Abstract
The enormous wealth of information available today from optical microscopy measurements on living samples is often underexploited. We argue that spatiotemporal analysis of fluorescence fluctuations using multiple detection channels can enhance the performance of current nanoscopy methods and provide further insight into dynamic molecular processes of high biological relevance.
Main Text
A major challenge of present (and future) biophysics is to quantitatively study how biomolecules dynamically fulfill their physiological role in living cells, tissues, or entire organisms. Over the last few decades, many biophysical approaches have been developed to study crucial molecular parameters (e.g., localization, diffusion, binding, and oligomerization state) in living samples with high accuracy.
In particular, the perceived spatial-resolution limit of far-field optical microscopy has dramatically changed over the last 25 years (1). New experimental methodologies were introduced that are able to unveil details on a length scale that is a tiny fraction of the wavelength of light, thus moving spatial resolution far beyond the diffraction limit set by Ernst Abbe’s equation (2). Available imaging methodologies can be grouped into three large families. The first family was proposed two decades ago and exploits the spectral properties of the fluorescent molecules under study to circumvent the diffraction limit, and comprises stimulated emission depletion (STED) (3), ground-state depletion (4), and reversible saturable optical fluorescence transition (5) microscopies. The second family includes strategies that improve spatial resolution by suitably shaping the excitation-light beam, as in the case of structured illumination microscopy (SIM) (6, 7, 8, 9). The third approach affords superresolution by combining selected spectral properties of the fluorescent molecules (e.g., photoactivation and on/off switching) with single-molecule localization (SML) methods (also known as pointillistic strategies) such as stochastic optical reconstruction microscopy (10, 11), photoactivated localization microscopy (PALM) (12), and point accumulation for imaging in nanoscale topography (13, 14). All of these techniques are rapidly spreading and yielding exciting new discoveries in molecular/cellular biology and related fields. As compared with classical methods to approach the nanoscale, such as transmission electron microscopy, these strategies offer a pivotal advantage: they do not require fixed samples and consequently have the potential to reveal the nanoscale dynamic behavior of single molecules directly within living samples (see notable examples in Refs. (15, 16, 17, 18, 19, 20, 21)). In fact, several important biological applications can be found for each one of these nanoscopy families. For instance, in the context of SML methods, by combining PALM with single-particle tracking (SPT), one can build mobility maps by measuring the trajectories of many individual molecules at the same time (22). This multiplexed capability was successfully used, for instance, to dissect the dynamic behavior of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptors at the postsynaptic density of live cells (23). Also, steady-state cross-correlation analysis of PALM-based superresolution images yielded a model-independent and robust quantification of protein dynamics and codistributions both in vesicles and in live cells (24, 25, 26). Again in the context of SML approaches, it is worth mentioning the application of stochastic optical reconstruction microscopy in live samples to reveal the ultrastructural dynamics of various organelles (27), as well as in vitro to probe the molecular exchange pathways in one-dimensional supramolecular fibers (28). Concerning SIM-based nanoscopy methods, their recent combination with ultrahigh numerical aperture (NA ∼ 1.7) and reversibly photoswitchable fluorescent proteins (FPs) provided access to the sub-60-nm-resolution regime in applications at the plasma membrane level (29). This in turn allowed researchers to investigate with high accuracy a number of crucial molecular processes involved in the regulation of endocytic and cytoskeletal dynamics (29). Lastly, STED-based microscopy (applied in both single-point and scanning modes) was successfully used in combination with fluctuation spectroscopy to probe the dynamic behavior of single molecules at the appropriate temporal (microsecond-to-millisecond) and spatial (30–50 nm) resolution in several biological applications (30, 31, 32, 33, 34, 35, 36, 37, 38). Among others, in 2009, Eggeling and co-workers (30) used noninvasive optical recordings of fluorescence fluctuations at tunable spatial scales to study the dynamics of biomolecules on live-cell membranes. In particular, by tuning the probed area ∼70 times below the diffraction limit, they could directly observe sphingolipids and glycosylphosphatidylinositol-anchored proteins transiently trapped in cholesterol-mediated molecular complexes dwelling within <20-nm-diameter areas. Interestingly, it was recently shown that the combination of raster-image correlation spectroscopy with STED offers an enhanced multiplexing capability because of the increased achievable spatial resolution (as compared with conventional raster-image correlation spectroscopy) as well as access to 10–100 times higher fluorophore concentrations (39). This combined approach was used to accurately map molecular dynamics on both model membranes and live cells (39).
