Abstract
Background
Epidemiological findings suggest a relationship between multiple sclerosis (MS) and cardiovascular disease (CVD) risk factors, although the nature of this relationship is not well understood.
Objective
We used genome-wide association study (GWAS) data to identify shared genetic factors (pleiotropy) between MS and CVD risk factors.
Methods
Using summary statistics from large, recent GWAS (total n > 250,000 individuals), we investigated overlap in single nucleotide polymorphisms (SNPs) associated with MS and a number of CVD risk factors including triglycerides (TG), low density lipoproteins cholesterol (LDL), high density lipoproteins cholesterol (HDL), body mass index, waist-hip-ratio, type 2 diabetes, systolic blood pressure and C-reactive protein level.
Results and Conclusion
Using conditional enrichment plots we found 30-fold enrichment of MS SNPs for different levels of association with LDL and TG SNPs, with a corresponding reduction in conditional False Discovery Rate. We identified 133 pleiotropic loci outside the extended Major Histocompatibility Complex with conditional False Discovery Rate < 0.01, of which 65 are novel. These pleiotropic loci were located on 21 different chromosomes. Our findings point to overlapping pathobiology between clinically diagnosed MS and cardiovascular risk factors and identify novel common variants associated with increased MS risk.
Keywords: multiple sclerosis, pleiotropy, gene discovery, cardiovascular risk factors
INTRODUCTION
Multiple sclerosis is an autoimmune disease characterized by demyelination of the central nervous system1. A recent systematic review of 6 databases suggests that CVD risk is increased among patients with MS2. However, it is unclear whether an increased CVD risk is secondary to lifestyle and environmental variables, such as medication use, dietary factors and physical activity or due to overlapping pathobiology between CVD risk factors and MS.
Large genome-wide association studies (GWAS) provide valuable insights into the role of biologic pathways in disease pathogenesis and have identified genetic polymorphisms associated with a range of human disorders and phenotypes3, 4. Recent GWAS have identified a total of 110 single nucleotide polymorphisms (SNPs) associated with MS5, 6. Combining GWAS from multiple disorders and phenotypes provides insights into genetic pleiotropy (defined as a single gene or variant being associated with more than one distinct phenotype) and could elucidate shared pathobiology. Using this approach, we have recently reported genetic overlap between a number of diseases and phenotypes and identified novel common variants associated with schizophrenia, bipolar disorder, prostate cancer, hypertension, primary sclerosing cholangitis, and Alzheimer’s disease7–15. Here, taking advantage of several large GWASs, we evaluated genetic overlap between MS and a number of CVD risk factors, including systolic blood pressure (SBP)16, low density lipoprotein (LDL) cholesterol17, high density lipoprotein (HDL) cholesterol17, triglycerides (TG)17, type 2 diabetes (T2D)18, body mass index (BMI)19, waist to hip ratio (WHR)20, and C-reactive protein level (CRP)21.
MATERIALS and METHODS
Ethics Statement
The relevant institutional review boards or ethics committees approved the research protocol of the individual GWAS used in the current analysis and all human participants gave written informed consent.
Participant Samples
We utilized summary statistics GWAS data (p-values and odds ratios) from the International Multiple Sclerosis Genetics Consortium (IMSGC), n=27,148)6 and from GWAS evaluating systolic blood pressure (SBP; n=203,056)16, low density lipoprotein (LDL; n=188,577)17, high density lipoprotein (HDL; n=188,577)17, triglycerides (TG; n=188,577)17, type 2 diabetes (T2D; n=22,044)18, body mass index (BMI; n=123,865)19, waist to hip ratio (WHR; n=77,167)20 and C-reactive protein level (CRP; n=66,185)21 (for details please see Supplementary Table 1). All studies were approved by the respective ethical committees and institutional review boards.
Statistical analysis
Using recently developed statistical methods to evaluate pleiotropic effects7–11, we evaluated genetic overlap between MS and CVD risk factors. For given associated phenotypes A and B, pleiotropic ‘enrichment’ of phenotype A with phenotype B exists if the proportion of SNPs or genes associated with phenotype A increases as a function of increased association with phenotype B (see Supplementary Text for details). To assess for pleiotropic enrichment, we constructed fold-enrichment plots of empirical quantiles of nominal −log10(p) values for SNP association with MS for all SNPs, and for subsets of SNPs determined by the nominal p-values of their association with CVD factors (BMI, CRP, HDL, LDL, SBP, T2D, TG and WHR). In fold-enrichment plots, the presence of enrichment is reflected as an upward deflection of the curve for phenotype A if the degree of deflection from the expected null line is dependent on the degree of association with phenotype B. To assess for polygenic effects below the standard GWAS significance threshold, we focused the fold-enrichment plots on SNPs with nominal −log10(p) < 7.3 (corresponding to p > 5×10−8). The nominal p-values (−log10(p)) are plotted on the x-axis, and cumulative relative fold enrichment in MS is plotted on the y-axis (Figure 1).
Figure 1. Pleiotropic enrichment of MS and CVD factors.
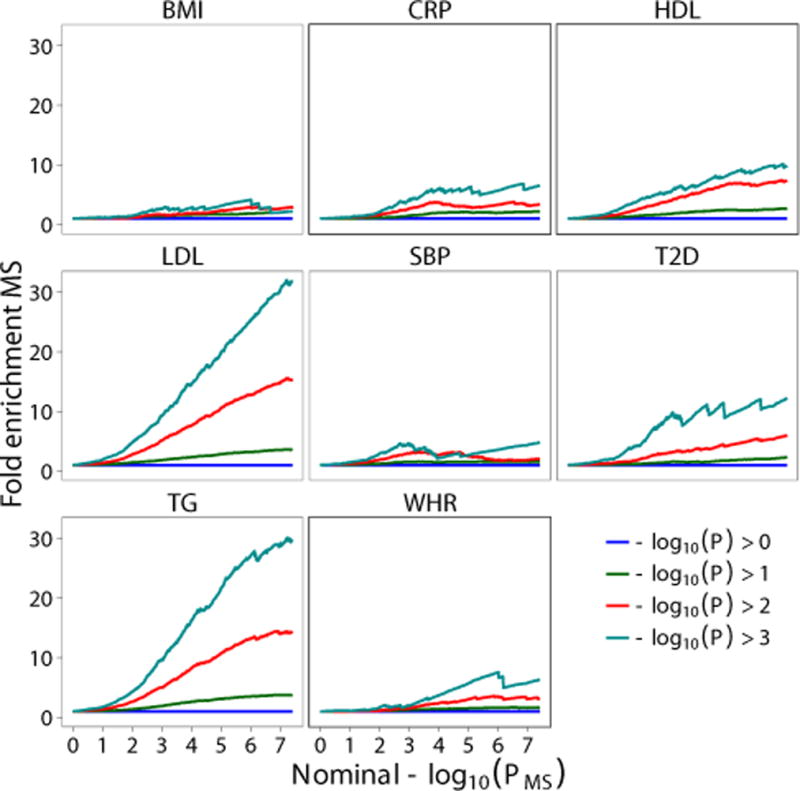
Fold enrichment plots of enrichment versus nominal −log10 p-values in multiple sclerosis (MS) below the standard GWAS threshold of p < 5×10−8 as a function of the association level with body mass index (BMI), C-reactive protein level (CRP), High density Lipoprotein cholesterol (HDL), Low density Lipoprotein cholesterol (LDL), systolic blood pressure (SBP), type 2 diabetes (T2D), triglycerides (TG) and Waist Hip Ratio (WHR) at the level of −log10(p) ≥ 0, −log10(p) ≥ 1, −log10(p) ≥ 2, −log10(p) ≥ 3 corresponding to p ≤ 1, p ≤ 0.1, p ≤ 0.01, p ≤ 0.001, respectively. Successive upward elevation in terms of all SNPs (−log10(p) ≥ 0, blue horizontal line) demonstrate pleiotropic enrichment of MS association conditioned CVD factors. The figure also shows that the fold enrichment of MS (y axis is also a monotonic increasing function of the nominal P value (x axis). All data are first genome corrected by intergenic SNPs.
