Background
Pathological examination of Cutaneous Squamous Cell Carcinoma (cSCC) can identify up to 12 distinct features of this disease (1). An outstanding question in the field is how so many different flavors of cSCC can arise. Direct sequencing of bulk tumors has uncovered generally 10 consistent gain or loss of function mutations common to SCC, with Ras and p53 mutations overrepresented (2). Years of effort have implicated Hras and Kras mutations in the etiology of SCC (3–5), and murine models of SCC strongly suggest that Ras activation is critical to SCC formation (5). Work from our lab demonstrated that Hair Follicle Stem Cells (HFSCs) can serve as cells of origin for SCC, and that gain of KrasG12D mutation and loss of p53 is sufficient to generate high grade SCC typical of Spindle Cell Carcinoma (6). These results suggest that SCC can be generated by mutations in Ras and p53 in HFSCs, but this particular model does not yield the full spectrum of phenotypes known to be present in human SCC.
Question Addressed
We sought to identify sources of tumor heterogeneity by fixing both the cell of origin and the initial oncogenic driver, while altering the identity of the second genomic hit, the tumor suppressor. In doing so, we could identify whether a specific population of cells within the epidermis (HFSCs) has the ability to generate different types of tumors, while also demonstrating the role tumor suppressors play in tumor heterogeneity.
Experimental Design
We used mice transgenic for lox-stop-lox-KrasG12D to enable Cre recombinase mediated induction of constitutively active Ras expressed from its endogenous locus. These mice were crossed to mice expressing inducible Cre (Cre-PR) under control of a promoter active specifically in HFSCs of the skin (Keratin 15). Double transgenic mice were then crossed to mice with floxed alleles of either p53, Rb, or Pten to produce mice that would induce expression of constitutively active Kras coupled with loss of a single tumor suppressor only upon treatment with a progesterone antagonist (Mifepristone). These experiments have potential clinical relevance since mutations in KRAS, TP53, RB1, and PTEN have all been shown to occur in human cutaneous SCC at varying rates (Fig 1A) (3, 7). Mice were allowed to reach adulthood, and Cre-mediated recombination was induced just prior to the second adult hair cycle which is synchronized at postnatal day 50.
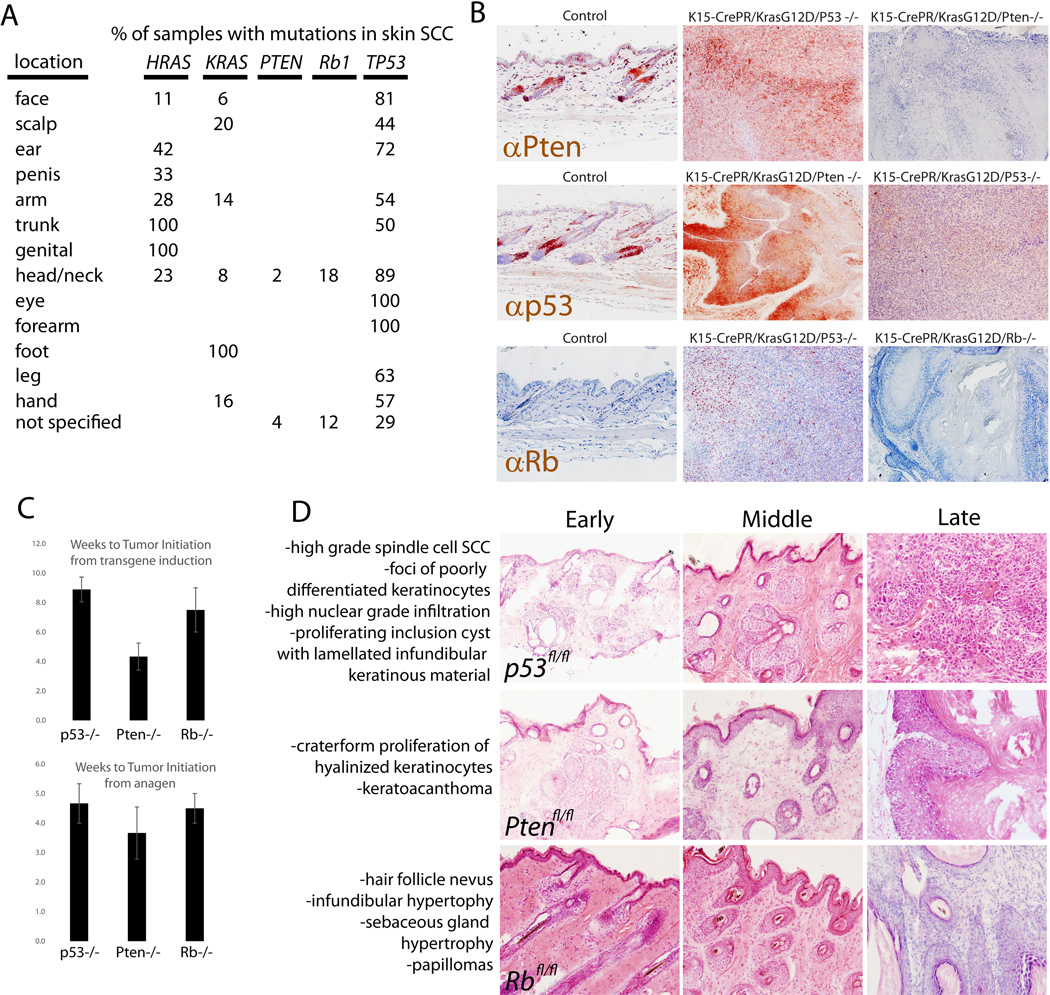
Figure 1. Hair follicle stem cells can drive different phenotypes in SCC depending on tumor suppressor identity.