From static to dynamic resolution
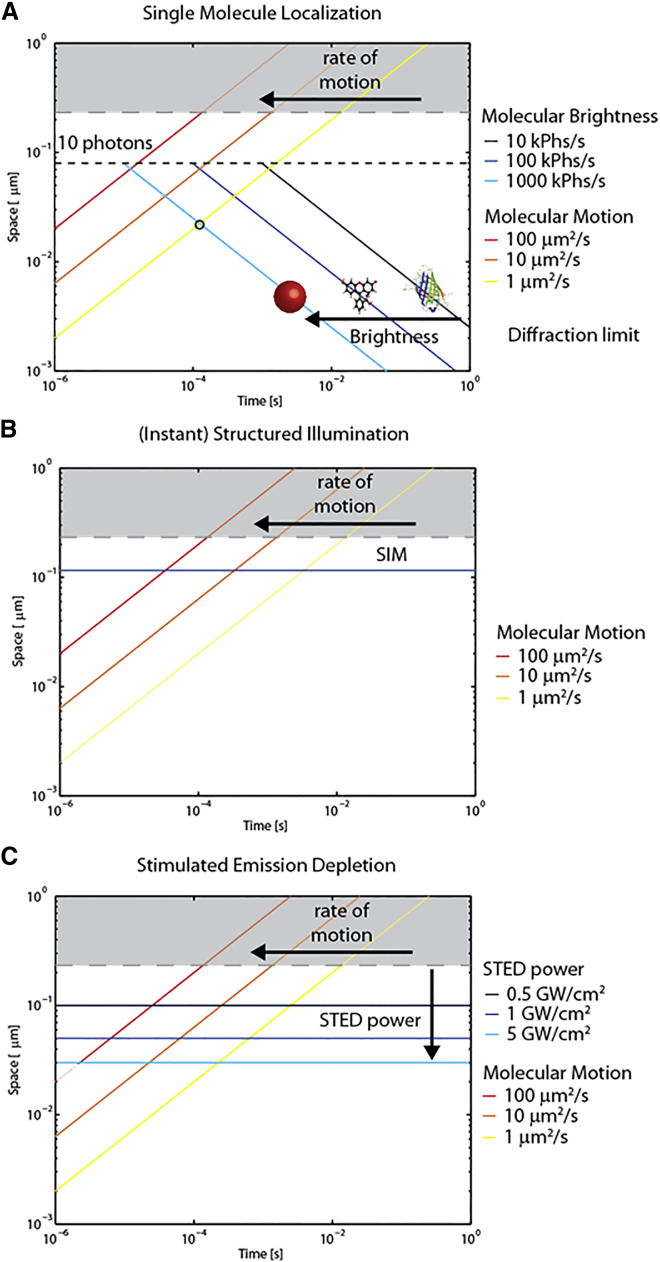
In all of the examples discussed so far, the actual resolution of the measurement is determined by both the spatial and temporal scales at which the dynamic molecular process is actually probed. As recently noted by several experts in the field (40, 41, 42), there is a need for a paradigm shift in the classical concept of measurement resolution, i.e., a shift from the capability to distinguish two different molecules separated in space but fixed in time (hereafter referred to as static resolution) to the capability to distinguish two positions of the same molecule that are separated both in space and in time (hereafter termed dynamic resolution). In other words, in the presence of a dynamic process, the goal of the measurement would be to probe the spatiotemporal displacement of single molecules. From this new perspective, it is of interest to discuss the characteristic potential of each of the abovementioned nanoscopy families in detail. In a conventional SML-based experiment, for instance, a minimum number of photons (typically ∼10 photons) is required to properly perform single-molecule localization. Moreover, the localization precision , which is generally considered as the static spatial resolution of the measurement, depends on the square root of the number of collected photons (Nphs). As a consequence, the brightness of the label directly determines the relationship between the static spatial and temporal resolution of the measurement (43). This relationship is reported in Fig. 1 A (green solid lines) for three representative brightness values, ranging from 104 photons/s (typical of an FP) to 106 photons/s (typical of a bright inorganic chromophore, such as a quantum dot). For instance, the minimum time required to localize a single FP with a 20-nm static spatial resolution is ∼20 ms (dark blue line in Fig. 1 A). By contrast, one can achieve a similar performance in 100-fold less time by using a 100-fold brighter chromophore (cyan line in Fig. 1 A). This scenario changes in the case of dynamic processes. In this case, the spatial scale that can be accessed is strongly influenced by the details of the dynamics. Three representative rates of (Brownian) motion are plotted in Fig. 1 A as solid yellow-to-red lines. For instance, a molecule diffusing at ∼1 μm2/s (yellow solid line in Fig. 1 A) will move ∼20 nm in 0.1 ms. As a consequence, the trajectory of the molecule cannot be described with a 20-nm dynamic resolution using FPs, as the time required for FP localization with such a resolution (i.e., ∼100 ms) is much longer than the time it takes the molecules to move 20 nm. Please note that a single FP provides, on average, <10 photons in 0.1 ms (threshold indicated by dashed black line in Fig. 1 A), thus preventing molecular localization. By contrast, one can achieve a 20-nm dynamic spatial resolution by using an ∼100-fold brighter label (cyan solid line in Fig. 1 A). In fact, in this case, a 20-nm static spatial resolution can be achieved before the molecule has moved ∼20 nm. Interestingly, in an intermediate case (e.g., an organic chromophore; blue solid line in Fig. 1 A), the molecule can be technically localized in 0.1 ms, but the achieved (static) spatial resolution is ∼80 nm. As a consequence, in this case, the molecular trajectory cannot be described with a 20-nm dynamic resolution. Note, however, that in principle, one