Abstract
Human genetics research employs the two opposing approaches of forward and reverse genetics. While forward genetics identifies and links a mutation to an observed disease etiology, reverse genetics induces mutations in model organisms to study their role in disease. In most cases, causality for mutations identified by forward genetics is confirmed by reverse genetics through the development of genetically engineered animal models and an assessment of whether the model can recapitulate the disease. While many technological advances have helped improve these approaches, some gaps still remain. CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated) system, which has emerged as a revolutionary genetic engineering tool, holds great promise for closing such gaps. By combining the benefits of forward and reverse genetics, it has dramatically expedited human genetics research. We provide a perspective on the power of CRISPR-based forward and reverse genetics tools in human genetics and discuss its applications using some disease examples.
Forward and Reverse Genetics
The use of forward and reverse genetic strategies in research has contributed significantly toward understanding the genetic basis of thousands of human diseases. These approaches are complementary but opposing: forward genetics identifies the genetic basis of a disease, while reverse genetics investigates if and how a gene function is related to a disease phenotype.
Forward genetics links a disease phenotype to a genetic etiology. The genetic factor is tracked by systematic analyses of mutations in families or populations through linkage analysis or genome-wide association studies (GWAS), respectively. A series of molecular methodologies are then used to map the specific critical region to a locus on a chromosome. Such loci often encompass hundreds of candidate genes, which are then investigated for a mutation that is directly linked to the disease.
Conversely, reverse genetics uses classical genetic engineering techniques to induce the candidate mutation in a model system as a way of testing whether the mutation leads to the hypothesized disease phenotype. Given that reverse genetics has canonically targeted a particular mutation, it relies on prior knowledge about specific genes and possible links to diseases. Such hypotheses may arise from observations of a differential expression pattern of the gene(s) under the disease condition of interest, another similar gene or gene family that is known to be linked to the disease, or another protein in the same biochemical pathway that is known to be associated with the disease. This knowledge is acquired from many fields of biomedical research, including forward genetic studies.
Genetically engineered animal models have been an invaluable reverse genetics approach for demonstrating causality of a mutation in the development of a phenotype. The laboratory mouse has been the preferred genetic model because there is less conservation between humans and other commonly used genetic model organisms such as yeast, flies, worms, and zebrafish. The development of mouse models has depended on the availability of critical methods such as genome engineering via homologous recombination in mouse embryonic stem (ES) cells, which has been the predominant technique over the last few decades (Bedell et al. 1997).
Traditional Forward and Reverse Genetics Technologies
Technological advances have expanded the repertoire of forward and reverse genetics approaches available to human genetics researchers. For example, improvements in cytogenetics, massively parallel DNA sequencing, and induced pluripotent stem cell and haploid ES cell techniques have expedited forward genetics research (Moresco et al. 2013). Conversely, advances in ES cell culturing, precise homologous recombination-mediated gene targeting, and assisted reproduction technologies have contributed significantly to reverse genetics (Bedell et al. 1997).
There are both merits and limitations to using forward and reverse genetic strategies. While forward genetics depends on making phenotypic observations and collecting genetic information from a population which is both time consuming and expensive, it brings great potential for a clear and unbiased understanding of the link between a mutation and the disease. On the other hand, while reverse genetic approaches require a shorter amount of time, their limitations arise from the complexity of human diseases. Although ES cell-based approaches using the laboratory mouse have been used extensively to create disease models, they cannot be routinely used for creating precise genetic changes such as point mutations, short insertion or deletions (indels), large deletions and chromosomal rearrangements. This is especially problematic given that the majority of human disease-causing mutations are subtle genetic changes, such as point mutations involving only one or a few base pairs that lead to alterations in protein expression and/or function (Lodish H et al. 2000). While genetically engineering small changes in animal models can be attempted, ES cell-based approaches inevitably require integration of additional genetic elements, termed “footprints,” into the genome, such as positive selection markers or recombinase recognition sites (Figure 1). It is difficult to assess if such footprints affect the phenotype in the animal model created. In addition, not all diseases are monogenic, and multiple mutations may need to be engineered before a disease phenotype is apparent. Applying reverse genetics to complex diseases has required technological advances beyond the capabilities of ES cell-based gene targeting.
Figure 1. Comparison of traditional and CRISPR-based genome editing technologies for inserting a point mutation.