A, using data from the COSMIC database, the percentage of tumors with mutations in the genes described here are shown. These data are for SCCs from the indicated locations on the body. B, immunostaining with the indicated antibodies showed that transgenic deletion of each tumor suppressor was successful as shown in the resulting tumors. C, quantification of the time to visible tumor induction across different genotypes, either from the time in which the transgene was activated (by administration of mifepristone) (Top panel) or the time from the subsequent anagen initiation (Bottom panel). D, Animals of the indicated genotype were induced to drive expression of oncogenic Ras or ablate a tumor suppressor, and phenotypes were allowed to develop. Representative images of some phenotypes are shown, and complete blinded pathological diagnoses were made based on a large number of animals analyzed (Fig S1).
Results
Between 8–10 weeks following delivery of genetic hits and activation of the hair cycle, tumors became apparent macroscopically. To demonstrate the efficiency of this transgenic system to delete various tumor suppressors, we immunostained each of the targeted tumor suppressors on skin from animals of all genotypes (Fig 1B). In addition, there were not large differences in the rate of tumor initiation from either the time transgene expression was induced or from the beginning of the next hair cycle (Fig 1C). This is important, as we previously showed that initiation of phenotype by induction of KrasG12D depends on the onset of the telogen to anagen transition (8).
Strikingly, we observed a profound difference in the type of squamous tumors formed between the animals with different deletions of tumor suppressors (Fig 1D and Fig S1). While we ideally would look to human SCC samples for a similar correlation, mutations in some of the genes examined here are rare in human SCC (Fig 1A), and human SCC is thought to derive from the interfollicular epidermis, not the hair, precluding simple identification of a similar correlation in human.
As described previously (8), K15CrePR;KrasG12D;p53fl/fl mice developed significant phenotypes and eventually progressed to high grade SCC (n > 15) (Fig 1D). In mice where Pten was deleted instead of p53 (K15CrePR;KrasG12D;Ptenfl/fl), hyperplasia developed over a similar time course but did not progress to the point of SCC within 15 weeks after induction (the time by which all animals had to be sacrificed due to tumor/phenotype burden) (n = 6) (8). Deletion of Pten along with induction of oncogenic Kras led to a different type of squamous hyperplasia (Fig S1). While we previously characterized phenotypes of K15CrePR;KrasG12D;p53fl/fl mice (6, 8), we provide a more complete description of phenotypes in K15CrePR;KrasG12D;Ptenfl/fl and K15CrePR;KrasG12D;Rbfl/fl mice in Fig S2.
In K15CrePR;KrasG12D;Rbfl/fl mice, the hyperplasia generated appeared to move upwards towards the infundibulum and the interfollicular epidermis (n = 4). This hyperplasia also did not progress to the point of bona fide SCC, but instead produced benign papilloma (Fig 1D and Fig S3). In summary, when controlling for the cell of origin and the initial oncogenic stimulus, the identity of the tumor suppressor appeared to be significantly related to the type of squamous phenotype that developed.
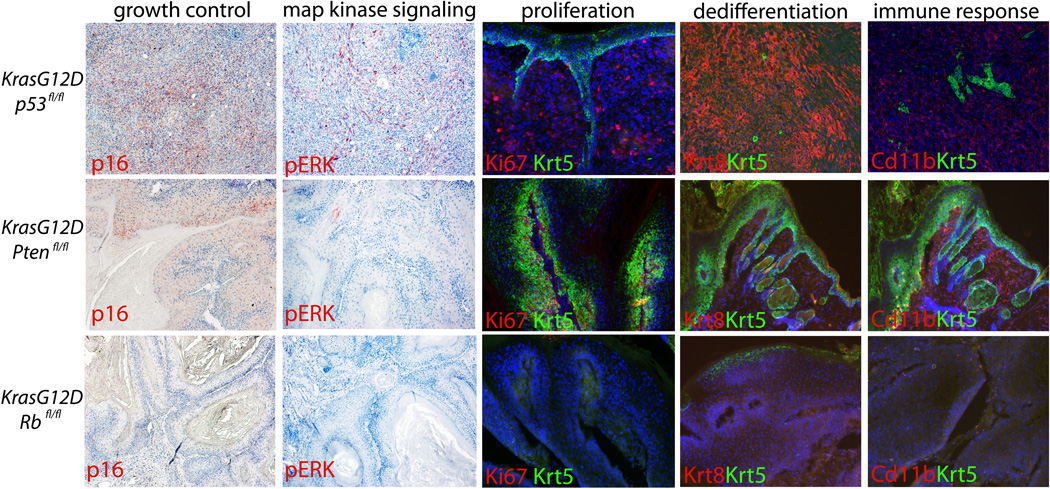
To characterize the phenotypic disparity between the genotypes tested, we assessed proliferation, growth control, map kinase signaling, dedifferentiation, and immune response in each genotype. Staining for Ki67 showed that tumors without p53 or Pten had extensive proliferation, whereas those made without Rb had significantly less proliferation once the tumors were formed. Coincident with this, staining for p16, a protein known to be induced where growth control is needed, was induced in tumors without p53 and Pten, but less so in tumors lacking Rb. In addition, staining for phospho-Erk, a marker of activated map kinase signaling showed significant activity only in tumors lacking p53. Keratin 8 is only expressed in post-natal keratinocytes upon dedifferentiation to SCC. Staining for Keratin 8 across the various genotypes presented demonstrated that only mice with deletion of p53 showed significant dedifferentiation (Fig 2).