To identify specific loci associated with MS we computed conditional False Discovery Rates (FDRs). The standard FDR framework is based on a mixture model of SNPs associated with the phenotype (either associated; non-null SNPs, or not; null SNPs). The conditional FDR is an extension of the standard FDR, which incorporates information from GWAS summary statistics of a second phenotype. Specifically, MS SNPs were stratified on the basis of p values of each of the CVD factors, separately. Then based on the combination of p values for SNPs in MS and each of the CVD factors, we assigned a conditional FDR value (FDRMS|CVD, CVD represent one of BMI, CRP, LDL, HDL, T2D, SBP, TG and WHR) to each SNP for MS by interpolating into a 2-D lookup table (Supplementary Figure 1). We used a conditional FDR threshold of 0.01, which means 1 false discovery per hundred. Loci thus identified can be visualized by a conditional Manhattan plot (Figure 2). It is important to note that ranking SNPs by FDR or by p-values both give the same ordering of SNPs, whereas the conditional FDR re-orders SNPs resulting in a different p-value based ranking if the primary and secondary phenotype are genetically related.
Figure 2. ‘Conditional FDR Manhattan plot’ of multiple sclerosis (MS) on cardiovascular disease risk factors.
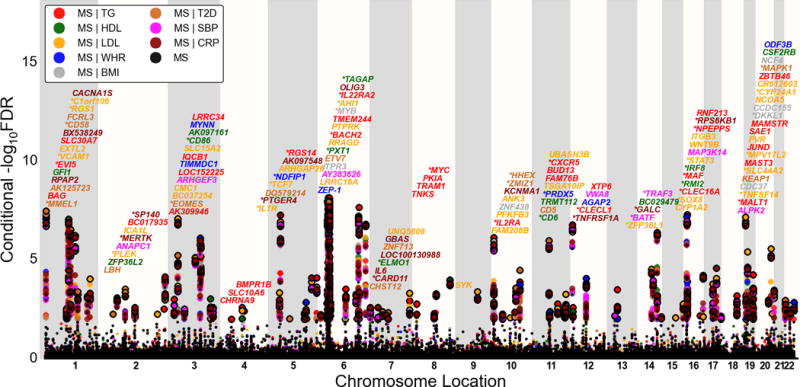
The unconditioned −log10 (FDR) values for multiple sclerosis (MS) alone (black) and conditioned on given cardiovascular disease risk factors triglycerides (TG; MS|TG), Low density Lipoprotein cholesterol (LDL; MS|LDL), High density Lipoprotein cholesterol (HDL; MS|HDL), systolic blood pressure (SBP; MS|SBP), body mass index (BMI; MS|BMI), waist hip ratio (WHR; MS|WHR), type 2 diabetes (T2D; MS|T2D) and C-reactive protein (CRP; MS|CRP) were plotted against the genomic locations of SNPs. SNPs with conditional −log10 FDR > 2 (i.e. FDR < 0.01) are shown with large points. A black line around the large points indicates the most significant SNP in each LD block and this SNP was annotated with the closest gene which is listed above the symbols in each locus (except for the xMHC region on chromosome 6). Genes replicated in this study were marked by stars (‘*’). Details for not previously reported non-MHC loci with −log10 FDR > 2 (i.e. FDR < 0.01) are shown in Table 1.
Low conditional FDR values can be driven by association with both phenotypes or with the primary phenotype only. To detect true pleiotropic signal (association with both phenotypes) we computed the conjunctional FDR, computed as the maximum of the two conditional FDR values (i.e., MS conditional on CVD factors and CVD risk factors conditional on MS). Similar to conditional FDR, we assigned to each MS SNP a conjunctional FDR value using a 2-D lookup table (Supplementary Figure 2). We used overall a conjunctional FDR threshold of 0.05, which means 5 expected false discoveries per hundred. To illustrate the genomic location of significant loci we constructed the conjunctional Manhattan plots based on the ranking of conjunctional FDR (Figure 3).
Figure 3. ‘Conjunctional FDR Manhattan plot’ for multiple sclerosis (MS) and cardiovascular disease risk factors.
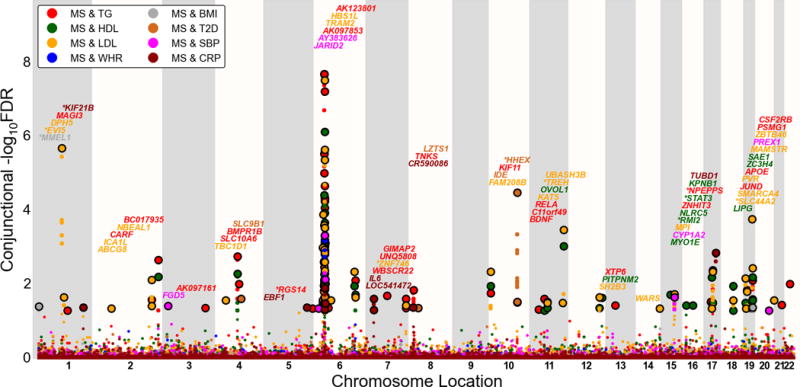
Conjunctional −log10 (FDR) values for multiple sclerosis (MS) given the cardiovascular disease risk factors triglycerides (TG; MS&TG), Low density Lipoprotein cholesterol (LDL; MS&LDL), High density Lipoprotein cholesterol (HDL, MS&HDL), systolic blood pressure (SBP, MS&SBP), body mass index (BMI, MS&BMI), waist hip ratio (WHR, MS&WHR), type 2 diabetes (T2D, MS&T2D) and C-reactive protein level (CRP, MS&CRP). SNPs with conditional −log10 FDR > 1.3 (i.e. FDR < 0.05) are shown with large points. A black line around the large points indicates the most significant SNP in each LD block and this SNP was annotated with the closest gene which is listed above the symbols in each locus (except for the MHC region on chromosome 6). The figure shows the localization of 60 loci on a total of 21 chromosomes. Genes previously reported for MS are marked by stars (“*”) and details for the not previously reported non-MHC loci with −log10 FDR > 1.3 (i.e. FDR < 0.05) are shown in Table 2.