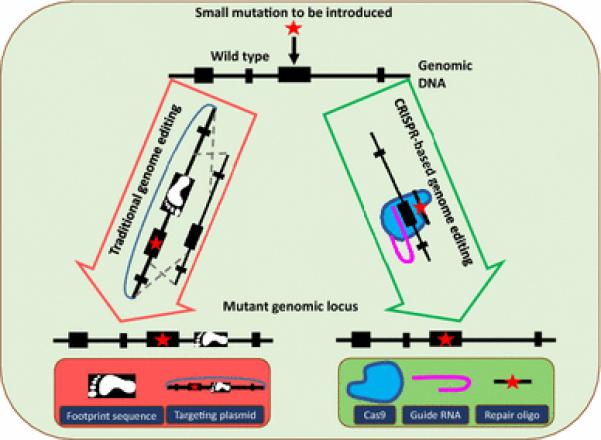
The ES cell-based traditional methods (shown on left) require the use of a targeting vector that contains additional elements (such as a positive selection marker) which get inserted and leave a footprint in the genome. Alternatively, the CRISPR approach (shown on right) enables scarless genome editing. The Cas9 protein and guide RNA help insert the mutation (provided as a repair oligo) at the target site.
Genome Editing Technologies Provide New Tools for Forward and Reverse Genetics
Genome engineering tools developed during the last decade, such as Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs) and CRISPR/Cas programmable nucleases, have made a tremendous impact on reverse genetics approaches (Kim and Kim 2014). Specifically, the ability to induce double strand breaks (DSB) directly into the genomes of embryos relieves major technical bottlenecks of previous approaches. Without the need for ES cells, reverse genetics can now be applied to species where ES cells cannot be established and in mouse strains where germline-competent ES cells are not available. Furthermore, the scalability of the CRISPR/Cas tool, the simplest among the programmable nuclease systems, can also be used as a forward genetics tool in constructing genome-wide mutation libraries to screen for new disease mutations.
The CRISPR/Cas system consists of a few simple components: i) a guide RNA with a unique stretch of 20 nucleotides that are complementary to the target genomic DNA site, and ii) Cas9 nuclease that creates the DSB. The guide RNA directs Cas9 to the desired site to induce a precise DSB event. The cleaved DNA is then repaired using normal cellular DNA repair processes. Non-homologous end joining (NHEJ) may lead to imperfect repair, which could induce frameshift mutations that disrupt the targeted coding sequence. If a repair template is provided, precise genetic changes at the DSB site, such as point mutations or insertion of fusion tags or expression cassettes, can also be achieved by homology-directed repair (HDR).
CRISPR: A Revolutionary Tool for Forward and Reverse Genetics
CRISPR as a forward genetics tool
In addition to observation-based forward genetics approaches in human populations, high throughput screening assays also apply forward genetics in that they take an unbiased approach to identify disease candidate genes. While RNAi has been one of the most commonly used tools for systematically inactivating gene expression in cell cultures, its limitations include incomplete and/or temporary inactivation. Interventions at the DNA level, as opposed to the RNA level, overcome such limitations. Because of its easy scalability (among the programmable nucleases), CRISPR has been readily adapted as a forward genetics tool. Indeed, there are already many reports on CRISPR and its use in high throughput screening and GWAS (Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014; Chen et al. 2015). Unlike the previously used forward genetics approaches, such as mutagenic chemicals or radiation, that induce random mutations and require subsequent identification of these alterations, CRISPR represents the first-ever reported mutagenic tool that can be used for creating libraries of mutations at known sites in the genome (Shalem et al. 2015).
CRISPR as a reverse genetics tool
The CRISPR system has become the most popular among the programmable nucleases because it is simple to design, inexpensive, and highly versatile for many biological applications and a variety cell types and organisms (Shen et al. 2013; Harms et al. 2014). CRISPR has also led to a series of paradigm shifts in animal genome engineering methods (Gurumurthy et al 2016, in press). For example, it can generate knock-out (KO) or knock-in (KI) models in many organisms that were previously inaccessible for genome engineering, point mutations without any other genetic disruptions, homozygous mutant mice in the G0 generation, multiple genetic mutations in one microinjection experiment(Wang et al. 2013), chromosomal translocations (Maddalo et al. 2014; Jiang et al. 2016) and large scale genomic deletions, insertions(Fujii et al. 2013; Zhang et al. 2015) or replacements of several hundred kilobase stretches (Yoshimi et al. 2016). Within less than half a decade, CRISPR has facilitated the creation of hundreds of disease models.