Figure 2. Tumor suppressor deletion correlates with distinct phenotypes in regards to proliferation, dedifferentiation, and immune response.
Immunostaining for Ki67, a marker of proliferation showed that tumors made in the absence of p53 or Pten continue to proliferate, whereas those made in the absence of Rb show little proliferation once fully formed. Immunostaining for p16 showed that only tumors deleted for p53 or Pten showed induction of this Cdk inhibitor. Staining for phospho-ERK marks cells with active map kinase signaling in tumors lacking p53. Keratin 8 is only expressed in adult epidermal cells upon dedifferentiation and tumorigenesis. Only tumors with deletion of p53 progressed enough to show expression of Keratin 8. Myeloid Derived Suppressor Cells (MDSCs) are known to facilitate tumor progression by suppressor inhibitory signals during tumorigenesis. Staining for MDSCs with Cd11b showed that only tumors with deletion of p53 or Pten recruited these immune cells. These same tumors also stained positively for Gr-1, another marker of MDSCs (not shown).
The skin is highly sensitive to inflammation and numerous inflammatory cells migrate to the skin in cases of wounding, infection or tumorigenesis. Myeloid Derived Suppressor Cells (MDSCs) are thought to suppress proliferation and contain tumor growth (9). MDSCs are defined by immunostaining with antibodies for Gr1 and Cd11b. Attempts to identify MDSCs with these markers only reliably uncovered them in tumors formed from K15CrePR;KrasG12D;p53fl/fl mice (Fig 2). To further characterize the immune response in tumors generated by loss of various tumor suppressors, we performed a complete pathological examination of immune cells present in the tumors characterized in Fig S1 (Fig S4).
Summary
While some driver mutations have been identified consistently across SCCs, it is unclear whether the heterogeneity of phenotypes is due to variability across mutations, cell of origin, immune response, or variable genomic instability. Our data show that HFSCs can drive a variety of SCC phenotypes that correlate strictly with the type of tumor suppressor deleted.
Supplementary Material
Acknowledgments
This work was supported by NIH (5R01AR057409-05, and TCBT32CA09056), a CIRM Training Grant (TG2-01169), a fellowship from the Broad Center for Regenerative Medicine (UCLA, AF), MARC program (UCLA, WG).
Footnotes
Author Contributions
AF, WG, ACW, PS and RT performed the research and analyzed the data. WL wrote the manuscript.
References
- 1.Rapini RP. Practical dermatopathology. Philadelphia: Elsevier Mosby; 2005. [Google Scholar]
- 2.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickering CR, Zhou JH, Lee JJ, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierceall WE, Goldberg LH, Tainsky MA, et al. Ras gene mutation and amplification in human nonmelanoma skin cancers. Mol Carcinog. 1991;4:196–202. doi: 10.1002/mc.2940040306. [DOI] [PubMed] [Google Scholar]
- 5.van der Schroeff JG, Evers LM, Boot AJ, et al. Ras oncogene mutations in basal cell carcinomas and squamous cell carcinomas of human skin. J Invest Dermatol. 1990;94:423–425. doi: 10.1111/1523-1747.ep12874504. [DOI] [PubMed] [Google Scholar]
- 6.White AC, Tran K, Khuu J, et al. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108:7425–7430. doi: 10.1073/pnas.1012670108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nassar D, Latil M, Boeckx B, et al. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat Med. 2015;21:946–954. doi: 10.1038/nm.3878. [DOI] [PubMed] [Google Scholar]
- 8.White AC, Khuu JK, Dang CY, et al. Stem cell quiescence acts as a tumour suppressor in squamous tumours. Nat Cell Biol. 2014;16:99–107. doi: 10.1038/ncb2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husain Z, Huang Y, Seth P, et al. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells. Journal of immunology. 2013;191:1486–1495. doi: 10.4049/jimmunol.1202702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.