pivotal roles in the formation of abdominal aortic aneurysms (AAA). These miRNAs post-transcriptionally regulate the expression of multiple targets with a function in extra-cellular matrix, such as collagen and fibrillin, and thus affect vascular wall stability and aneurysm formation.2,12,17,26,28,29,31 The identification of specific miRNAs implicated in cerebral aneurysms, and their association with gene expression is particularly appealing, because these molecules can be silenced, providing potential therapeutic targets. A recent study20 investigating the miRNA profile of ruptured cerebral aneurysms lacked control for multiple comparisons and a separate analysis of corresponding gene expression.
In order to address these shortcomings, we performed a prospective case-control study in human subjects to measure the gene and miRNA expression in unruptured cerebral aneurysm tissue. Gene expression was investigated with the use of whole genome deep sequencing. In addition, we attempted to identify statistically significant miRNA-mRNA pairs with correlated expression changes consistent with miRNA regulation.
METHODS
Study design
The current study was approved by the Dartmouth Committee for the Protection of Human Subjects, and every participant signed an informed consent. We followed a prospective case-control design to investigate the expression profiles of miRNA and mRNA, as well as their correlation in unruptured human cerebral aneurysm tissue. Cerebral aneurysm samples were obtained from patients undergoing craniotomy for unruptured cerebral aneurysm clipping in our institution from 2012-2014.
Control tissue was collected from the superficial temporal artery (STA) of patients undergoing pterional craniotomies (for aneurysm clipping or other treatments) or patients undergoing temporal artery biopsies to rule out temporal arteritis (used only after the regular pathology was negative for temporal arteritis). STA has been used in prior comparative studies of cerebral aneurysm tissue as a control.34 Clinical history was obtained from all patients preoperatively on the day of the surgery. No postoperative follow-up data were collected for the purpose of this study.
Specimen collection
After successful clipping of the aneurysm, the dome was resected with the use of micro-scissors and bayonetted forceps (not used before during the case) and was placed immediately into a vial containing RNAlater solution (Ambion, Austin, TX) to protect RNA from degradation. The vials were immediately transported to the Genomics and Molecular Biology Shared Resource (GMBSR) in Geisel School of Medicine at Dartmouth, and frozen to −80 °C. The same process was followed for the control tissue.
RNA isolation and library preparation
RNA was harvested from patient samples using mirVana miRNA isolation kit (Ambion, Austin, TX), allowing total RNA to be isolated containing the miRNA population. RNA quality and quantity were assessed by Fragment Analyzer (Advanced Analytical, Ames, IA) and Qubit (Invitrogen, Carlsbad, CA), respectively. The mRNA-sequencing (RNA-seq) libraries were prepared using the Ion Total RNA-seq kit v2 (Life Technologies, Carlsbad, CA) from 100ng of total RNA.
RNA was fragmented prior to adaptor ligation, reverse transcription was performed, and the resulting cDNA was purified and amplified. This protocol allows for strand information to be preserved so that all mapped reads are aligned in the direction of transcription relative to the chromosomal strand. Each biological sample was generated with a unique INDEX sequence to allow multiple samples to be pooled together (Ion Xpress RNA barcodes provided by Life Technologies, Carlsbad, CA). Library quality was assessed with the Fragment Analyzer (Advanced Analytical, Ames, IA) and Qubit (Invitrogen, Carlsbad, CA) prior to sequencing.
Ion Torrent Sequencing
RNA sequencing was performed on the Ion Proton sequencer (Life Technologies, Carlsbad, CA), using the PI chip, according to the manufacturers protocol. The sequencing template was prepared from 6pM of pooled libraries (n=3) by emulsion PCR using the One Touch 2 Instrument (Life Technologies, Carlsbad, CA) and the Ion PI Template OT2 200 kit v2 chemistry (Life Technologies, Carlsbad, CA). Following enrichment on the One Touch ES Instrument, the sample was loaded on the PI chip and sequenced using the Ion PI Sequencing v2 chemistry with the pre-existing Ion RNA-whole transcriptome run plan, utilizing 500 flows. The Proton server is pre-loaded with base calling and adapter trimming software. FASTQ files were generated and used for data analysis. On average (across the 17 samples) 24 million reads were generated for each barcoded biological sample, with a median read length of 90 base pairs.
Reads were aligned using TopHat version 2.0.10 disallowing novel splice junctions, and statistically significant transcripts were identified by comparing gene expression in cerebral aneurysm and STA tissues with Cuffdiff version 2.2.1, using each sample as a replicate and an FDR of 0.05. Gene Ontology (GO) Terms that were statistically enriched in up- and down-regulated genes at this level of significance were identified with GOEAST, using a Hypergeometric test and a Yekutieli FDR of 0.01.
MicroRNA expression
Affymetrix miRNA 4.0 microarrays (Affymetrix, Santa Clara, CA) were used to analyze the miRNA expression levels from the same RNA isolates. Following the manufacturer’s protocol, 130ng of total RNA was labeled with the FlashTag Biotin HSR RNA labeling kit (Affymetrix, Santa Clara, CA). To confirm efficient labeling the ELOSA QC assay (Affymetrix, Santa Clara, CA) was performed, and samples were hybridized on the Affymetrix 4.0 microarrays overnight. Washing, staining and scanning of the arrays was performed using the Command Console, and the spike-in analysis of control oligonucleotides was evaluated using the Expression Console. Data were analyzed using the recommended Transcriptome Analysis Console (TAC) software from Affymetrix.
The miRNA and mRNA expression was quantified and normalized for each patient so that they could be compared across patients. We quantified the differences between cerebral aneurysms and control tissues by computing an ANOVA p-value for each miRNA across the different individuals, and using a Benjamini-Hochberg multiple test correction with an FDR of 0.15 to identify statistically significant changes in expression. The FDR of 0.15 was used to account for the inherent variability expected in clinical samples.