CRISPR's versatility has made it a powerful tool that represents the most promising technology capable of converging and advancing forward and reverse genetic approaches for performing human disease research (Figure 2).
Figure 2. CRISPR is a powerful tool for both forward and reverse genetics.
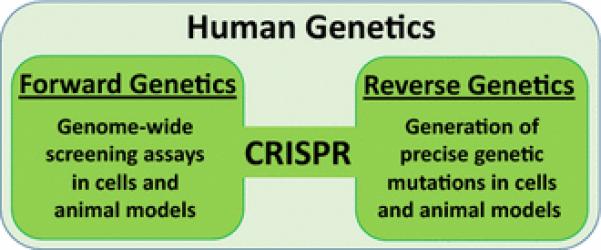
A few examples of forward genetics include genome-wide forward screens for independent replication of previously discovered genes and identification of new genetic factors in human cells ((Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014) and animals (Chen et al. 2015). As a reverse genetics tool, CRISPR has facilitated germline correction of a genetic mutation (Mianné et al. 2016), somatic gene editing (Nelson et al. 2016; Yang et al. 2016b), generation of inducible Cas9 knock-in models (Platt et al. 2014), large genome modifications (Yoshimi et al. 2016), and ex vivo genome editing in human cells (Orack et al. 2015; Claussnitzer et al. 2015).
Examples of CRISPR Applications to Human Disease Research
CRISPR has already emerged as an invaluable tool for human genetics research. We discuss here a few disease models that exemplify the utility of CRISPR for forward and reverse genetics research.
CRISPR as a forward and reverse genetics tool for cancer research
Cancer is largely a genetic disease, as hundreds of mutations have been attributed to its development (Hanahan and Weinberg 2000) which were largely identified through traditional forward and reverse genetics approaches. CRISPR's ability to create libraries of thousands of precise genetic mutations (Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014) has made possible the identification of new tumor modulating genes, in addition to validating the previously known tumor suppressor and oncogenes (Sánchez-Rivera and Jacks 2015). Elegant Cas9 expressing animal models have also been generated that are suitable for forward genetics approaches (Xue et al. 2014; Dow et al. 2015; Chen et al. 2015).
As a reverse genetics tool for cancer research, the CRISPR system has drastically accelerated the process of studying functional consequences of mutations in vivo by enabling direct genome editing in somatic cells (reviewed in Sánchez-Rivera and Jacks 2015) and the development of cancer mutation models in mice including leukemia (Heckl et al. 2014), liver cancer (Xue et al. 2014), lung cancer brain cancer (Zuckermann et al. 2015) and pancreatic cancer (Chiou et al. 2015), just to name a few. These studies demonstrate the power of CRISPR as a reverse genetics tool for rapidly generating cancer models in animals.
CRISPR as a reverse genetics tool for deafness research and as a potential therapeutic tool for hearing loss
The CRISPR system is used as a reverse genetics tool for developing animal models for various disease areas. Described here is deafness, as an example of a disease system, where CRISPR is not only been used as a reverse genetics tool but is also shown to be potential gene therapy tool for correcting hearing loss.
Nonsyndromic Hearing Loss (NSHL) is the most common sensory deficit in humans, affecting more than 28 million Americans, with a prevalence of at least 1.9 per 1,000 infants at birth (Morton and Nance 2006). NSHL is highly genetically heterogeneous; it is inherited in an autosomal recessive mode (ARNSHL) in 77% of cases, an autosomal dominant manner (ADNSHL) in 22% of cases, and X-linked and mitochondrial forms in 1% of cases. Human ARNSHL forms are frequently characterized by an early onset and severe phenotype that generally affects the in utero development and maturation of key inner ear compartments or cell types (Yan and Liu 2010). On the other hand, many ADNSHL forms are commonly characterized by late onset progressive phenotypes, such as the P2RX2 p.Val60Leu mutation (Yan et al. 2013) or the MYO3A p.Gly488Glu mutation (Grati et al. 2016). These ADNSHL diseases have a major impact on our understanding of the biology of hearing and deafness and the late onset of their phenotype provides a potential for reverse genetics-based intervention (Liu and Yan 2007). The developmental course, complex anatomy, and diversity of cell types in the cochlea further adds to the challenge of studying these genetic cases (Angeli et al. 2012). The wide spectrum of NSHL genes and the scarcity of tools for their prevention or reversion have provided a unique opportunity for CRISPR to help understand the etiology and pathophysiology of such mutations.