Annotation of new MS associated loci
The list of significant SNPs identified by conditional and conjunctional FDR were binned into independent loci using the LD structure of the European subpopulation from 1000 Genomes Project at the LD-r2 > 0.2 level and a radius of 1 mega base. In addition, the extended MHC region (chr6:25652429–33368333) was considered as a single locus and SNPs close to the same genes were also binned into a single locus. These loci are numbered (locus #) in Tables 1 and 2 and Supplementary Tables 2, 3 and 4. Genes at or closest to each SNP locus were obtained from the HGNC gene database. Any loci that did not contain previously reported MS associated SNPs or genes were deemed as findings adding to the currently known associations in MS (Tables 1 and 2).
Table 1.
Conditional FDR; Independent MS loci given CVD risk factors (MS|CVD) in non-MHC regions excluding previously reported MS loci
| Locus# | SNPa | Chr_locb | Pos | Gene | OR_L | OR_U | P | FDR | Min condFDRc | P expression | Driving pheno |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs4465231 | 1p36.22 | 9347278 | SPSB1 | 1.0427 | 1.1274 | 4.93E-05 | 3.07E-02 | 9.66E-03 | NAd | HDL |
| 2 | rs17519972 | 1p22.1 | 92828505 | RPAP2 | 1.0719 | 1.1489 | 9.42E-08 | 1.33E-04 | 3.25E-05 | 5.96E-03 | CRP |
| 3 | rs6662618 | 1p22 | 92935411 | GFI1 | 1.0965 | 1.1754 | 2.76E-10 | 4.69E-07 | 3.09E-07 | 1.10E-02 | T2D |
| 4 | rs12756986 | 1 | 101604753 | S1PR1 | 1.068 | 1.1594 | 3.52E-06 | 3.05E-03 | 1.97E-03 | NA | HDL |
| rs17123757 | 1 | 101622960 | S1PR1 | 1.0664 | 1.1551 | 3.21E-06 | 3.05E-03 | 1.75E-03 | NA | CRP | |
| 5 | rs3761959 | 1q21 | 157669278 | FCRL3 | 0.8596 | 0.9422 | 9.97E-07 | 1.08E-03 | 3.46E-04 | 1.45E-03 | LDL |
| 6 | rs6733372 | 2p23.1 | 30510444 | LBH | 1.0373 | 1.1008 | 3.47E-05 | 2.12E-02 | 5.42E-03 | 2.47E-02 | T2D |
| 7 | rs1439287 | 2q13 | 111871897 | ACOXL | 1.0367 | 1.1148 | 9.46E-05 | 5.26E-02 | 8.82E-03 | NA | TG |
| 8 | rs12373588 | 2q12.1 | 112466265 | ANAPC1 | 0.8779 | 0.9557 | 1.52E-05 | 1.19E-02 | 3.69E-03 | NA | SBP |
| 9 | rs2052401 | 2q23 | 162962045 | DPP4 | 0.884 | 0.9615 | 5.88E-05 | 3.68E-02 | 7.15E-03 | NA | LDL |
| 10 | rs2036927 | 2q33 | 203640713 | ICA1L | 0.8718 | 0.9596 | 1.08E-04 | 5.26E-02 | 3.50E-03 | NA | HDL |
| 11 | rs2943633 | 2 | 227054881 | MIR548AR | 0.8839 | 0.9614 | 5.83E-05 | 3.68E-02 | 2.08E-03 | NA | TG |
| 12 | rs9821630 | 3 | 16970938 | PLCL2 | 0.8628 | 0.9484 | 7.50E-06 | 6.75E-03 | 1.75E-03 | NA | HDL |
| rs433317 | 3 | 28060456 | CMC1 | 1.0393 | 1.1025 | 1.97E-05 | 1.45E-02 | 8.43E-03 | 8.16E-01 | LDL | |
| 13 | rs338610 | 3p24.1 | 28081442 | CMC1 | 0.8376 | 0.9303 | 2.84E-07 | 3.86E-04 | 2.11E-04 | 8.16E-01 | HDL |
| 14 | rs1500710 | 3p14.3 | 56914065 | ARHGEF3 | 0.8841 | 0.9526 | 6.45E-06 | 5.57E-03 | 2.02E-03 | 8.78E-02 | TG |
| 15 | rs771767 | 3 | 101748638 | ZPLD1 | 1.0713 | 1.1401 | 1.02E-08 | 1.57E-05 | 1.02E-05 | NA | CRP |
| rs1398607 | 3 | 101755738 | ZPLD1 | 1.0794 | 1.1687 | 1.03E-08 | 1.57E-05 | 3.91E-06 | NA | TG | |
| 16 | rs1920296 | 3q21.1 | 121543577 | IQCB1 | 0.8523 | 0.9336 | 7.21E-08 | 1.08E-04 | 5.67E-06 | 5.80E-02 | TG |
| rs4285028 | 3q21.1 | 121660664 | SLC15A2 | 1.0701 | 1.1419 | 3.75E-08 | 5.75E-05 | 2.28E-05 | NA | HDL | |
| 17 | rs10936599 | 3q26.31 | 169492101 | MYNN | 0.8514 | 0.9451 | 9.93E-06 | 8.16E-03 | 2.69E-03 | NA | TG |
| rs1997392 | 3q26.2 | 169509652 | LRRC34 | 0.8561 | 0.9468 | 1.02E-05 | 8.16E-03 | 1.42E-03 | 3.54E-01 | TG | |
| 18 | rs6832151 | 4p14 | 40303633 | CHRNA9 | 0.8724 | 0.9562 | 3.58E-05 | 2.56E-02 | 5.65E-03 | NA | TG |
| 19 | rs13106574 | 4q22.1 | 87769929 | SLC10A6 | 1.0477 | 1.1304 | 4.81E-05 | 3.07E-02 | 1.69E-03 | NA | TG |
| 20 | rs6819188 | 4q23 | 95694977 | BMPR1B | 0.8694 | 0.9561 | 4.72E-05 | 3.07E-02 | 1.69E-03 | 3.74E-01 | TG |
| 21 | rs12515731 | 5 | 79666489 | ZFYVE16 | 1.0801 | 1.2586 | 8.35E-05 | 4.40E-02 | 9.18E-03 | NA | T2D |
| 22 | rs853158 | 5q31 | 142605172 | NR3C1 | 1.0412 | 1.1149 | 5.88E-05 | 3.68E-02 | 9.31E-03 | NA | HDL |
| 23 | rs11755724 | 6 | 7118990 | RREB1 | 1.0413 | 1.1099 | 2.83E-05 | 2.12E-02 | 5.71E-03 | NA | WHR |
| 24 | rs9358854 | 6p22.1 | 25411464 | LRRC16A | 1.041 | 1.1175 | 2.88E-05 | 2.12E-02 | 6.72E-03 | NA | TG |
| rs9358858 | 6p22.1 | 25446308 | LRRC16A | 1.0428 | 1.111 | 1.91E-05 | 1.45E-02 | 7.71E-03 | NA | LDL | |
| 25 | rs932316 | 6p22.3 | 25641200 | SCGN | 0.8613 | 0.9538 | 5.16E-05 | 3.07E-02 | 2.43E-03 | NA | HDL |
| 26 | rs210131 | 6p21.31 | 33535466 | BAK1 | 1.1096 | 1.2146 | 2.33E-08 | 3.76E-05 | 1.92E-05 | 4.78E-01 | SBP |