NanoString validation
To validate miRNA array expression data, expression of approximately 800 miRNAs was determined with the NanoString nCounter Human v2 miRNA Expression Assay (NanoString Technologies, Seattle, WA), using 100 ng of total RNA as input. The protocol was carried out as described in the nCounter miRNA Expression Assay Manual, Sample Preparation and Hybridization Protocols (11/15/2012 version). Incubations were performed in a Veriti 96-well Thermocycler (Applied Biosystems, Foster City California); the duration of the overnight hybridization step was 13 hours. Samples were processed and applied to the NanoString sample cartridge using the nCounter Prep Station, and data were collected with the nCounter Digital Analyzer.
Data generated in this study were submitted to the Gene Expression Omnibus (GEO) under the accession GSE66240.
RESULTS
Patient characteristics
During the study period, a total of 7 samples of unruptured cerebral aneurysm tissue (mean age 47 years, ranging from 17-68, 2 males) and 10 samples of control tissue (mean age 63.4 years, ranging from 50-77 years, 1 male) were collected. The aneurysms collected were in the following locations: 3 in the middle cerebral artery, 2 in the posterior communicating artery, one in the internal carotid artery terminus, and one in the pericallosal artery. The mean aneurysm size was 11 mm (ranging from 4 to 25 mm).
Three of the control specimens were harvested from aneurysm patients, whose aneurysms were also collected, whereas 2 belonged to aneurysm patients, whose aneurysm domes were judged intraoperatively to be too small (mean size 4 mm) to be safely resected. Four of the control specimens were harvested during STA biopsies, which eventually were non-pathologic on histology. The remaining control specimen was collected during a craniotomy for tumor resection. The relative vascular comorbidities of our 2 cohorts are detailed in Supplemental Table I.
Gene expression profile
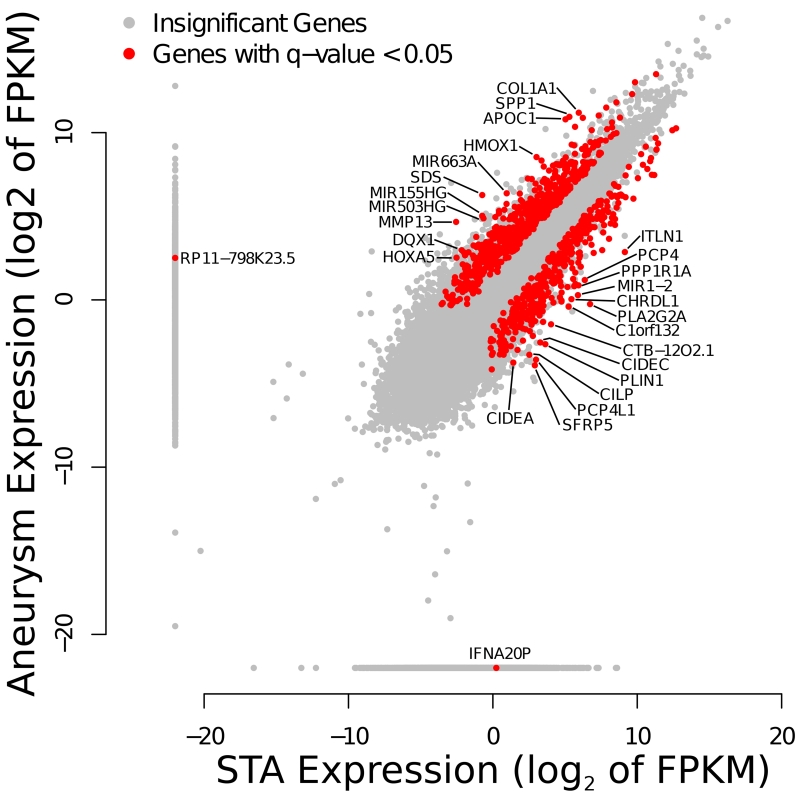
We identified 1,028 genes (Figure 1) with differential expression between aneurysm and control tissue, at an FDR of 0.05. From these 623 were upregulated and 405 downregulated. The top 20 upregulated and top 20 downregulated genes with the largest fold change are listed in Table 1. The protein-coding gene with the largest fold change in expression was a member of the matrix metalloproteinase family (MMP-13, Fold change 7.21), which codes for a metalloproteinase involved in extracellular matrix remodeling and collagenase activity. We also identified a number of host genes for microRNAs, suggesting that some of the differential expression observed in the mature microRNAs could occur before processing by Dicer and Drosha. In particular, MIR143HG, the host gene for both miR-143 and miR-145, is downregulated in cerebral aneurysms with a highly significant 5-fold reduction, likely explaining their decreased expression. Among the most statistically enriched GO Terms (Supplemental Table II) found in this dataset were “extracellular matrix,” “collagen metabolic process,” and “response to stress”.
Figure 1.
Differential expression by RNA-Sequencing of genes in unruptured cerebral aneurysm compared to STA tissues using Cuffdiff. Insignificant changes in gene expression are shown as gray dots; statistical significant changes in expression at an FDR of 0.05 are shown in red. Genes with a log2 fold change of 4 or greater are labeled.
