Abstract
While a variety of powerful tools exist for analyzing RNA structure, identifying long-range and intermolecular base pairing interactions has remained challenging. Recently, three groups introduced a high-throughput strategy that uses psoralen-mediated cross-linking to directly identify RNA-RNA duplexes in cells. Initial application of these methods highlights the preponderance of long-range structures within and between RNA molecules and their widespread structural dynamics.
The role of RNA as a central functional molecule in biology has become increasingly clear. Long non-coding RNAs (lncRNAs), small nuclear and nucleolar RNAs, microRNAs, and the untranslated regions of messenger RNAs (mRNAs) regulate integral cellular processes including transcription, splicing, localization, ribosome assembly, and translation [1]. Even within coding regions of mRNAs, regulatory information is layered on top of the protein message via differential usage of synonymous codons. Correspondingly, the list of human diseases associated with mutations in and misregulation of RNA molecules is growing at an accelerating pace [2].
RNA molecules mediate diverse functions in part by folding into unique structures consisting of complementary base pairing and higher order tertiary interactions. A classical example is the L-shaped structure of transfer RNA, which is essential to its role in peptide synthesis. Because underlying structure is inextricably linked to the complex activities of most RNAs, the ability to accurately identify intracellular RNA structure is paramount, and multiple technologies have been developed for RNA structure analysis (Fig. 1). Two long-used methods are sequence covariation analysis and thermodynamic free-energy minimization. Covariation analysis is limited by the need for a diverse multiple sequence alignment, and thermodynamic approaches, which work well for short RNAs, struggle when used to explore biological hypotheses for large and complex RNAs. For most RNAs, structure analysis driven by experimental information is therefore essential. NMR, X-ray crystallography, and cryo-electron microscopy (cryo-EM) provide unparalleled resolution but are low-throughput and limited to “well-behaved” molecules. The most widely useful class of methods are single-nucleotide resolution structure probing experiments, which use a chemical or enzymatic reagent that react with RNA in a quantitative and structurally informative way [3].
Figure 1. Direct duplex detection methods join a host of powerful tools for RNA structure analysis.
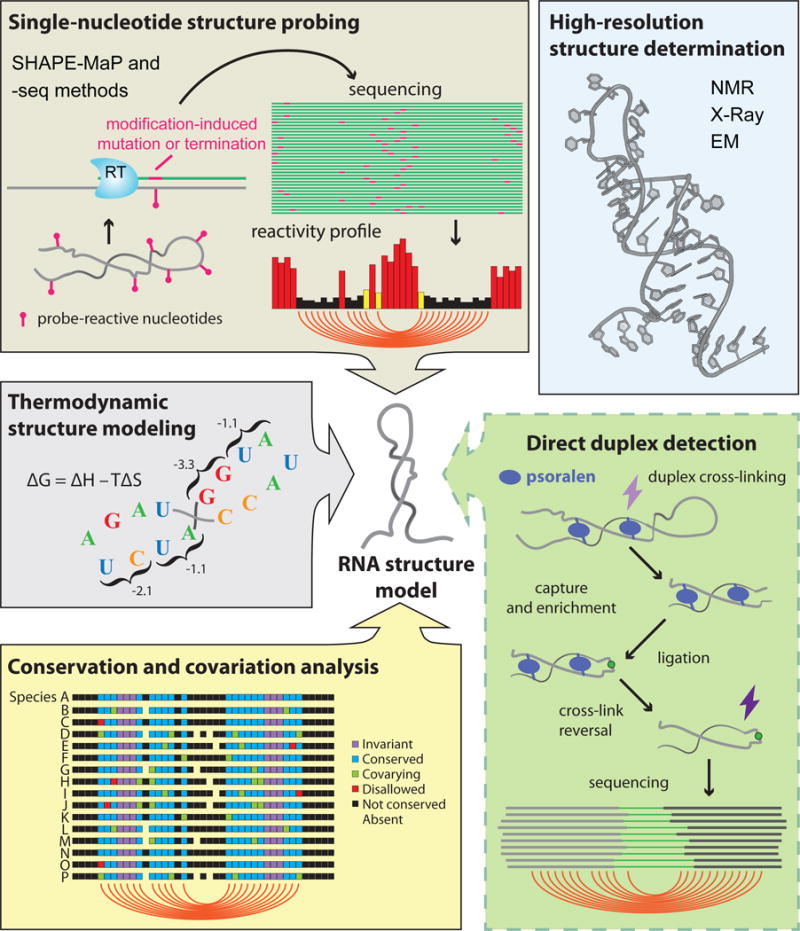
NMR, X-Ray crystallography, and cryo-EM can be used to determine atomic-resolution structures for “well-behaved” RNAs (blue box). For most other RNAs, a suite of complementary methods facilitates secondary structure determination. RNA structure can be probed with chemical or enzymatic reagents that, when read out by sequencing, provide a nucleotide-resolution profile of RNA flexibility (SHAPE-MaP and -seq methods, brown box). Thermodynamic stability parameters can be used to estimate the minimum energy structure for an RNA sequence (gray box). Base-paired nucleotides can be inferred from conservation patterns observed in sequence alignments of homologous RNAs (yellow box). This existing toolset is complemented by new direct duplex detection methods, in which paired RNA strands are cross-linked with psoralen and ligated together before detection via sequencing (green box).