can obtain the mean-square displacement (MSD) at 20-nm dynamic resolution by averaging multiple steps of a single-molecule trajectory and/or many single-molecule trajectories. All of these considerations are far more important with regard to molecules diffusing, for instance, in a three-dimensional intracellular environment, where molecular short-range motion shows diffusion coefficients of ∼100 μm2/s (red solid line in Fig. 1 A) (44). In these conditions, even a bright inorganic chromophore cannot afford a 20-nm dynamic resolution to describe the molecular trajectory or MSD. These limitations justify current intense efforts to search for improved chromophores and labeling methodologies (21, 45, 46, 47, 48, 49, 50).
Figure 1.
Comparison between the static resolution of superresolution approaches and the temporal scale of molecular motion. (A) In SML methods, the relationship between spatial and temporal resolution is set by the brightness of the chromophore (dark blue-to-cyan solid lines; see Supporting Materials and Methods for further details). In fact, the brighter the chromophore, the shorter is the minimum time required to localize the single molecule. On the other side, the space explored by molecular motion increases in time according to the law of motion of the molecule (which is simplified here as a Brownian motion). The characteristic spatial scale of molecular displacement is identified as the square root of the expected MSD. Three representative diffusivities spanning from 0.1 to 100 μm2/s are pictured as yellow-to-red lines. Please note that 0.1 μm2/s well represents the slow diffusivity of membrane proteins, and 100 μm2/s represents the fast diffusivity of soluble proteins. The maximum dynamic resolution of each selected label in describing a dynamic system is represented by the intercept between the corresponding brightness and rate-of-motion curves. (B) In a typical fluorescence-based SIM experiment, the sample is illuminated with a defined light pattern and the image is collected for each illumination structure. The illumination pattern defines where the sample is illuminated, and the image thus formed provides spatial information about the sample emission. This information provides a gain in resolution by a factor of ∼2. On the other hand, the temporal resolution is set solely by the acquisition protocol that is applied. (C) In STED microscopy, the spatial resolution is set by the efficiency of the depletion of peripheral chromophores and, in the simplest case, it depends only on the power of the depletion beam. Also in this case, the temporal resolution is set by the acquisition protocol that is applied.
As opposed to SML, both SIM- and STED-based strategies afford a static spatial resolution that is constant in time (Fig. 1, B and C). As an example, in the case of STED, the spatial scale at which the molecular dynamic behavior can be probed is invariably set by the efficiency of the depletion process, which, in the simplest case, depends on the power of the depletion beam (30). This in turn sets the size of the observation spot and hence the static resolution limit (Fig. 1 C) (30). Based on these general considerations, it is easy to understand why the dynamic spatial resolution of a SIM- or STED-based measurement can be limited only by the temporal resolution of the acquisition, which is constantly improving thanks to innovative technical solutions (Fig. 1, B and C) (42, 51).