Table 1. Differentially expressed genes.
| Significantly upregulated genes with the highest fold change | ||||||
|---|---|---|---|---|---|---|
| Gene Name | STA FPKM | Cerebral Aneurysm FPKM |
Fold Change (CA/STA) |
P-Value | Q-Value | Description |
| RP11-798K23.5 | 0 | 5.69579 | inf | 5.00E-05 | 0.00257671 | processed pseudogene |
| MMP13 | 0.171024 | 25.309 | 147.9850781 | 0.0007 | 0.0196391 | matrix metallopeptidase 13 (collagenase 3) |
| SDS | 0.600028 | 77.034 | 128.3840087 | 5.00E-05 | 0.00257671 | serine dehydratase |
| MIR155HG | 0.599021 | 32.6115 | 54.4413301 | 5.00E-05 | 0.00257671 | MIR155 host gene (non-protein coding) |
| APOC1 | 32.6523 | 1776.04 | 54.39249302 | 5.00E-05 | 0.00257671 | apolipoprotein C-I |
| SPP1 | 39.359 | 1972.19 | 50.10772631 | 5.00E-05 | 0.00257671 | secreted phosphoprotein 1 |
| MIR503HG | 0.638055 | 29.3629 | 46.01938704 | 0.0026 | 0.049529 | MIR503 host gene (non-protein coding) |
| HMOX1 | 8.13042 | 371.704 | 45.7176874 | 5.00E-05 | 0.00257671 | heme oxygenase (decycling) 1 |
| MIR663A | 1.94222 | 82.861 | 42.66303508 | 5.00E-05 | 0.00257671 | microRNA 663a |
| COL1A1 | 61.7918 | 2337.76 | 37.83285161 | 5.00E-05 | 0.00257671 | collagen, type I, alpha 1 |
| DQX1 | 0.22374 | 7.84051 | 35.04295164 | 0.0007 | 0.0196391 | DEAQ box RNA- dependent ATPase 1 |
| HOXA5 | 0.174839 | 5.74981 | 32.88631255 | 0.00185 | 0.0391237 | homeobox A5 |
| RGS1 | 10.3331 | 322.062 | 31.16799412 | 5.00E-05 | 0.00257671 | regulator of G- protein signaling 1 |
| SLC16A10 | 0.450357 | 13.573 | 30.13831249 | 0.00015 | 0.00623662 | solute carrier family 16 (aromatic amino acid transporter), member 10 |
| KIAA1199 | 1.34223 | 39.8339 | 29.67740253 | 5.00E-05 | 0.00257671 | KIAA1199 |
| GREM1 | 0.249676 | 7.14698 | 28.62501802 | 5.00E-05 | 0.00257671 | gremlin 1, DAN family BMP antagonist |
| ACP5 | 5.36724 | 152.303 | 28.37640948 | 5.00E-05 | 0.00257671 | acid phosphatase 5, tartrate resistant |
| APOC2 | 1.92664 | 53.409 | 27.72131794 | 5.00E-05 | 0.00257671 | apolipoprotein C-II |
| PLA2G7 | 1.12637 | 30.951 | 27.47853725 | 5.00E-05 | 0.00257671 | phospholipase A2, group VII (platelet- activating factor acetylhydrolase, plasma) |
| Significantly downregulated genes with the highest absolute fold change | |||||
|---|---|---|---|---|---|
| Gene Name | STA FPKM | Cerebral Aneurysm FPKM |
Log Fold | Q-Value | Description |
| IFNA20P | 1.18 | 0.00 | −inf | 2.26E-02 | interferon, alpha 20, pseudogene |
| PLA2G2A | 106.23 | 0.84 | −6.98 | 2.58E-03 | phospholipase A2, group IIA (platelets, synovial fluid) |
| SFRP5 | 7.50 | 0.07 | −6.82 | 2.95E-02 | secreted frizzled-related protein 5 |
| PCP4L1 | 7.94 | 0.08 | −6.56 | 2.87E-02 | Purkinje cell protein 4 like 1 |
| PLIN1 | 12.42 | 0.16 | −6.28 | 7.63E-03 | perilipin 1 |
| ITLN1 | 565.18 | 7.26 | −6.28 | 2.58E-03 | intelectin 1 (galactofuranose binding) |
| CIDEC | 9.74 | 0.17 | −5.83 | 2.58E-03 | cell death-inducing DFFA-like effector c |
| CILP | 5.76 | 0.10 | −5.81 | 2.58E-03 | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| C1orf132 | 37.97 | 0.76 | −5.65 | 2.58E-03 | chromosome 1 open reading frame 132 |
| MIR1-2 | 59.43 | 1.22 | −5.61 | 2.58E-03 | microRNA 1-2 |
| CTB-12O2.1 | 16.33 | 0.36 | −5.49 | 2.58E-03 | Uncharacterized noncoding RNA |
| CHRDL1 | 43.49 | 1.03 | −5.40 | 2.58E-03 | chordin-like 1 |
| PCP4 | 82.39 | 2.28 | −5.18 | 2.58E-03 | Purkinje cell protein 4 |
| CIDEA | 2.67 | 0.07 | −5.17 | 1.75E-02 | cell death-inducing DFFA-like effector a |