| rs394199 | 6 | 33553580 | GGNBP1 | 0.8625 | 0.9356 | 1.67E-08 | 2.44E-05 | 6.58E-06 | NA | SBP | |
| rs942637 | 6p21.31 | 33653111 | ITPR3 | 1.0883 | 1.1837 | 1.64E-07 | 2.04E-04 | 9.16E-05 | 1.07E-02 | BMI | |
| rs471942 | 6p21.31 | 33696785 | IP6K3 | 1.0918 | 1.2145 | 5.11E-06 | 4.57E-03 | 2.99E-03 | 6.00E-01 | WHR | |
| 27 | rs854917 | 6q15 | 90127390 | RRAGD | 1.0463 | 1.1195 | 1.85E-05 | 1.45E-02 | 8.33E-03 | NA | HDL |
| 28 | rs6933404 | 6q23.3 | 137959235 | OLIG3 | 1.0482 | 1.1257 | 2.24E-05 | 1.45E-02 | 5.62E-03 | 8.09E-01 | CRP |
| 29 | rs632057 | 6 | 139834012 | CITED2 | 0.8891 | 0.9674 | 2.34E-04 | 1.02E-01 | 7.42E-03 | NA | TG |
| 30 | rs6952809 | 7p22 | 2448493 | CHST12 | 1.0403 | 1.1085 | 3.39E-05 | 2.12E-02 | 7.57E-03 | 9.67E-01 | T2D |
| 31 | rs2066992 | 7p21-p15 | 22768249 | IL6 | 0.7198 | 0.9039 | 1.90E-05 | 1.45E-02 | 2.55E-03 | NA | CRP |
| 32 | rs921911 | 7 | 50241812 | C7orf72 | 1.0523 | 1.1361 | 2.31E-05 | 1.75E-02 | 5.22E-03 | NA | LDL |
| 33 | rs10271662 | 7p11.2 | 55962907 | ZNF713 | 1.0461 | 1.1244 | 3.81E-05 | 2.56E-02 | 4.60E-03 | NA | T2D |
| rs4543497 | 7p12 | 56047215 | GBAS | 0.863 | 0.954 | 4.71E-05 | 3.07E-02 | 8.87E-03 | NA | TG | |
| 34 | rs10111980 | 8p23.1 | 9418167 | TNKS | 1.0428 | 1.1142 | 3.06E-05 | 2.12E-02 | 1.11E-03 | 3.97E-04 | TG |
| 35 | rs10106461 | 8q13.1 | 71405288 | TRAM1 | 0.8702 | 0.9528 | 1.41E-05 | 9.87E-03 | 6.14E-03 | NA | LDL |
| 36 | rs290986 | 9q22 | 93563536 | SYK | 1.0641 | 1.1436 | 1.04E-06 | 1.08E-03 | 2.18E-04 | NA | HDL |
| 37 | rs2275774 | 10p15.1 | 5799613 | FAM208B | 1.0416 | 1.1233 | 1.29E-04 | 6.26E-02 | 3.94E-03 | NA | TG |
| 38 | rs7905327 | 10p15.1 | 6176112 | PFKFB3 | 1.0463 | 1.129 | 1.81E-05 | 1.45E-02 | 9.20E-03 | NA | HDL |
| 39 | rs1442539 | 10q21 | 62481380 | ANK3 | 1.0417 | 1.1147 | 4.86E-05 | 3.07E-02 | 4.60E-03 | 8.84E-03 | HDL |
| 40 | rs7912269 | 10q22 | 78727604 | KCNMA1 | 1.0776 | 1.1972 | 2.26E-05 | 1.75E-02 | 4.19E-03 | NA | CRP |
| 41 | rs12289836 | 11q13 | 65436888 | RELA | 0.8857 | 0.9669 | 2.70E-04 | 1.02E-01 | 9.86E-03 | NA | HDL |
| 42 | rs606978 | 11q13.1 | 65711517 | TSGA10IP | 1.0394 | 1.1062 | 3.44E-05 | 2.12E-02 | 8.16E-03 | 9.13E-01 | HDL |
| 43 | rs4409785 | 11q21 | 95311422 | FAM76B | 0.8432 | 0.9405 | 5.67E-06 | 5.57E-03 | 1.45E-03 | NA | LDL |
| 44 | rs491111 | 11q23.3 | 116238034 | BUD13 | 1.046 | 1.1239 | 1.02E-05 | 8.16E-03 | 2.69E-03 | 1.59E-01 | TG |
| 45 | rs7941030 | 11q24.1 | 122522375 | UBASH3B | 0.8691 | 0.9509 | 8.24E-06 | 6.75E-03 | 3.21E-04 | 3.50E-01 | HDL |
| 46 | rs2059405 | 12q13.11 | 46175469 | ARID2 | 0.8561 | 0.953 | 6.57E-05 | 3.68E-02 | 9.93E-03 | NA | LDL |
| 47 | rs17594362 | 13q14.11 | 42139245 | VWA8 | 1.0568 | 1.1499 | 3.15E-05 | 2.12E-02 | 6.32E-03 | NA | SBP |
| 48 | rs806321 | 13 | 50841323 | DLEU1 | 1.0466 | 1.1118 | 4.29E-06 | 3.74E-03 | 6.03E-04 | NA | TG |
| 49 | rs17119756 | 14 | 84634432 | LINC00911 | 0.8311 | 0.9439 | 4.77E-05 | 3.07E-02 | 4.23E-03 | NA | TG |
| 50 | rs4886406 | 15q24.1 | 75057203 | CYP1A2 | 0.8712 | 0.9609 | 1.59E-04 | 7.40E-02 | 5.71E-03 | 4.31E-01 | HDL |
| 51 | rs11864333 | 16p13.13 | 11475576 | PRM1 | 1.0444 | 1.1097 | 7.82E-06 | 6.75E-03 | 3.15E-03 | NA | T2D |
| 52 | rs2012068 | 17p13.3 | 2255351 | SGSM2 | 0.8805 | 0.9604 | 5.89E-05 | 3.68E-02 | 6.69E-03 | NA | LDL |
| 53 | rs4792814 | 17q21 | 43403005 | MAP3K14 | 0.8694 | 0.9496 | 4.80E-06 | 4.57E-03 | 9.55E-04 | NA | LDL |
| 54 | rs1373089 | 17q21 | 44915265 | WNT9B | 0.8937 | 0.9579 | 1.12E-05 | 8.16E-03 | 1.47E-03 | NA | HDL |
| 55 | rs12603582 | 17q21.32 | 45377577 | ITGB3 | 0.8608 | 0.9551 | 7.88E-05 | 4.40E-02 | 2.84E-03 | 2.79E-02 | HDL |
| 56 | rs8081176 | 17q25.3 | 78283987 | RNF213 | 0.8735 | 0.9543 | 1.59E-05 | 1.19E-02 | 6.68E-03 | 2.21E-08 | TG |
| 57 | rs12456021 | 18q21.31 | 56213390 | ALPK2 | 0.8663 | 0.9455 | 7.87E-06 | 6.75E-03 | 2.95E-03 | 2.86E-01 | SBP |
| 58 | rs243354 | 19p13.3 | 4412713 | CHAF1A | 0.8798 | 0.9651 | 2.53E-04 | 1.02E-01 | 9.86E-03 | NA | HDL |
| 59 | rs7255066 | 19q13.2 | 45146103 | PVR | 0.8566 | 0.9447 | 4.45E-06 | 3.74E-03 | 1.63E-04 | NA | HDL |
| 60 | rs307896 | 19q13.32 | 47661493 | SAE1 | 1.0574 | 1.1396 | 1.04E-06 | 1.08E-03 | 5.77E-05 | NA | TG |
| 61 | rs2032809 | 19q13.3 | 47736216 | MIR3191 | 0.8952 | 0.9697 | 2.79E-04 | 1.02E-01 | 9.84E-03 | NA | TG |
| 62 | rs281380 | 19q13.33 | 49214470 | MAMSTR | 0.8826 | 0.9607 | 5.32E-05 | 3.07E-02 | 1.78E-03 | NA | TG |
| 63 | rs7260291 | 19q13.33 | 49886010 | CCDC155 | 1.0525 | 1.1432 | 1.15E-05 | 9.87E-03 | 4.88E-03 | NA | BMI |
| 64 | rs967990 | 20 | 56335741 | PMEPA1 | 0.8876 | 0.9624 | 5.39E-05 | 3.07E-02 | 6.69E-03 | NA | HDL |