Improving dynamic resolution by spatiotemporal correlation analysis
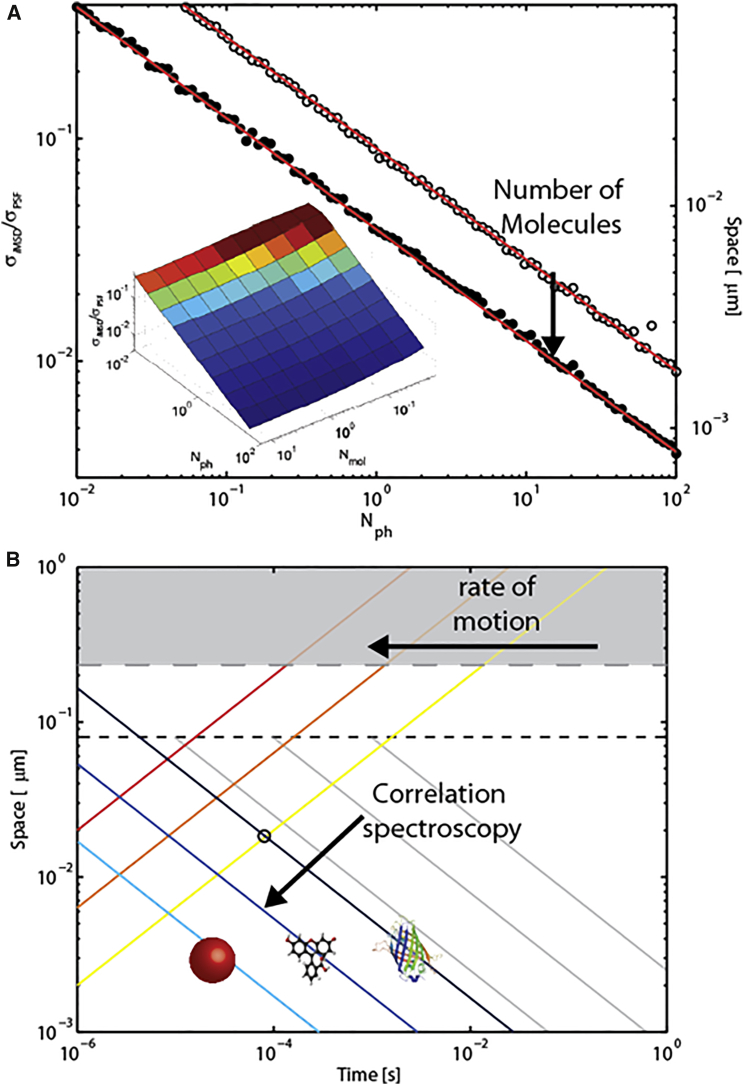
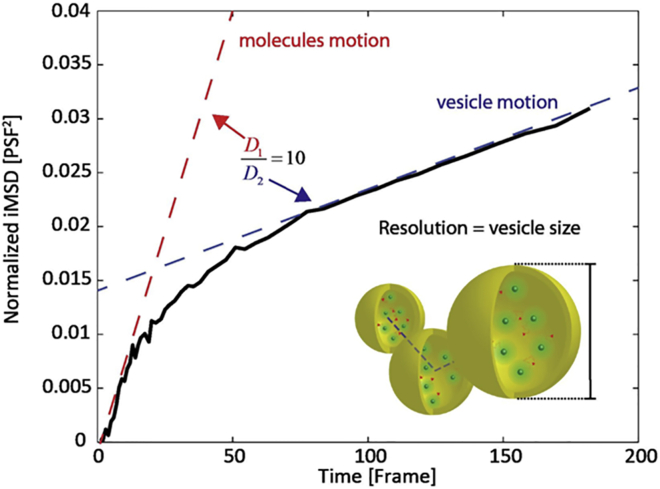
Despite the efforts described above, we argue that the arsenal of analytical tools at our disposal to measure the subresolution details of molecular motion in today’s optical microscopy measurements is not fully exploited. In this regard, it is worth mentioning that the ability to resolve the dynamic behavior of molecules well below the nominal imaging resolution of the measurement was demonstrated in recent years by the sole use of fluctuation analysis in standard diffraction-limited optical setups (29, 44, 52, 53, 54, 55, 56). For example, Shusterman and co-workers (52, 53) derived an expression of the single-point fluorescence correlation spectroscopy correlation function, which is directly related to the molecular MSD. Using this approach, they measured the cuvette molecular displacements of fluorescently labeled DNA molecules with a 30-nm dynamic spatial resolution. For a while now, the theoretical basis underlying these results has been the subject of debate in the field of correlation spectroscopy (57, 58). Interestingly, recent in silico and experimental results have further extended the possibility to probe the nanoscale dynamic behavior of molecules by means of spatiotemporal correlation spectroscopy in standard microscopy setups (44, 55). For instance, by using an FP-tagged variant of Transferrin Receptor (TfR), we recently probed the regulation of protein diffusion imparted by the cytoskeleton meshwork on the plasma membrane of live cells (55). In particular, the diffusion law of TfR was reconstructed by spatiotemporal fluctuation spectroscopy at a temporal resolution of ∼0.1 ms. Thanks to this strategy, we were able to quantitatively describe subdiffraction confinement areas (with a linear length of ∼100 nm) in agreement with high-speed SPT (59). Of note, in our experiments, the achieved dynamic spatial resolution was not determined solely by the imaging technology used (i.e., a standard, diffraction-limited setup with static resolution of ∼250 nm), but rather by the temporal scale at which the fluorescence signal was measured and by the signal/noise ratio achieved in the description of the correlation function. To better elucidate this crucial point, let us consider the case of the so-called imaging-derived MSD (or iMSD) approach applied to spatiotemporal image correlation spectroscopy (STICS) (44, 55) (see Fig. S1 in the Supporting Material for further details). In STICS-iMDS, the average displacement of many single molecules is measured as the increase in width of the spatiotemporal correlation function in time. As a consequence, the smallest measurable average displacement is set by the precision that can be achieved in measuring the width of the correlation function . To quantitatively illustrate this concept, is quantified in Fig. 2 as a function of Nphs and molecular density (Nmol, the number of molecules per observation volume) in a classical time-lapse acquisition. First of all, by increasing Nmol from 0.01 to 10 molecules per point spread function (PSF), one can achieve a threefold improvement in the precision of the measurement regardless of the molecular brightness (open versus solid black dots in Fig. 2 A). Moreover, similarly to SML, decreases according to the square root of Nphs, independently