State-of-the-art strategies leverage high-throughput sequencing (-seq) and integrate the unique advantages of multiple tools. For example, in the SHAPE-MaP approach (Selective 2′-Hydroxyl Acylation analyzed by Primer Extension and Mutational Profiling), highly accurate structure models can be obtained by using nucleotide-resolution structure-probing data to restrain thermodynamic RNA modeling algorithms [4]. Typically, mutational analysis or comparative genomics are then used to validate de novo structure models. Nevertheless, it is challenging to identify long-range intra-molecular pairing interactions, and new strategies are needed to detect inter-molecular interactions.
Several groups have recently introduced high-throughput cross-linking-based structure analysis experiments that can detect RNA duplexes in cells. These approaches are Psoralen Analysis of RNA Interactions and Structures (PARIS), LIGation of interacting RNAs followed by high-throughput sequencing (LIGR-seq), and Sequencing of Psoralen cross-linked, Ligated, and selected Hybrids (SPLASH) [5–7]. These three duplex detection experiments are similar and employ five major steps: (i) a derivative of the chemical reagent psoralen is used to selectively cross-link duplexed RNA strands; (ii) cross-linked duplexes are enriched; (iii) the two strands of each duplex are ligated together; (iv) the cross-link is reversed; and (v) the ligated junctions are sequenced to identify the interacting RNA sequences (Fig. 1). The trapping of RNA duplexes through strand-to-strand cross-links distinguishes PARIS, LIGR-seq, and SPLASH from prior methods. For example, CLASH (Cross-linking, Ligation, And Sequencing of Hybrids) and hiCLIP (RNA hybrid and individual-nucleotide resolution ultraviolet Cross-Linking and ImmunoPrecipitation) can be used to detect RNA duplexes, but rely on cross-linking between RNA duplexes and duplex-specific RNA-binding proteins, thus excluding non-protein-bound duplexes [8,9]. RNA proximity ligation is another promising method but, since it forgoes cross-linking, only the subset of duplexes that persist throughout downstream biochemical processing steps are detected [10].
These new psoralen-mediated duplex detection methods feature important advances in RNA structural analysis. First, unlike structure probing, duplex detection provides direct evidence of specific pairing interactions. Lu et al. report that up to 40% of PARIS-identified duplexes in mammalian cells are separated by distances of greater than 200 nucleotides, and 5–10% span distances greater than 1000 nucleotides. The ability to detect very long-range interactions suggests it may soon be possible to examine potential functional roles of higher-order structure in mRNAs and lncRNAs. The additional ability to detect inter-molecular duplexes offers a much-needed method for studying higher-order structure in multi-RNA complexes. A second advantage is that duplex detection methods are single molecule experiments, with each ligated sequence read representing a single occurrence of an inter- or intra-RNA duplex. Thus, it is possible to directly observe RNA regions that participate in mutually exclusive duplex interactions, suggestive that an RNA adopts multiple structures.
PARIS, LIGR-seq, and SPLASH represent a novel class of tools for understanding RNA structure-function relationships, but it is important to recognize several current limitations. First, the methods are complex to implement and require multiple biochemical processing steps that are each likely to introduce significant biases. Further, ligated junctions represent only a small fraction of obtained sequencing reads, with most data discarded as waste. Third, the resolution of these approaches is on the scale of tens-of-nucleotides rather than individual-nucleotides. Finally, these methods primarily provide qualitative confirmation that a duplex exists, and the absence of a cross-link does not indicate an absence of structure. More quantitative data are needed for de novo RNA structure determination, and understanding dynamic RNAs requires quantitative information about the relative population of different states. Thus, as currently implemented, duplex detection methods are most useful for identifying interesting structures for subsequent in-depth follow-up, and for validating structures established by complementary approaches.
RNA structure analysis has matured to a state beyond what was imaginable just several years ago, with researchers now attempting to parse structure-function relationships in vivo for highly complex and dynamic RNA molecules. Direct duplex detection experiments are a welcome addition to the RNA structure analysis toolbox. As we push forward on the frontiers of RNA structural and mechanistic biology, we will need to leverage and integrate multiple innovative tools.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 3.Kwok CK, et al. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Siegfried NA, et al. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Z, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aw JGA, et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 7.Sharma E, et al. Global Mapping of Human RNA-RNA Interactions. Mol Cell. 2016;62:618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 8.Sugimoto Y, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudla G, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramani V, et al. High-throughput determination of RNA structure by proximity ligation. Nat Biotechnol. 2015;33:980–984. doi: 10.1038/nbt.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]