| 65 | rs2072711 | 22q13.1 | 37268555 | NCF4 | 1.0615 | 1.1455 | 3.96E-06 | 3.74E-03 | 1.94E-03 | 5.95E-01 | TG |
| rs2413436 | 22q12.2 | 37312561 | CSF2RB | 1.0437 | 1.1088 | 8.78E-06 | 6.75E-03 | 3.42E-03 | NA | LDL |
the most significant MS SNP in each LD block based on the minimum condFDR (min condFDR) for each phenotype.
chromosome location
the CVD risk factor which provided the signal, i.e., minimal condFDR (driving pheno).
NA indicates not available.
Table 2.
Conjunctional FDR; MS and CVD risk factors loci (MS & CVD) in non-MHC regions excluding previously reported MS loci
| Locus# | SNPa | A1 | A2 | Chr_locb | pos | Gene | Min cnjFDR | Driv phenoc |
|---|---|---|---|---|---|---|---|---|
| 1 | rs10909880 | G | C | 1p36 | 2727804 | TTC34 | 3.77E-02 | BMI |
| 2 | rs11588410 | G | C | 1p21.2 | 101486061 | DPH5 | 2.16E-02 | LDL |
| 3 | rs11102646 | G | A | 1p12 | 114068933 | MAGI3 | 4.98E-02 | TG |
| 4 | rs4148211 | A | G | 2p21 | 44071743 | ABCG8 | 4.32E-02 | LDL |
| 5 | rs2036927 | A | G | 2q33 | 203640713 | ICA1L | 7.20E-03 | LDL |
| rs17406900 | T | C | 2q33.3 | 203784202 | WDR12 | 2.51E-02 | TG | |
| rs7573079 | C | T | 2q33 | 203927551 | NBEAL1 | 3.65E-02 | LDL | |
| 6 | rs2943633 | G | A | 227054881 | MIR548AR | 2.08E-03 | TG | |
| 7 | rs13070927 | A | G | 3p25.1 | 14919646 | FGD5 | 3.63E-02 | SBP |
| 8 | rs907314 | A | T | 4p14 | 38254453 | PTTG2 | 2.57E-02 | LDL |
| 9 | rs13106574 | A | G | 4q22.1 | 87769929 | SLC10A6 | 1.69E-03 | TG |
| 10 | rs6819188 | T | A | 4q23 | 95694977 | BMPR1B | 9.29E-03 | TG |
| 11 | rs4698874 | G | A | 4q24 | 103930511 | SLC9B1 | 2.35E-02 | T2D |
| 12 | rs4704963 | T | C | 5q34 | 158247378 | EBF1 | 4.07E-02 | CRP |
| 13 | rs1267499 | G | A | 6p24 | 14715882 | JARID2 | 4.26E-02 | SBP |
| 14 | rs2076890 | G | A | 6p22.1 | 25420744 | LRRC16A | 3.05E-02 | LDL |
| 15 | rs394199 | T | C | 6 | 33553580 | GGNBP1 | 7.03E-03 | SBP |
| 16 | rs9381257 | T | C | 6 | 43798902 | VEGFA | 4.14E-02 | TG |
| 17 | rs9367490 | T | C | 6 | 52459940 | TRAM2-AS1 | 2.57E-02 | LDL |
| 18 | rs632057 | C | T | 6 | 139834012 | CITED2 | 7.42E-03 | TG |
| 19 | rs7776857 | C | A | 6 | 22754768 | IL6 | 4.83E-02 | CRP |
| rs2066992 | G | C | 7p21 | 22768249 | IL6 | 2.37E-02 | CRP | |
| 20 | rs2293489 | C | T | 7q11.23 | 73107279 | WBSCR22 | 1.98E-02 | TG |
| 21 | rs6944136 | G | C | 7q36.1 | 150370546 | GIMAP2 | 4.14E-02 | TG |
| 22 | rs13262031 | C | A | 8 | 9389241 | TNKS | 1.64E-02 | TG |
| rs10111980 | T | C | 8p23.1 | 9418167 | TNKS | 1.39E-02 | TG | |
| 23 | rs2616214 | T | C | 8p22 | 20611119 | LZTS1 | 4.12E-02 | T2D |
| 24 | rs2275774 | A | G | 10p15.1 | 5799613 | FAM208B | 4.32E-03 | LDL |
| 25 | rs10835211 | A | C | 11p14.1 | 27701365 | BDNF | 4.56E-02 | TG |
| 26 | rs11039035 | C | T | 11 | 46967415 | C11orf49 | 2.38E-02 | TG |
| 27 | rs2306365 | G | A | 11q13 | 65427346 | RELA | 2.86E-02 | TG |
| rs6591188 | T | C | 11q13 | 65467953 | KAT5 | 3.05E-02 | LDL | |
| rs557675 | G | A | 11q13 | 65566719 | OVOL1 | 4.21E-02 | HDL | |
| 28 | rs7941030 | T | C | 11q24.1 | 122522375 | UBASH3B | 3.21E-04 | LDL |
| 29 | rs3184504 | G | A | 12q24.12 | 111884608 | SH2B3 | 2.14E-02 | LDL |
| 30 | rs940904 | C | T | 12q24.31 | 123491572 | PITPNM2 | 2.20E-02 | HDL |
| 31 | rs12871645 | G | A | 13 | 50931565 | DLEU1 | 3.56E-02 | TG |
| rs2400899 | G | A | 14q32.2 | 100828487 | WARS | 4.32E-02 | LDL | |
| 32 | rs2306791 | C | T | 15q21 | 59453384 | MYO1E | 1.85E-02 | HDL |
| 33 | rs4886406 | G | A | 15q24.1 | 75057203 | CYP1A2 | 2.14E-02 | SBP |
| rs6495126 | T | C | 15q22-qter | 75175026 | MPI | 1.77E-02 | LDL | |
| 34 | rs158481 | A | T | 16q13 | 57075253 | NLRC5 | 3.61E-02 | HDL |
| 35 | rs2306589 | G | T | 17q21.1 | 34848874 | ZNHIT3 | 2.86E-02 | TG |
| 36 | rs1292053 | T | C | 17q23.1 | 57963537 | TUBD1 | 1.35E-03 | CRP |
| 37 | rs8090363 | G | T | 18q21.1 | 47133828 | LIPG | 1.08E-02 | HDL |
| rs4939883 | C | T | 18q21.1 | 47167214 | LIPG | 2.57E-02 | LDL | |
| 38 | rs8102273 | G | C | 19p13.3 | 11180047 | SMARCA4 | 4.32E-02 | LDL |
| 39 | rs12608504 | C | T | 19p13.2 | 18389135 | JUND | 1.39E-02 | TG |
| 40 | rs7255066 | C | A | 19q13.2 | 45146103 | PVR | 1.63E-04 | LDL |
| 41 | rs405509 | T | C | 19q13.31 | 45408836 | APOE | 4.14E-02 | TG |
| 42 | rs10408163 | G | A | 19q13.33 | 47597102 | ZC3H4 | 1.11E-02 | TG |
| rs307896 | A | G | 19q13.32 | 47661493 | SAE1 | 6.22E-03 | HDL | |
| rs466477 | G | A | 19q13.32 | 47679798 | SAE1 | 7.42E-03 | TG | |
| 43 | rs281380 | C | A | 19q13.33 | 49214470 | MAMSTR | 3.45E-03 | LDL |
| 44 | rs926629 | C | T | 20q13.13 | 47335736 | PREX1 | 4.97E-02 | SBP |
| 45 | rs6010669 | G | A | 20q13.33 | 62445688 | ZBTB46 | 2.57E-02 | LDL |