of Nmol (solid lines in Fig. 2 A). In general, a of >20 nm can be obtained in a wide range of conditions, even if individual molecules provide less than one photon per frame (cyan-to-blue portion of the surface plot in the inset of Fig. 2 A). This is possible thanks to spatiotemporal correlation spectroscopy, because the motion of many single molecules is averaged before (not after, as in SML-based experiments) their displacement is measured. This difference makes it possible to explore a temporal scale that is inaccessible to classical SML. By following this general strategy, in principle, one can push the dynamic spatial resolution of any optical microscopy measurement well below the limits imposed by its static spatial resolution (dashed gray line in Fig. 2 B) and by the temporal resolution needed for single-molecule localization (solid gray lines in Fig. 2 B). Such an approach may make it possible to probe TfR-GFP motion at the plasma membrane with a dynamic spatial resolution of ∼20 nm and a temporal resolution of ∼0.1 ms (55). These considerations regarding correlation spectroscopy are quite general and are not restricted to diffraction-limited setups. Therefore, we are asking, Can superresolution methods, which are intrinsically endowed with high static spatial resolution properties, take advantage of the resolution improvements that are accessible through spatiotemporal fluctuation analysis to describe dynamical processes? The answer is yes! We do believe that the time is ripe to push nanoscale investigations of dynamic processes to an entirely new level. To corroborate this statement, in the following we shall consider a recently reported set of superresolution experiments on dynamic biological processes. In particular, Schneider and co-workers (42) successfully combined STED-based imaging with electro-optical scanning technologies to obtain an unprecedented line-scanning frequency of 250 kHz. Using SPT and taking advantage of the 70-nm static resolution provided by STED, these authors investigated the dynamics of fluorescently labeled vesicles in living Drosophila or HIV-1 particles in cells with a temporal resolution of 5–10 ms (42). Such a platform appears ideal to show how the application of spatiotemporal fluctuation analysis can push the dynamic spatial resolution of a measurement well below the imaging nominal resolution, thus shedding light on unexplored dynamic phenomena such as the behavior of fluorescently labeled molecules trapped inside a vesicle. To illustrate this, in Fig. 3 we reproduce a set of analogous data from a simulated experiment in which fluorescent molecules can move within a vesicle that is comparable in size to the static spatial resolution of the measurement. The simulated image series is used to reconstruct the iMSD profile of fluorescent molecules (as described in Ref. (55); see also Fig. S1). Fig. 3 shows that by means of spatiotemporal fluorescence correlation spectroscopy, one can access the motion of fluorescent molecules trapped within moving vesicles and thus fully exploit information collected on timescales up to microseconds, which can be easily reached in a line-scanning acquisition with the aforementioned technology. In more detail, the iMSD plot displays two different diffusive regimes: a short-range diffusion that quantitatively describes the motion of molecules within the vesicle, and a 10-fold slower, long-range diffusion that reflects the movement of the entire vesicle. Both of these regimes match the imposed values. What is of importance here is that by applying spatiotemporal fluctuation analysis, we can resolve the dynamic behavior of molecules within a submicrometric environment with a dynamic spatial resolution of ∼7 nm, a limit that is actually 10 times smaller than the nominal static spatial resolution set by the STED experimental conditions.