| PPP1R1A | 61.02 | 1.83 | −5.06 | 2.58E-03 | protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| FAM180B | 6.82 | 0.22 | −4.93 | 2.58E-03 | family with sequence similarity 180, member B |
| CCL21 | 26.95 | 0.93 | −4.86 | 2.58E-03 | chemokine (C-C motif) ligand 21 |
| SBSPON | 49.80 | 1.76 | −4.82 | 4.50E-03 | somatomedin B and thrombospondin, type 1 domain containing |
| MIR4453 | 27.00 | 0.97 | −4.80 | 2.58E-03 | microRNA 4453 |
| AZGP1 | 11.11 | 0.40 | −4.79 | 2.58E-03 | alpha-2-glycoprotein 1, zinc-binding |
FPKM: Fragments Per Kilobase of transcript per million fragments mapped; BA: brain aneurysm; STA: superficial temporal artery
MiRNA expression profile
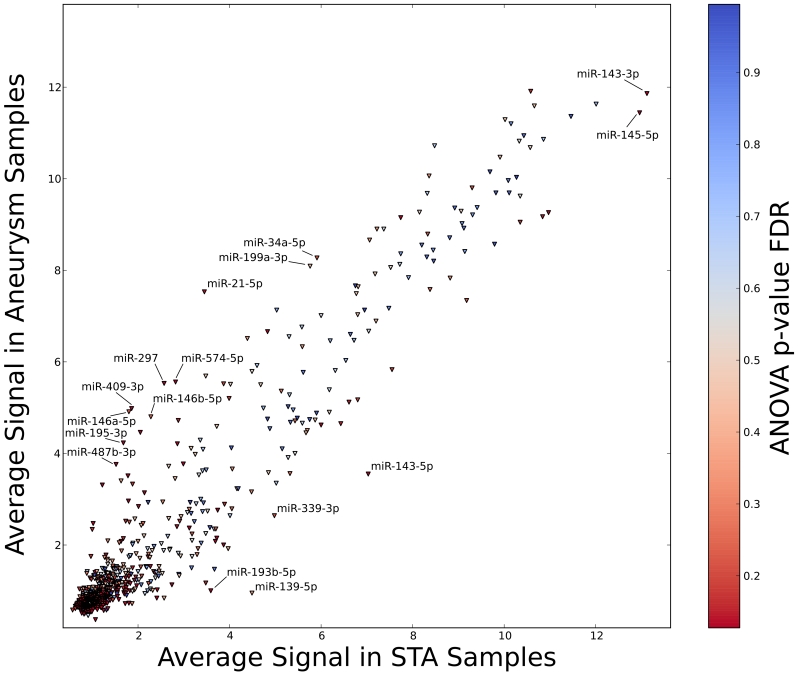
We identified 1,338 miRNA (Figure 2) with differential expression between aneurysm and control tissue, at an FDR of 0.15. Among these, 19 have a log fold change of 2 or greater, and 5 have a log fold change of less than −2. The 15 most upregulated and the 15 most downregulated miRNA are presented in Table 2. The mature miRNAs with the largest absolute fold change were miR-21 (Fold change 16.97), and miR-143-5p (Fold change −11.14). The 3-prime product of miR-143, which has conserved target sites, was also significantly downregulated (Fold change −5.27). Both miR-21 and miR-143 have been implicated in the pathogenesis of AAA. These results were confirmed with the use of Nanostring technology, with results that were correlated with the original microarray results (Supplemental Figure I).
Figure 2.
Differential expression of microRNAs with annotated targets by microarray. Each triangle corresponds to a microRNA, shaded by a color corresponding to the FDR at which it is significant. Highly significant microRNAs are labeled.
Table 2. Significantly differentially expressed microRNAs with the highest fold change.
| Mature miRNA Name |
Aneurysm Signal |
STA Signal |
Fold Change |
ANOVA p-value |
ANOVA FDR |
|---|---|---|---|---|---|
| UPREGULATED | |||||
| miR-21-5p | 7.53 | 3.45 | 16.97 | 0.075185 | 0.139337 |
| hsa-miR-1246 | 5.86 | 2.36 | 11.26 | 0.005104 | 0.127714 |
| hsa-miR-6875-3p | 5.87 | 2.43 | 10.81 | 0.001077 | 0.127714 |
| hsa-miR-6753-3p | 5.59 | 2.37 | 9.28 | 0.00051 | 0.127714 |
| hsa-miR-4685-3p | 5.32 | 2.17 | 8.88 | 0.000825 | 0.127714 |
| hsa-miR-409-3p | 4.98 | 1.86 | 8.68 | 0.004163 | 0.127714 |
| hsa-miR-297 | 5.53 | 2.57 | 7.75 | 0.000597 | 0.127714 |
| hsa-miR-4484 | 8.84 | 5.98 | 7.24 | 0.000192 | 0.127714 |
| hsa-miR-1972 | 4.8 | 2.03 | 6.85 | 0.000263 | 0.127714 |
| hsa-miR-574-5p | 5.56 | 2.82 | 6.69 | 0.003852 | 0.127714 |
| hsa-miR-6877-3p | 6.47 | 3.92 | 5.86 | 0.007492 | 0.127714 |
| hsa-miR-487b-3p | 4.23 | 1.68 | 5.85 | 0.022793 | 0.127714 |
| hsa-miR-619-5p | 8.87 | 6.35 | 5.72 | 0.00016 | 0.127714 |
| hsa-miR-195-3p | 4.46 | 2.05 | 5.33 | 0.000684 | 0.127714 |
| hsa-miR-4793-3p | 4.61 | 2.24 | 5.16 | 0.000186 | 0.127714 |
| hsa-miR-4440 | 5.37 | 3.07 | 4.93 | 0.000441 | 0.127714 |
| hsa-miR-382-5p | 3.76 | 1.52 | 4.72 | 0.009787 | 0.127714 |
| hsa-miR-432-5p | 3.31 | 1.22 | 4.25 | 0.010858 | 0.127714 |
| hsa-miR-3197 | 4.65 | 2.64 | 4.04 | 0.013788 | 0.127714 |
| hsa-miR-127-3p | 6.66 | 4.83 | 3.55 | 0.014006 | 0.127714 |
| DOWNREGULATED | |||||
| hsa-miR-143-5p | 3.55 | 7.03 | −11.14 | 0.006126 | 0.127714 |
| hsa-miR-3195 | 2.09 | 5.08 | −7.93 | 0.007712 | 0.127714 |
| hsa-miR-6068 | 3.5 | 6.18 | −6.39 | 0.00964 | 0.127714 |
| hsa-miR-193b-5p | 1 | 3.59 | −6.01 | 0.016744 | 0.127714 |
| hsa-miR-6848-5p | 2.34 | 4.61 | −4.81 | 0.058811 | 0.130779 |
| hsa-miR-4655-5p | 1.95 | 3.83 | −3.67 | 0.072579 | 0.137742 |
| hsa-miR-606 | 2 | 3.87 | −3.64 | 0.007971 | 0.127714 |