| 46 | rs2836878 | T | C | 21q22.3 | 40465534 | PSMG1 | 3.44E-02 | TG |
| 47 | rs5756391 | C | T | 22q12.2 | 37298344 | CSF2RB | 9.29E-03 | TG |
the most significant MS SNP in each LD block based on the minimum conjunctional FDR (min cnjFDR) for each phenotype.
chromosome location
the CVD risk factor which provided the signal, i.e., minimal cnjFDR (driving pheno).
The impact of MHC region on enrichment
To test the possibility that the observed enrichment may be driven by the large extended MHC region (chr6: 25652429–33368333, xMHC) we removed the xMHC region related SNPs, defined as SNPs located within the xMHC or SNPs within 1Mb and in LD (r2 > 0.2) with such SNPs, and then re-performed the analyses.
Non-genetic confounding
To investigate whether non-genetic confounders between MS and CVD risk factors contribute to the observed enrichment we used a permutation procedure. Specifically, we permuted the p values of each of the CVD risk factors 100 times, and reconstructed the fold enrichment plots using the average empirical cumulative distributions across all iterations.
Gene expression analysis of new MS loci
We used publicly available gene expression data for 170 MS patient and 60 controls22 (NCBI Gene Expression Ontology database (GSE41850)) and mapped the suggested genes from our conditional analysis of MS on CVD to the assigned genes in this dataset by gene symbols. We restricted our analyses to the baseline expression level data and applied a two-sided t-test to baseline expression levels of the mapped genes for patients and controls.
RESULTS
Pleiotropic enrichment of MS conditioned by association with related phenotypes
As illustrated by the conditional fold enrichment plots, we found a strong enrichment of MS SNPs conditioned on the nominal p-values of association with several CVD risk factors (Figure 1). Across all evaluated CVD phenotypes, we found that the polygenic pleiotropic enrichment was strongest for LDL and TG (approximately 30 fold with respect to whole genome SNPs). We additionally performed a normal test of the empirical cumulative distribution of MS SNP p values conditioned on the association level of CVD phenotypes (with −log10P(CVD) >1, 2, 3, and 4 versus the depleted category, −log10P(CVD) <1), and found that all four tests were significant (p < 0.05) for HDL and LDL but only 1 or 2 tests were significant for other the CVD phenotypes (Supplementary Table 5).
MS gene loci identified with conditional FDR
As shown in the conditional FDR Manhattan plot for MS and each of the related CVD risk factors (Figure 2), we identified a total of 133 non-MHC loci, of which 65 are novel compared to the GWAS (see Table 1 for not previously reported non MHC loci and Supplementary Table 2 for all loci).
Overlapping gene loci in MS and CVD risk factors identified with conjunctional FDR
As indicated by the ‘Conjunction FDR Manhattan plot’ (Figure 3), we detected loci significantly associated with both MS and the CVD risk factors on all chromosome (including chromosome 6) except chromosome 21(see Table 2 and Supplementary Tables 3 and 4). In general, we observed an opposite direction of effect between MS and TG, LDL, WHR and T2D and the same direction of effect between MS and BMI (Supplementary Figure 3 and Supplementary Table 4).
Differential impact of xMHC on pleiotropic enrichment
We found that removing the xMHC-related SNPs resulted in substantial attenuation of the enrichment of MS conditioned on TG and HDL (Supplementary Figure 4). For the strata with −log10P > 3, the fold enrichment for TG was reduced from 30 to about 2 fold and for HDL from 10 to 2 fold.
Control of non-genetic artifacts
Figure 4 shows the comparison of distributions of genotypic variance (2*(p*(1−p)), p: reference allele frequencies from the 1000 Genomes Project) of SNPs having conditional FDR < 0.05 for each conditioned trait with all SNPs analyzed. The majority of SNPs with conditional FDR < 0.05 are common SNPs, i.e., tagging more genotypic variances, for all conditional analysis. In the Supplementary Figure 5 we show the fold enrichment plot based on 100 permutations of the p values of each conditioned trait. The observed pleiotropic enrichment between MS and CVD risk factors disappeared after randomizing the genotype-phenotype relationship of the conditioned traits indicating that the observed enrichment between MS and CVD risk factors is not a result of confounders, such as sample overlap or technical artifacts.
Figure 4. Distribution of tagged genotypic variance of identified SNPs at conditional FDR < 0.05.