Figure 2.
Precision in measuring the molecular MSD by correlation spectroscopy. (A) , as defined in Supporting Materials and Methods, is quantified for a defined range of molecular brightness (Nph, number of photons per molecule per frame) and molecular density (Nmol, number of molecules per PSF) values. In detail, the measured is plotted against Nph for the two selected brightness values of 0.1 and 10 molecules per PSF (open and solid dots, respectively). The red lines underline the dependence of on the square root of Nph. In the inset, a surface plot for all tested conditions is shown. (B) The same plot as in Fig. 1A shows the contribution of spatiotemporal fluctuation analysis to the calculation of as obtained from simulated experiments for three representative molecular brightness levels of 10, 100, and 1000 kPhs/s (dark blue to cyan solid lines).
Figure 3.
Spatiotemporal fluctuation analysis can superresolve single-molecule dynamics: a simulated experiment. A three-dimensional moving spherical object (in this case, a vesicle) with a diameter corresponding to the nominal measurement resolution (PSF) is filled with fluorescent molecules (see drawing in the inset). Both the vesicle and the molecules are free to diffuse, but the latter are 10 times faster than the vesicle and cannot cross the imposed spherical boundary. By applying spatiotemporal analysis of fluorescence fluctuations, one can measure the motion of the molecules within the vesicle (red dashed line) and the motion of the vesicle (blue dashed line) concomitantly, even if both are significantly smaller than the nominal imaging resolution. Further details about the simulations and data analysis are reported in Supporting Materials and Methods.
In the words of Stefan Hell (2014 Nobel Laureate in Chemistry) and Steffen Sahl, “We envision several important developments going forward, of which the main thrusts are clear: we wish to move more and more toward real-time, four-dimensional (4D) molecular analysis not only in cells, but tissue-like preparations or tissues themselves” (1). Such an ambitious task will obviously entail efforts on many levels. We argue here that extending fluctuation analysis to the spatial dimension is the key to unraveling the dynamic behavior of molecules well below the nominal imaging resolution, down to the nanoscale. Using spatiotemporal fluctuation analysis, we can definitely integrate the arsenal of methods at our disposal to investigate living matter at the molecular level.
Author Contributions
C.D.R. performed research, contributed analytic tools, analyzed data, and wrote the manuscript. E.G. and F.B. designed research and wrote the manuscript. F.C. designed and performed research, analyzed data, and wrote the manuscript.
Acknowledgments
We thank Dr. Vincenzo Piazza for useful discussions.
Editor: Brian Salzberg.
Footnotes
Supporting Materials and Methods and one figure are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30578-1.