| hsa-miR-3613-3p | 10.58 | 12.39 | −3.51 | 0.014591 | 0.127714 |
| hsa-miR-4445-3p | 2.47 | 4.23 | −3.39 | 0.02922 | 0.127714 |
| hsa-miR-320a | 9.26 | 10.97 | −3.26 | 0.069366 | 0.135963 |
| hsa-miR-5010-5p | 0.95 | 2.64 | −3.21 | 0.037616 | 0.127714 |
| hsa-miR-320b | 9.17 | 10.84 | −3.19 | 0.088324 | 0.148826 |
| hsa-miR-139-3p | 1.13 | 2.74 | −3.06 | 0.001682 | 0.127714 |
| hsa-miR-933 | 2.08 | 3.69 | −3.06 | 0.014096 | 0.127714 |
| hsa-miR-940 | 2.15 | 3.72 | −2.97 | 0.028514 | 0.127714 |
| hsa-miR-452-5p | 0.84 | 2.41 | −2.96 | 0.02465 | 0.127714 |
| hsa-miR-4743-5p | 1.48 | 3.02 | −2.9 | 0.072175 | 0.137684 |
| hsa-miR-1468-3p | 1.73 | 3.25 | −2.86 | 0.003147 | 0.127714 |
| hsa-miR-145-5p | 11.44 | 12.96 | −2.86 | 0.040347 | 0.127714 |
| hsa-miR-6889-5p | 1.39 | 2.91 | −2.85 | 0.059015 | 0.130779 |
STA: superficial temporal artery; ANOVA: analysis of variance
MiRNA regulatory effects
We investigated our cohorts for pairs of miRNAs and their target genes that showed changes in expression that are consistent with miRNA regulation. We analyzed Targetscan Human Release 6.2, consisting of 72,770 miRNA family-gene pairs with annotated target sites in the 3’ UTR of at least one isoform. Since each miRNA family has multiple mature miRNA members, we had 165,560 distinct miRNA-gene pairs with measured expression data to investigate. Within this dataset, there were 495 miRNA-gene pairs with predicted conserved target sites that satisfied both FDR thresholds defined above. Further, we required that the sites have a probability of conserved target (PCT) of 0.25 or greater, which resulted in 370 miR-gene pairs, 68.7% of which show opposite changes in expression consistent with microRNA regulation.19
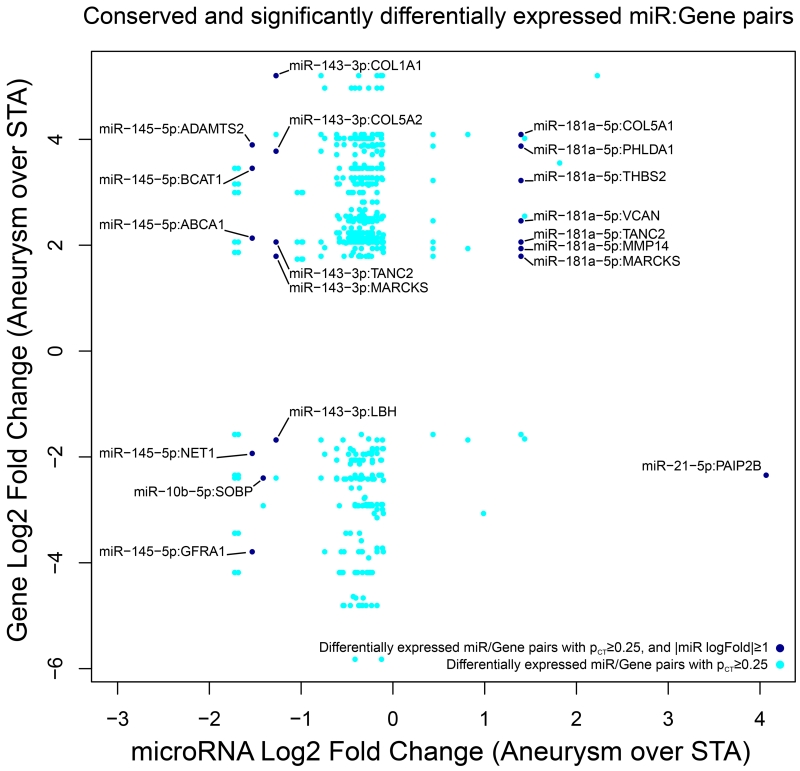
We then examined the effect of highly differentially expressed miRNA with a fold change of 2 or greater in this dataset. Only 5 microRNAs were differentially expressed at this level, targeting 17 distinct genes (Figure 3). Most of these miRNAs have significantly differentially expressed target genes with opposite changes in expression compared to the miRNA. The exceptions to this pattern are miR-10b and miR-181a. Although other regulatory effects could explain these differences, it is notable that both of these miRNAs have been shown to exhibit expression that is significantly correlated with their targets.16 Graphs demonstrating the correlation of these miRNAs with the expression of their targets are presented in Supplemental Figure II, III, IV, V, and VI.
Figure 3.
The most significant miRNA-gene pairs. Each dot corresponds to a miRNA-gene pair, where dark blue corresponds to pairs with a miRNA fold change of 2 or greater, and cyan for all other pairs. Each column of dots with the same x-value corresponds to a single miRNA. At this level of significance, the majority of targeted genes have opposite changes in expression compared with the targeting miRNA.
Specifically, miR-21 was upregulated with significant downregulation of its target gene PAIP2B, a translational inhibitor. MiR-143 was downregulated with significant upregulation of several of its targets, including multiple genes involved in the metabolism of collagen (COL1A1, COL5A1, COL5A2), MARCKS (encoding an actin filament crosslinking protein), and TANC2. MiR-145 was also downregulated with corresponding upregulation of ABCA1, ADAMTS2, and BCAT1. The significant differential expression of miR-21, miR-143, and miR-145 was corroborated with Nanostring (Supplemental Figure VII, and VIII).