The inner ear is an ideal organ system for undertaking proof-of-principle gene therapy experiments because of its anatomical size and its easy accessibility for treatment delivery. CRISPR-based reverse genetic approaches are being used to correct deafness alleles. Rescue trials have been initiated for delivering CRISPR reagents via the ear cavity into sensory hair cells of animal models for genes causing ADNSHL (Zuris et al. 2014; Zou et al. 2015), and a successful rescue from hearing loss in the Oblivion mouse (Spiden et al. 2008) has been recently reported (Chen ZY et al. 2016). Hair cell-targeted mutation reversion in other mouse models could serve as additional evidence for rescuing dominant hearing loss, with the potential for translation to deaf patients. Optimizing the delivery time point and the delivery vector into the targeted cell types is extremely critical for the success of the rescue and will ultimately determine the potential for effective patient treatment.
Current Challenges and Future Perspectives
Even though CRISPR has been widely adopted as a reverse genetics tool, its role as a forward genetics tool is still in its infancy, having been used in only in a handful of studies (Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014). Further development of robust forward genetics CRISPR systems will require future research to minimize off-target effects, target different splice variants or isoforms, and noncoding sequences including gene regulatory elements and microRNAs.
Challenges that remain for using CRISPR as a reverse genetics tool include poor HDR efficiency for creating knock-in models and limited availability of guide sequences for some target sites. These are currently being addressed by the development of a few strategies for improving HDR efficiency (Lin et al. 2014; Maruyama et al. 2015; Miura et al. 2015; Aida et al. 2015; Yoshimi et al. 2016) and discovery of additional Cas9-like nucleases (Zetsche et al. 2015; Shmakov et al. 2015) that possess different sequence requirements for guide RNA binding sites.
Off-target cleavage continues to be a concern for both forward and reverse genetics, and certainly will be a major concern as it evolves to be a therapeutic tool. Current research is tackling the effective detection as well as reduction of off-target breaks. A few unbiased and highly sensitive methods of off-target effects detection methods have recently been developed (Tsai et al. 2014; Kim et al. 2015). Additionally, optimization of the CRISPR system continues to improve its efficiency and reduce off-target effects (Slaymaker et al. 2016; Kleinstiver et al. 2016).
CRISPR's reputation as a robust and powerful tool for target identification, genome-wide high throughput screening, and target validation, has inspired investments from several pharmaceutical companies for applications to drug discovery research platforms and novel therapy development (Woolf and Gurumurthy et al., under review). There are already reports that demonstrate ex vivo gene conversion in cultured human patient iPS cells, with the potential to develop cell-based therapies. In addition, successful applications of CRISPR to therapeutic genome engineering in vivo have already been demonstrated in mice (Yang et al. 2016a; Yin et al. 2016). Similar strategies are being developed for human use, although many challenges including optimizing delivery methods, targeting specific tissues, improving Cas9 nuclease specificity, and managing ethical concerns still remain to be addressed (Savić and Schwank 2016). Future research will aim to tackle these challenges.
Acknowledgements
We thank Sidi Chen for his helpful comments on the manuscript. CBG's lab is partially supported by National Institute of General Medical Sciences of the National Institutes of Health under Grant no. P20GM103471 and the Center for Humanized Mice from ORIP/DPCPSI/NIH/1R24OD018546-01. XZL's lab is supported by R01 DC05575, R01 DC01246, and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders. SLPS is supported by the NSF Graduate Research Fellowship DGE1144152. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. We apologize to colleagues whose studies could not be cited because of space constraints.