Supporting Material
References
- 1.Hell S.W., Sahl S.J., Tinnefeld P. The 2015 super-resolution microscopy roadmap. J. Phys. D Appl. Phys. 2015;48:443001. [Google Scholar]
- 2.Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch. F. Microsc. Anat. 1873;9:413–418. [Google Scholar]
- 3.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 4.Hell S.W., Kroug M. Ground-state-depletion fluorscence microscopy: a concept for breaking the diffraction resolution limit. Appl. Phys. B. 1995;60:495–497. [Google Scholar]
- 5.Hell S., Jakobs S., Kastrup L. Imaging and writing at the nanoscale with focused visible light through saturable optical transitions. Appl. Phys. A Mater. Sci. Process. 2003;77:859–860. [Google Scholar]
- 6.Heintzmann R., Cremer C.G. Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. Proc. SPIE. 1999;3568:185–196. [Google Scholar]
- 7.Chung E., Kim D., So P.T. Two-dimensional standing wave total internal reflection fluorescence microscopy: superresolution imaging of single molecular and biological specimens. Biophys. J. 2007;93:1747–1757. doi: 10.1529/biophysj.106.097907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustafsson M.G., Shao L., Sedat J.W. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Axelrod D. Evanescent excitation and emission in fluorescence microscopy. Biophys. J. 2013;104:1401–1409. doi: 10.1016/j.bpj.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukamel E.A., Babcock H., Zhuang X. Statistical deconvolution for superresolution fluorescence microscopy. Biophys. J. 2012;102:2391–2400. doi: 10.1016/j.bpj.2012.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 13.Sharonov A., Hochstrasser R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannone G., Hosy E., Cognet L. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao L., Kner P., Gustafsson M.G. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat. Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- 16.Jones S.A., Shim S.H., Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shroff H., Galbraith C.G., Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiuchi T., Higuchi M., Watanabe N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods. 2015;12:743–746. doi: 10.1038/nmeth.3466. [DOI] [PubMed] [Google Scholar]
- 19.Fölling J., Bossi M., Hell S.W. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat. Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 20.Willig K.I., Kellner R.R., Hell S.W. Nanoscale resolution in GFP-based microscopy. Nat. Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari D.K., Arai Y., Nagai T. A fast- and positively photoswitchable fluorescent protein for ultralow-laser-power RESOLFT nanoscopy. Nat. Methods. 2015;12:515–518. doi: 10.1038/nmeth.3362. [DOI] [PubMed] [Google Scholar]
- 22.Manley S., Gillette J.M., Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 23.Hoze N., Nair D., Holcman D. Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc. Natl. Acad. Sci. USA. 2012;109:17052–17057. doi: 10.1073/pnas.1204589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semrau S., Schmidt T. Particle image correlation spectroscopy (PICS): retrieving nanometer-scale correlations from high-density single-molecule position data. Biophys. J. 2007;92:613–621. doi: 10.1529/biophysj.106.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semrau S., Holtzer L., Schmidt T. Quantification of biological interactions with particle image cross-correlation spectroscopy (PICCS) Biophys. J. 2011;100:1810–1818. doi: 10.1016/j.bpj.2010.12.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone M.B., Veatch S.L. Steady-state cross-correlations for live two-colour super-resolution localization data sets. Nat. Commun. 2015;6:7437. doi: 10.1038/ncomms8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim S.-H., Xia C., Zhuang X. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA. 2012;109:13978–13983. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albertazzi L., van der Zwaag D., Meijer E.W. Probing exchange pathways in one-dimensional aggregates with super-resolution microscopy. Science. 2014;344:491–495. doi: 10.1126/science.1250945. [DOI] [PubMed] [Google Scholar]
- 29.Storti B., Di Rienzo C., Beltram F. Unveiling TRPV1 spatio-temporal organization in live cell membranes. PLoS One. 2015;10:e0116900. doi: 10.1371/journal.pone.0116900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eggeling C., Ringemann C., Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 31.Vicidomini G., Moneron G., Hell S.W. Sharper low-power STED nanoscopy by time gating. Nat. Methods. 2011;8:571–573. doi: 10.1038/nmeth.1624. [DOI] [PubMed] [Google Scholar]