We additionally examined the role of strong conservation of microRNA target sites by requiring a PCT of 0.75 or greater for the miRNA-genes pairs that match our FDR significance thresholds. At this conservation level, 76.1% of all miRNA-gene pairs show opposite changes in expression. This list consisted of 83 target sites for 33 miRNA within 78 genes. The list of genes with 10 or more highly conserved target sites included only 5 genes including BCAT1, TANC2, ABCA1, SIK1, and FOXF2. We found that all of the miRNAs targeting these genes showed opposite changes in expression, suggesting that the regulation of these genes could be the aggregate effect of a number of differentially expressed miRNAs. These results are summarized in Supplemental Table III.
DISCUSSION
In this prospective study we describe the gene and miRNA expression in unruptured cerebral aneurysm tissue in human subjects undergoing surgical clipping. Additionally, we detected gene-miRNA pairs with anti-correlated expression changes consistent with microRNA regulation. Our analysis addresses methodological drawbacks of prior studies, and utilizes deep sequencing to investigate the gene profile of unruptured human cerebral aneurysm tissue for the first time. Although changes in aneurysmal gene expression have been identified in microarray studies, the role of miRNA in these changes is unknown. These molecules post-transcriptionally regulate mRNAs with complementary seed matches, hence could play a significant role in gene regulation.10 In addition, they are highly conserved among eukaryotic organisms, making them ideal for reverse translational research in animal models of disease.
We identified a large number of differentially expressed genes in aneurysmal tissue. The most upregulated protein-coding gene was MMP-13, several members of which have been implicated6,7,21,22,36,37 in extracellular matrix (ECM) remodeling of intracranial blood vessels. This results in loss of structural support and higher propensity for cerebral aneurysm formation. Additionally, this process can be mediated by most other highly upregulated genes in our cohort, which were involved in inflammatory regulation, ECM remodeling (SPP1), lipid metabolism (APOC1), and collagen formation (COL1A1, COL5A1, COL5A2, and others).3-5,8,9,24 Several members of the collagen family were overexpressed, some of which have been identified in prior microarray studies.25,27,30,35 This is likely a response to the degradation of ECM and thinning of the arterial wall, resulting in fibrosis. The downregulated genes in our cohort had a complimentary effect to the overexpressed ones, with potential role in defective ECM (CILP), inadequate lipid metabolism and storage (ITLN1, PLIN1 and its target CIDEC), as well as defective apoptosis (CIDEC, SFRP5), and intracellular signaling (PLA2G2A).
The miRNA expression profile of cerebral aneurysms was also investigated. In order to limit our analysis to functionally significant miRNA, which result in post-transcriptional mRNA degradation, we investigated our sample for anti-correlated miRNA-mRNA pairs. MiR-21 demonstrated the highest expression in our cohort, and was associated with multiple downregulated target genes, with PAIP2B (a translation repressor) being the most significant. Overexpression of miR-21 has been identified in human subjects and animal models of AAA, but not in human cerebral aneurysms before.1,11,23,28,38 There is controversy in regards to its role in AAA pathogenesis. Some animal AAA models28 have demonstrated that upregulation of miR-21 results in a protective cellular proliferation, in an attempt to repair the defective vascular wall. Others1,11,23,38 have supported that miR-21 contributes to AAA formation through downregulation of ECM remodeling genes. Given the differences in pathophysiology between AAA and cerebral aneurysms, further research to unravel the role of miR-21 in the latter is needed.
MiR-143 is the most significantly downregulated miRNA in our patients, and has three upregulated target collagen genes, along with MARCKS and TANC2. Additionally, miR-143 downregulation has been identified as a central step contributing to VSMC phenotypic modulation, which is crucial in cerebral aneurysm formation.11,38 During this process, VSMC lose their contractile apparatus and acquire a synthetic phenotype. As a result, they dissociate from each other and migrate into the intima, producing myointimal hyperplasia.11,38 Several authors have demonstrated that this process results in loss of structural integrity of the media, the layer providing support to the vessel wall.11,38 Downregulation of miR-145 plays a complementary role in VSMC homeostasis,13,15,17,39 and has been implicated in the pathophysiology of AAA in prior studies.11,38 In addition, it has 3 significant upregulated targets with potential involvement in the pathophysiology of cerebral aneurysms. ADAMTS2 is a metalloproteinase involved in collagen synthesis and ECM remodeling. BCAT1 contributes to cellular proliferation, and ABCA1 is a cholesterol efflux regulatory protein. ABCA1 and BCAT1 also had highly conserved target sites for more than 10 distinct miRNAs, all downregulated in expression, suggesting that other miRNAs could also play a role in their upregulation.
A prior investigation of miRNA expression patterns by Jiang et al20 focused on ruptured cerebral aneurysms, contrary to our study investigating only unruptured aneurysms. The generalized inflammation associated with aneurysm rupture and subarachnoid hemorrhage, might mask the tissue miRNA expression patterns, and make it difficult to recognize which changes are attributable to inflammation and which are part of the aneurysm formation cascade. In order to address this, we elected to focus on unruptured aneurysms only. MiR143 and miR145 were found to be downregulated by Jiang et al,20 in agreement with our study. However, they did not identify any upregulated miRNAs, contrary to our investigation. Jiang et al20 did not demonstrate anti-correlated pairs of miRNA and mRNA in order to identify functionally significant miRNAs. This limitation was addressed in our study.