References
- Aida T, Chiyo K, Usami T, et al. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 2015 doi: 10.1186/s13059-015-0653-x. doi: 10.1186/s13059-015-0653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli S, Lin X, Liu XZ. Genetics of Hearing and Deafness. Anat Rec Adv Integr Anat Evol Biol. 2012;295:1812–1829. doi: 10.1002/ar.22579. doi: 10.1002/ar.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell MA, Jenkins NA, Copeland NG. Mouse models of human disease. Part I: techniques and resources for genetic analysis in mice. Genes Dev. 1997;11:1–10. doi: 10.1101/gad.11.1.1. [DOI] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Tao Y, Gao X, et al. Restoration of hearing by CRISPR/Cas9-mediated genome editing in the Pmca2 deafness mouse model by protein delivery. Assoc Res Otolaryngol Abs. 2016;39:138–139. [Google Scholar]
- Chiou S-H, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Fisher J, O'Rourke KP, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013;41:e187–e187. doi: 10.1093/nar/gkt772. doi: 10.1093/nar/gkt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'hamed Grati, Yan D, Raval MH, et al. Myo3a Causes Human Dominant Deafness And Interacts With Protocadherin 15-Cd2 Isoform. Hum Mutat. 2016 doi: 10.1002/humu.22961. doi: 10.1002/humu.22961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy CB, Quadros R, Sato M, Mashimo T, Lloyd KCK, Ohtsuka M. CRISPR/Cas9 and the Paradigm Shift in Mouse Genome Manipulation Technologies. Genome Editing, Book Chapter Editor, Kursad Turksen. doi: 10.1007/978-3-319-34148-4. [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harms DW, Quadros RM, Seruggia D, et al. Mouse Genome Editing Using the CRISPR/Cas System. Curr Protoc Hum Genet Editor Board Jonathan Haines Al. 2014;83:15.7.1–15.7.27. doi: 10.1002/0471142905.hg1507s83. doi: 10.1002/0471142905.hg1507s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang L, Zhou X, et al. Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci Rep. 2016;6:21918. doi: 10.1038/srep21918. doi: 10.1038/srep21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, et al. Mutations: Types and Causes. In Molecular Cell Biology. 4th ed. W. H. Freeman and Company; New York, NY, USA: 2000. [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014 doi: 10.7554/eLife.04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mianné J, Chessum L, Kumar S, et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med. 2016 doi: 10.1186/s13073-016-0273-4. doi: 10.1186/s13073-016-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H, Gurumurthy CB, Sato T, et al. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci Rep. 2015;5:12799. doi: 10.1038/srep12799. doi: 10.1038/srep12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EMY, Li X, Beutler B. Going Forward with Genetics. Am J Pathol. 2013;182:1462–1473. doi: 10.1016/j.ajpath.2013.02.002. doi: 10.1016/j.ajpath.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orack JC, Deleidi M, Pitt D, et al. Concise Review: Modeling Multiple Sclerosis With Stem Cell Biological Platforms: Toward Functional Validation of Cellular and Molecular Phenotypes in Inflammation-Induced Neurodegeneration. Stem Cells Transl Med. 2015;4:252–260. doi: 10.5966/sctm.2014-0133. doi: 10.5966/sctm.2014-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR–Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR–Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiden SL, Bortolozzi M, Di Leva F, et al. The novel mouse mutation Oblivion inactivates the PMCA2 pump and causes progressive hearing loss. PLoS Genet. 2008;4:e1000238. doi: 10.1371/journal.pgen.1000238. doi: 10.1371/journal.pgen.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33:187–197. doi: 10.1038/nbt.3117. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Liu X-Z. Modifiers of Hearing Impairment in Humans and Mice. Curr Genomics. 2010;11:269–278. doi: 10.2174/138920210791233054. doi: 10.2174/138920210791233054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016a;34:334–338. doi: 10.1038/nbt.3469. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016b;34:334–338. doi: 10.1038/nbt.3469. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Yin H, Song C-Q, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kunihiro Y, Kaneko T, et al. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun. 2016;7:10431. doi: 10.1038/ncomms10431. doi: 10.1038/ncomms10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jia R, Palange NJ, et al. Large Genomic Fragment Deletions and Insertions in Mouse Using CRISPR/Cas9. PLOS ONE. 2015;10:e0120396. doi: 10.1371/journal.pone.0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zou B, Mittal R, M'hamed Grati, et al. The application of genome editing in studying hearing loss. Hear Res. 2015;327:102–108. doi: 10.1016/j.heares.2015.04.016. doi: 10.1016/j.heares.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2014;33:73–80. doi: 10.1038/nbt.3081. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]