- 32.Mueller V., Ringemann C., Eggeling C. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 2011;101:1651–1660. doi: 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Göttfert F., Wurm C.A., Hell S.W. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 2013;105:L01–L03. doi: 10.1016/j.bpj.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honigmann A., Mueller V., Eggeling C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 2014;5:5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 35.Vicidomini G., Ta H., Eggeling C. STED-FLCS: an advanced tool to reveal spatiotemporal heterogeneity of molecular membrane dynamics. Nano Lett. 2015;15:5912–5918. doi: 10.1021/acs.nanolett.5b02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrade D.M., Clausen M.P., Eggeling C. Cortical actin networks induce spatio-temporal confinement of phospholipids in the plasma membrane—a minimally invasive investigation by STED-FCS. Sci. Rep. 2015;5:11454. doi: 10.1038/srep11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benda A., Ma Y., Gaus K. Self-calibrated line-scan STED-FCS to quantify lipid dynamics in model and cell membranes. Biophys. J. 2015;108:596–609. doi: 10.1016/j.bpj.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianchini P., Cardarelli F., Bizzarri R. Nanoscale protein diffusion by STED-based pair correlation analysis. PLoS One. 2014;9:e99619. doi: 10.1371/journal.pone.0099619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedde P.N., Dörlich R.M., Nienhaus G.U. Stimulated emission depletion-based raster image correlation spectroscopy reveals biomolecular dynamics in live cells. Nat. Commun. 2013;4:2093. doi: 10.1038/ncomms3093. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie K., Shan X.Y., Kusumi A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 2005;88:2266–2277. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kusumi A., Shirai Y.M., Fujiwara T.K. Hierarchical organization of the plasma membrane: investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. FEBS Lett. 2010;584:1814–1823. doi: 10.1016/j.febslet.2010.02.047. [DOI] [PubMed] [Google Scholar]
- 42.Schneider J., Zahn J., Hell S.W. Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics. Nat. Methods. 2015;12:827–830. doi: 10.1038/nmeth.3481. [DOI] [PubMed] [Google Scholar]
- 43.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Rienzo C., Piazza V., Cardarelli F. Probing short-range protein Brownian motion in the cytoplasm of living cells. Nat. Commun. 2014;5:5891. doi: 10.1038/ncomms6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimm J.B., English B.P., Lavis L.D. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. 3, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinney S.A., Murphy C.S., Looger L.L. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dempsey G.T., Vaughan J.C., Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith C. Keeping tabs on fluorescent tags. Nat. Methods. 2007;4:755–761. [Google Scholar]
- 49.Durisic N., Laparra-Cuervo L., Lakadamyali M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat. Methods. 2014;11:156–162. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- 50.Wombacher R., Heidbreder M., Sauer M. Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 51.York A.G., Chandris P., Shroff H. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat. Methods. 2013;10:1122–1126. doi: 10.1038/nmeth.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shusterman R., Alon S., Krichevsky O. Monomer dynamics in double- and single-stranded DNA polymers. Phys. Rev. Lett. 2004;92:048303. doi: 10.1103/PhysRevLett.92.048303. [DOI] [PubMed] [Google Scholar]
- 53.Shusterman R., Gavrinyov T., Krichevsky O. Internal dynamics of superhelical DNA. Phys. Rev. Lett. 2008;100:098102. doi: 10.1103/PhysRevLett.100.098102. [DOI] [PubMed] [Google Scholar]
- 54.Hebert B., Costantino S., Wiseman P.W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Rienzo C., Gratton E., Cardarelli F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci. USA. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Rienzo C., Gratton E., Beltram F., Cardarelli F. From fast fluorescence imaging to molecular diffusion law on live cell membranes in a commercial microscope. J. Vis. Exp. 2014;92:e51994. doi: 10.3791/51994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enderlein J. Polymer dynamics, fluorescence correlation spectroscopy, and the limits of optical resolution. Phys. Rev. Lett. 2012;108:108101. doi: 10.1103/PhysRevLett.108.108101. [DOI] [PubMed] [Google Scholar]
- 58.Krichevsky O. Comment on “Polymer dynamics, fluorescence correlation spectroscopy, and the limits of optical resolution”. Phys. Rev. Lett. 2013;110:159801. doi: 10.1103/PhysRevLett.110.159801. [DOI] [PubMed] [Google Scholar]
- 59.Kusumi A., Nakada C., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.