The current study has several limitations. First, the selection of STA as the control tissue is not ideal. Although both the STA and the intracranial vessels arise from the same parent artery (common carotid artery), extracranial vessels can potentially have different baseline genetic profile (given the presence of adventitia) in comparison to intracranial vessels. However, collecting control tissue from normal intracranial vessels is not possible, and STA has been a well-established control in prior literature.34,35 In an attempt to further refine this control tissue, we utilized specimens not only from patients with aneurysms, but also from patients with unrelated intracranial pathology, or no pathology at all.
Second, the present observational study design cannot establish whether the identified profiles resulted in aneurysm formation or were an epiphenomenon, secondary to the presence of the aneurysm. In addition, we cannot identify the crucial molecular pathways, the inhibition of which could halt or reverse aneurysm progression. However, the current data create a framework for further investigation in an animal model. The first step in this process would involve the creation of genetically modified endothelial cell lines, where the identified miRNA targets would be up- or down- regulated, in an effort to identify the downstream effects of these interventions, and whether the latter agree with our study. Subsequently, we would utilize rodent cerebral aneurysm models, where we would silence upregulated miRNAs and monitor aneurysm progression and rupture. The latter would establish whether the identified targets are involved in crucial steps of aneurysm pathophysiology, and would delineate the mechanism behind cerebral aneurysm progression and rupture.
Third, there is a plethora of cells that are collected and analyzed together with the aneurysm tissue, although their concentration is not expected to be different among different samples. Fourth, our results are limited to aneurysms that were large enough to allow safe resection during surgery. The molecular profiles of smaller aneurysms might be different, and our findings do not necessarily apply to that population. Fifth, we collected aneurysms for which treatment was deemed necessary, either due to patient preference or because the aneurysm harbored some dangerous features. Though these concerns limit the generalization of our results, acquiring tissue from aneurysms that are deemed appropriate for observation is not feasible. Sixth, our sample size is limited. However, this is a preliminary single center study, which can be used as the basis for further multicenter investigations.
Conclusions
Although there is still much work to be done to fully understand the molecular mechanisms behind cerebral aneurysm formation and rupture, we have uncovered some likely facets of gene regulation in these tissues. Our analysis identified several significantly differentially expressed genes and miRNAs in unruptured human cerebral aneurysms. The strongest changes in expression were observed for miR-21, miR-143, miR-145 and their target genes. The majority of these genes are involved in collagen formation, inflammation regulation, lipid metabolism, smooth muscle phenotypic modification, and extracellular matrix remodeling, processes which have been implicated in cerebral aneurysm formation. Further research to separate causative and responsive gene expression differences may identify potential therapeutic targets among these miRNA-mRNA pairs.
Supplementary Material
Acknowledgments
Funding. This study was supported in part by the Dandy Fellowship Award (KB) by the Congress of Neurological Surgeons and by a Hitchcock Foundation Pilot Grant (KB) by the Hitchcock Foundation. The funders had no participation in the study design, data analysis and interpretation, or the manuscript creation.
Footnotes
Disclosures: none
REFERENCES
- 1.Adam M, Raaz U, Spin JM, Tsao PS. MicroRNAs in Abdominal Aortic Aneurysm. Curr Vasc Pharmacol. 2013 doi: 10.2174/15701611113119990015. published online ahead of print May 13, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Atheroscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki T, Kataoka H, Ishibashi R, Nakagami H, Nozaki K, Morishita R, et al. Pitavastatin suppresses formation and progression of cerebral aneurysms through inhibition of the nuclear factor kappaB pathway. Neurosurgery. 2009;64:357–365. doi: 10.1227/01.NEU.0000336764.92606.1D. [DOI] [PubMed] [Google Scholar]
- 4.Aoki T, Kataoka H, Ishibashi R, Nozaki K, Hashimoto N. Cathepsin B, K, and S are expressed in cerebral aneurysms and promote the progression of cerebral aneurysms. Stroke. 2008;39:2603–2610. doi: 10.1161/STROKEAHA.107.513648. [DOI] [PubMed] [Google Scholar]
- 5.Aoki T, Kataoka H, Ishibashi R, Nozaki K, Hashimoto N. Simvastatin suppresses the progression of experimentally induced cerebral aneurysms in rats. Stroke. 2008;39:1276–1285. doi: 10.1161/STROKEAHA.107.503086. [DOI] [PubMed] [Google Scholar]
- 6.Aoki T, Kataoka H, Morimoto M, Nozaki K, Hashimoto N. Macrophage-derived matrix metalloproteinase-2 and -9 promote the progression of cerebral aneurysms in rats. Stroke. 2007;38:162–169. doi: 10.1161/01.STR.0000252129.18605.c8. [DOI] [PubMed] [Google Scholar]
- 7.Aoki T, Kataoka H, Moriwaki T, Nozaki K, Hashimoto N. Role of TIMP-1 and TIMP-2 in the progression of cerebral aneurysms. Stroke. 2007;38:2337–2345. doi: 10.1161/STROKEAHA.107.481838. [DOI] [PubMed] [Google Scholar]
- 8.Aoki T, Kataoka H, Shimamura M, Nakagami H, Wakayama K, Moriwaki T, et al. NF-kappaB is a key mediator of cerebral aneurysm formation. Circulation. 2007;116:2830–2840. doi: 10.1161/CIRCULATIONAHA.107.728303. [DOI] [PubMed] [Google Scholar]