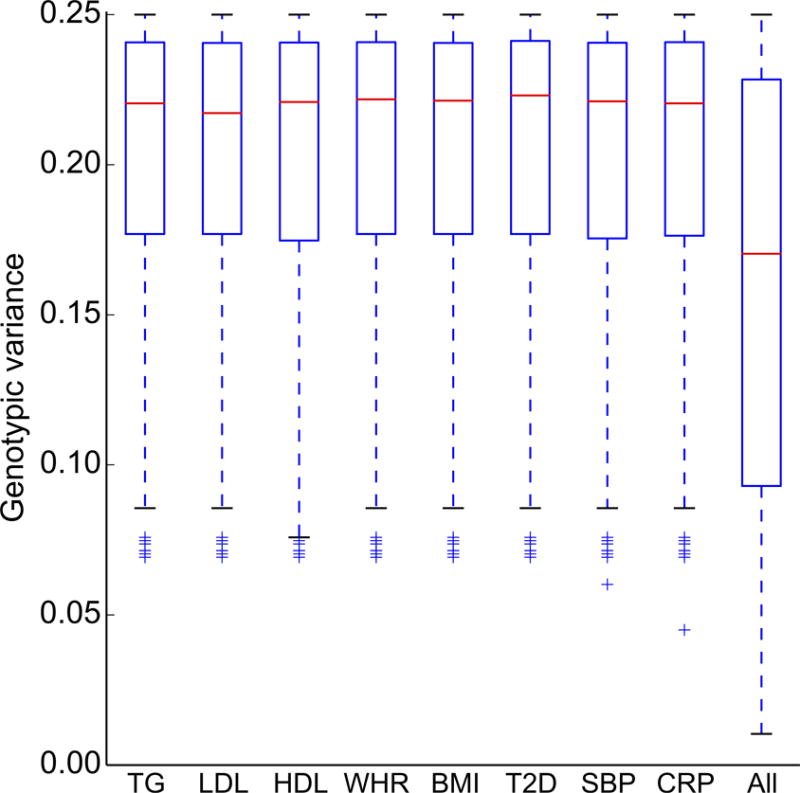
Comparison of the distribution of tagged genotypic variance (y axis) by SNPs identified by conditional FDR < 0.05 of MS conditional on cardiovascular risk factors (x axis): triglycerides (TG), Low density Lipoprotein cholesterol (LDL), High density Lipoprotein cholesterol (HDL), Waist Hip Ratio (WHR), body mass index (BMI), type 2 diabetes (T2D), systolic blood pressure (SBP) and C-reactive protein level (CRP), with all SNPs analyzed (All).
Gene expression analysis of new MS loci
Out of the 279 unique genes suggested by the conditional analysis of MS on CVD at the level condFDR (MS|CVD) < 0.01, we found available baseline expression data for 129 genes. Across all 129 genes, we found a significant association between baseline expression level and MS status for 28 genes (p < 0.05, Table 1).
Pathway analysis for conjunction loci
We investigated the probable pathways involving the loci identified by the conjunctional analysis of MS with CVD factors using PANTHER23 and Reactome24. The most enriched biological pathways were metabolic process (GO:0008152), cellular process (GO:0009987) and immune system process (GO:0002376) (see Supplementary Figures 6 and 7 and Supplementary Table 6 for details). We found that 32 genes mapped to known pathways in PANTHER, among which the Apoptosis signaling pathway, the Integrin signaling pathway, the Inflammation mediated by chemokine and cytokine signaling pathway and the T cell activation pathway showed 3 hits and others showed 1 or 2 hits (Supplementary Figure 6). Consistent with the PANTHER results, we observed that the immune related pathways were significant (p < 0.05) by Reactome (Supplementary Table 6). Moreover, several signaling pathways within the immune system, such as Interferon alpha/beta signaling and cytokine signaling, were also detected.
DISCUSSION
Here, we observed polygenic pleiotropy between MS and several CVD risk factors, identifying 133 independent loci associated with MS conditioned on CVD risk factors. Further, we identified 60 genes associated with both MS and CVD risk factors. Considered together, our findings implicate overlapping genetic factors between MS and several CVD risk factors.
The current results suggest that multiple loci in the xMHC region are overlapping between MS, and TG and HDL. These loci seem mainly located in the xMHC region, and due to the high and complex LD pattern in this region, it is difficult to interpret the results in a functional setting. Interestingly, the polygenic overlap observed between MS and LDL, and also some of the overlap between MS and the other CVD factors seem less dependent on the xMHC region (Supplementary Figure 4). This strongly suggest that there are also non-MHC genes shared between MS and CVD risk factors. This may further indicate that genetic factors may play a role in the immune activation found in several cardiovascular diseases. On a methodological note, unlike epidemiological studies, co-heritability analyses25, 26 or LD regression27, one strength of our current approach is the ability to detect genetic effects even when there is no correlation of the signed effects (mixed directionality of effect); the method presented in this work can detect SNPs that have a non-null effect in one trait and that also tend to have a non-null effect in another trait, independent of directionality14, 28. Taken collectively, these findings illustrate that the genetic relationship between cardiovascular disease risk factors and MS may not be straightforward; considerable work will be required to carefully characterize the biological mechanisms underlying how each cholesterol-associated genetic variant influences MS pathobiology.
Although the method robustly identifies new variants, the functional mechanisms behind SNP associations to disease remain elusive. However, the fact that these polymorphisms influence both MS and CVD risk suggests the possibility of shared mechanisms for these shared variants. Such functional effects should be studied for each risk polymorphism individually and strategies for such investigations are dependent on the genes that are putatively affected by this polymorphism. The current results suggest a complex pattern of pathological pathways, involving both xMHC and other parts of the genome. Further, the results of the conjunctional FDR identify specific overlapping gene variants between MS and CVD risk factors. Inflammatory processes play an important role in MS, and associations to both HLA class I and II loci are well established29. Recently, a large number of non-HLA markers have also been associated with MS risk, and immunologically relevant genetic loci were significantly overrepresented among these6. Most of the pleiotropic loci between MS and the CVD risk factors were located on chromosome 6, suggesting involvement of HLA genes also in several CVD risk factors. This is in line with previous findings of the involvement of immunological mechanisms also in CVD. On the other hand, immune related mechanisms have been implicated in the pathology of several CVD30–32 and vascular pathology33, 34. Our approach further elucidates other possible common mechanisms between MS and CVD risk factors. For instance, this may be related both to vascular and lipid biology or inflammatory processes shared between MS etiology and CVD risk factors, although the exact mechanisms may vary between polymorphisms. The interesting recent reports of a relation between body mass index and MS susceptibility35, 36 also support the existence of common mechanisms between this CVD risk factor and MS.
The current findings of new genetic variants in MS conditional on CVD risk factors show the feasibility of using a genetic epidemiology framework that leverages overlap in genetic signals from independent GWASs to improve statistical power for gene discovery. In the original MS GWAS sample, > 52 loci were significantly associated with MS susceptibility. By combining the original MS sample with independent GWAS of selected CVD risk factors, we identified abundant pleiotropic signal (total of 133 loci). Several of these genetic risk loci have not been previously reported in MS, whereas one of the loci not reported in the GWAS was genome wide significant in the later immunoChip analysis. Our findings demonstrate the increased power of the combined analytical approach. It is important to note that by applying the conjunctional FDR method37, we minimized concerns that our results were solely driven by a strong signal in one phenotype.