- 9.Aoki T, Nishimura M, Matsuoka T, Yamamoto K, Furuyashiki T, Kataoka H, et al. PGE(2) -EP(2) signalling in endothelium is activated by haemodynamic stress and induces cerebral aneurysm through an amplifying loop via NF-κB. Br J Pharmacol. 2011;163:1237–1249. doi: 10.1111/j.1476-5381.2011.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boon RA, Dimmeler S. MicroRNAs and aneurysm formation. Trends Cardiovasc Med. 2011;21:172–177. doi: 10.1016/j.tcm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 13.Boucher JM, Peterson SM, Urs S, Zhang C, Liaw L. The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286:28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broderick JP, Sauerbeck LR, Foroud T, Huston Jr, Pankratz N, Meissner I, et al. The Familial Intracranial Aneurysm (FIA) study protocol. BMC Med Genet. 2005;26:6–17. doi: 10.1186/1471-2350-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 17.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foroud T, Sauerbeck L, Brown R, Anderson C, Woo D, Kleindorfer D, et al. Genome screen to detect linkage to intracranial aneurysm susceptibility genes: the Familial Intracranial Aneurysm (FIA) study. Stroke. 2008;39:1434–1440. doi: 10.1161/STROKEAHA.107.502930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Zhang M, He H, Chen J, Zeng H, Li J, et al. MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med Genomics. 2013;6:36. doi: 10.1186/1755-8794-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin D, Sheng J, Yang X, Gao B. Matrix metalloproteinases and tissue inhibitors of metalloproteinases expression in human cerebral ruptured and unruptured aneurysm. Surg Neurol. 2007;68(Suppl 2):S11–16. doi: 10.1016/j.surneu.2007.02.060. [DOI] [PubMed] [Google Scholar]
- 22.Kim SC, Singh M, Huang J, Prestigiacomo CJ, Winfree CJ, Solomon RA, et al. Matrix metalloproteinase-9 in cerebral aneurysms. Neurosurgery. 1997;41:642–666. doi: 10.1097/00006123-199709000-00027. [DOI] [PubMed] [Google Scholar]
- 23.Kin K, Miyagawa S, Fukushima S, Shirakawa Y, Torikai K, Shimamura K, et al. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc. 2012;1:e000745. doi: 10.1161/JAHA.112.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosierkiewicz TA, Factor SM, Dickson DW. Immunocytochemical studies of atherosclerotic lesions of cerebral berry aneurysms. J Neuropathol Exp Neurol. 1994;53:399–406. doi: 10.1097/00005072-199407000-00012. [DOI] [PubMed] [Google Scholar]
- 25.Krischek B, Kasuya H, Tajima A, Akagawa H, Sasaki T, Yoneyama T, et al. Network-based gene expression analysis of intracranial aneurysm tissue reveals role of antigen presenting cells. Neuroscience. 2008;154:1398–1407. doi: 10.1016/j.neuroscience.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 26.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Yang X, Jiang F, Dusting GJ, Wu Z. Transcriptome-wide characterization of gene expression associated with unruptured intracranial aneurysms. Eur Neurol. 2009;62:330–337. doi: 10.1159/000236911. [DOI] [PubMed] [Google Scholar]
- 28.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. 2012;4:122ra122. doi: 10.1126/scitranslmed.3003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. 2012;122:497–506. doi: 10.1172/JCI61598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchese E, Vignati A, Albanese A, Nucci CG, Sabatino G, Tirpakova B, et al. Comparative evaluation of genome-wide gene expression profiles in ruptured and unruptured human intracranial aneurysms. J Biol Regul Homeost Agents. 2010;24:185–195. [PubMed] [Google Scholar]
- 31.Merk DR, Chin JT, Dake BA, Maegdefessel L, Miller MO, Kimura N, et al. miR-29b participates in early aneurysm development in Marfan syndrome. Circ Res. 2012;110:312–324. doi: 10.1161/CIRCRESAHA.111.253740. [DOI] [PubMed] [Google Scholar]
- 32.Molyneux A, Kerr R, Stratton I, Sandercock P, Clarke M, Shrimpton J, et al. International Subarachnoid Aneurysm Trial (ISAT) of neurosurgical clipping versus endovascular coiling in 2143 patients with ruptured intracranial aneurysms: a randomised trial. Lancet. 2002;360:1267–1274. doi: 10.1016/s0140-6736(02)11314-6. [DOI] [PubMed] [Google Scholar]
- 33.Pera J, Korostynski M, Krzyszkowski T, Czopek J, Slowik A, Dziedzic T, et al. Gene expression profiles in human ruptured and unruptured intracranial aneurysms: what is the role of inflammation? Stroke. 2010;41:224–231. doi: 10.1161/STROKEAHA.109.562009. [DOI] [PubMed] [Google Scholar]
- 34.Roder C, Kasuya H, Harati A, Tatagiba M, Inoue I, Krischek B. Meta-analysis of microarray gene expression studies on intracranial aneurysms. Neuroscience. 2012;201 doi: 10.1016/j.neuroscience.2011.10.033. [DOI] [PubMed] [Google Scholar]
- 35.Shi C, Awad IA, Jafari N, Lin S, Du P, Hage ZA, et al. Genomics of human intracranial aneurysm wall. Stroke. 2009;40:1252–1261. doi: 10.1161/STROKEAHA.108.532036. [DOI] [PubMed] [Google Scholar]
- 36.Takemura Y, Hirata Y, Sakata N, Nabeshima K, Takeshita M, Inoue T. Histopathologic characteristics of a saccular aneurysm arising in the non-branching segment of the distal middle cerebral artery. Pathol Res Pract. 2010;206:391–396. doi: 10.1016/j.prp.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Tronc F, Mallat Z, Lehoux S, Wassef M, Esposito B, Tedgui A. Role of matrix metalloproteinases in blood flow-induced arterial enlargement: interaction with NO. Arterioscler Thromb Vasc Biol. 2000;20:E120–126. doi: 10.1161/01.atv.20.12.e120. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Schober A, Weber C. Pathogenic arterial remodeling: the good and bad of microRNAs. Am J Physiol Heart Circ Physiol. 2013;304:H1050–1059. doi: 10.1152/ajpheart.00267.2012. [DOI] [PubMed] [Google Scholar]
- 39.Xin M, S EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.