Our analysis is based on results from GWAS of different phenotypes, and there might be some overlapping individuals included in several of the primary studies. Since the analysis is restricted to summary statistics, we could not identify the specific individuals. However, we performed standard single phenotype GWAS genomic corrections38 for genetic stratification before our pleiotropy analysis. The fact that the pleiotropic loci were located at different sites for different CVD risk factors, suggest that our findings are not driven by conditional genetic effects and rather by true increases in risk for MS or association to the CVD risk factors. Moreover, when the genotype-phenotype association in CVD risk factors were perturbed, the observed enrichment disappeared (Supplementary Figure 5) indicating that the identified pleiotropic structure is not the result of overlapping samples or non-genetic confounders. It is known that relative rare variants suffer more from technical artifacts, however the SNPs we identified are concentrated in common variants (Figure 4) further suggesting that our results are not artifacts. It is also important to note that our conditional FDR is capable of identifying the majority of the established MS risk loci, thereby showing the power and specificity of the method. The current study only analyzed SNPs reported by both MS and CVD risk factors GWAS studies which excluded the large number of SNPs analyzed in the latest immunoChip study of MS5. Thus the low replication rates of the SNPs reported by immunoChip study of MS is reasonable. Finally we hypothesize that when new and larger GWAS data appear for these phenotypes, more pleiotropic loci are expected to be identified.
This work has clinical implications. The present results revealed a large number of genetic loci associated with MS. Careful work will be required to further characterize the candidate genes detected in this study and how these impact MS risk on an individual basis. Although no single variant may be informative clinically, identifying shared loci with cardiovascular risk factors will elucidate more of the polygenic architecture of a complex disease and may offer novel insights into lipid-lowering primary and secondary prevention trials in MS.
Supplementary Material
Acknowledgments
The authors would like to thank the International Multiple Sclerosis Genetics Consortium (IMSGC), DIAGRAM Consortium and The Blood Pressure GWAS Consortium for the summary statistics GWAS data.
FUNDING
This work was supported by the Research Council of Norway (www.forskningsradet.no), the South East Norway Health Authority (www.helse-sorost.no), the Kristian Gerhard Jebsen Foundation, and the National Institutes of Health [T32 EB005970, R01AG031224, R01EB000790, and RC2DA29475].
Footnotes
AUTHORS’ CONTRIBUTION
AMD and OAA conceived the study and participated in its design and help to draft the manuscript; SD, HFH and SDB contributed the data and help to draft the manuscript; YW, WKT, AJS, SB, FB, AW, WL, RSD performed the data analysis and draft the manuscript; WL, BAL, LKM, AMD, OAA, SD, HFH, YW and RSD helped interpretation of the analysis. All authors have read and approved the manuscript.
CONFLICT of INTEREST
The authors declare that there is no conflict of interest.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–17. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Wens I, Dalgas U, Stenager E, Eijnde BO. Risk factors related to cardiovascular diseases and the metabolic syndrome in multiple sclerosis–a systematic review. Mult Scler. 2013;19:1556–64. doi: 10.1177/1352458513504252. [DOI] [PubMed] [Google Scholar]
- 3.Feero WG, Guttmacher AE, Collins FS. Genomic medicine–an updated primer. N Engl J Med. 2010;362:2001–11. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 5.Consortium IMSG. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreassen O, Harbo H, Wang Y, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Molecular psychiatry. 2014:1–8. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreassen OA, McEvoy LK, Thompson WK, et al. Identifying Common Genetic Variants in Blood Pressure Due to Polygenic Pleiotropy With Associated Phenotypes. Hypertension. 2014;63:819–26. doi: 10.1161/HYPERTENSIONAHA.113.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreassen OA, Thompson WK, Dale AM. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophrenia Bull. 2014;40:13–7. doi: 10.1093/schbul/sbt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JZ, Hov JR, Folseraas T, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–5. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreassen OA, Zuber V, Thompson WK, et al. Shared common variants in prostate cancer and blood lipids. International Journal of Epidemiology. 2014:1205–14. doi: 10.1093/ije/dyu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desikan RS, Schork AJ, Wang Y, et al. Polygenic Overlap Between C-Reactive Protein, Plasma Lipids and Alzheimer’s Disease. Circulation. 2015 doi: 10.1161/CIRCULATIONAHA.115.015489. CIRCULATIONAHA.115.015489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desikan R, Schork AJ, Wang Y, et al. Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Molecular psychiatry. 2015 doi: 10.1038/mp.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International Consortium For Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehghan A, Dupuis J, Barbalic M, et al. Meta-analysis of genome-wide association studies in> 80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–8. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickles D, Chen HP, Li MM, et al. Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls. Hum Mol Genet. 2013;22(20):194–205. doi: 10.1093/hmg/ddt267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi-Tope G, Gillespie M, Vastrik I, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D32. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G-B, Lee SH, Brion M-JA, et al. Estimation and partitioning of (co) heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet. 2014;23:4710–20. doi: 10.1093/hmg/ddu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schork AJ, Wang Y, Thompson WK, Dale AM, Andreassen OA. New statistical approaches exploit the polygenic architecture of schizophrenia—implications for the underlying neurobiology. Curr Opin Neurobiol. 2016;36:89–98. doi: 10.1016/j.conb.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemmer B, Kerschensteiner M, Korn T. Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 2015;14:406–19. doi: 10.1016/S1474-4422(14)70305-9. [DOI] [PubMed] [Google Scholar]
- 30.Marchant DJ, Boyd JH, Lin DC, Granville DJ, Garmaroudi FS, McManus BM. Inflammation in myocardial diseases. Circ Res. 2012;110:126–44. doi: 10.1161/CIRCRESAHA.111.243170. [DOI] [PubMed] [Google Scholar]
- 31.Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–12. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- 32.Norata GD, Pirillo A, Ammirati E, Catapano AL. Emerging role of high density lipoproteins as a player in the immune system. Atherosclerosis. 2012;220:11–21. doi: 10.1016/j.atherosclerosis.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 33.Manetti M, Guiducci S, Ibba-Manneschi L, Matucci-Cerinic M. Mechanisms in the loss of capillaries in systemic sclerosis: angiogenesis versus vasculogenesis. J Cell Mol Med. 2010;14:1241–54. doi: 10.1111/j.1582-4934.2010.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’haeseleer M, Cambron M, Vanopdenbosch L, De Keyser J. Vascular aspects of multiple sclerosis. Lancet Neurol. 2011;10:657–66. doi: 10.1016/S1474-4422(11)70105-3. [DOI] [PubMed] [Google Scholar]
- 35.Munger KL, Bentzen J, Laursen B, et al. Childhood body mass index and multiple sclerosis risk: a long-term cohort study. Mult Scler. 2013;19:1323–9. doi: 10.1177/1352458513483889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correale J, Aguirre MEB, Farez M. Body Mass Index and Multiple Sclerosis Risk. The Role of Leptin (S24. 004) Neurology. 2014;82 [Google Scholar]
- 37.Nichols T, Brett M, Andersson J, Wager T, Poline J-B. Valid conjunction inference with the minimum statistic. Neuroimage. 2005;25:653–60. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


