Abstract
Transcription activator-like effector nucleases (TALENs) are one of several types of programmable, engineered nucleases that bind and cleave specific DNA sequences. Cellular machinery repairs the cleaved DNA by introducing indels. In this review, we emphasize the potential, explore progress, and identify challenges in using TALENs as a therapeutic tool to treat HIV infection. TALENs have less off-target editing and can be more effective at tolerating HIV escape mutations than CRISPR/Cas-9. Scientists have explored TALEN-mediated editing of host genes such as viral entry receptors (CCR5 and CXCR4), and a protein involved in proviral integration (LEDGF/p75). Viral targets include the proviral DNA, particularly, and the long terminal repeats. Major challenges with translating gene therapy from bench to bedside are improving cleavage efficiency and delivery, while minimizing off-target editing, cytotoxicity, and immunogenicity. However, rapid improvements in TALEN technology are enhancing cleavage efficiency and specificity. Therapeutic testing in animal models of HIV infection will help determine whether TALENs are a viable HIV treatment therapy. TALENs or other engineered nucleases could shift the therapeutic paradigm from life-long antiretroviral therapy towards eradication of HIV infection.
Keywords: gene editing, TALEN, delivery, animal model, HIV, off-target
Introduction
Transcription activator-like effector (TALE), proteins from the Xanthomonas pathogen, activate transcription of specific plant host proteins (Bogdanove et al. 2010). TALEs contain a nuclear localization signal, a transcriptional activation domain, and a DNA binding domain (Fig. 1). The DNA binding domain contains modular repeats, with each monomer bound to one base in the DNA target. The number of, and order of repeats in the DNA binding domain varies and determine the binding specificity for the DNA sequence. The cipher encoding TALE recognition for specific DNA sequences identified the recognition determinants as the repeat variable di-residues (RVDs) found at amino acid positions 12 and 13 of each monomer (Boch et al. 2009; Moscou and Bogdanove 2009). The race to develop this system as a tool to target specific DNA sequences ensued after cracking of the cipher.
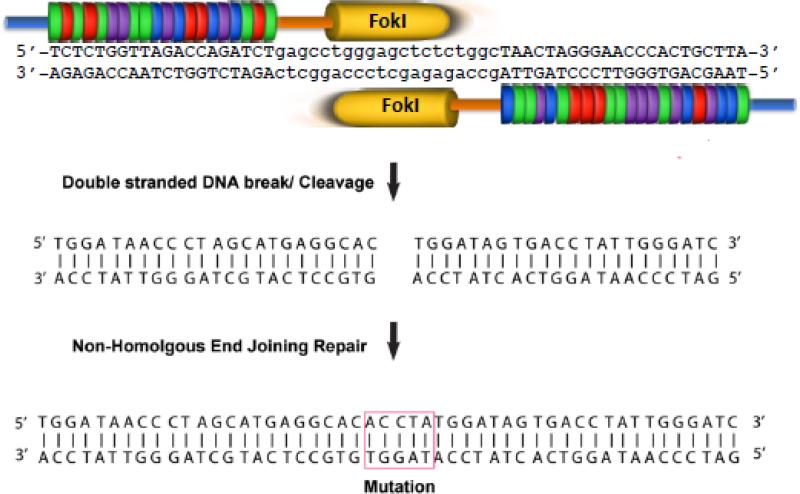
Fig. 1. TALEN pair targeting HIV proviral DNA.
Example of TALEN binding, cleavage and repair of double strand breaks. TALENs bind and cleave a DNA target sequence; the HIV LTR sequence is the example shown (Strong et al. 2015). Endogenous NHEJ repair of double strand breaks introduces indels.
Researchers took advantage of past successes fusing the catalytic domain of FokI endonuclease to zinc fingers to design TALE/FokI chimera called Transcription activator-like effector nucleases (TALENs). These TALENs, in the form of functional protein pairs, bind and cleave their designated DNA targets (Li et al. 2011; Mahfouz et al. 2011; Cermak et al. 2011). The resulting double stranded break triggers the host cell's DNA repair systems. Cellular non-homologous end joining repair (NHEJ) or homology-directed repair/recombination (HDR) repair the lesion (Budhagatapalli et al. 2015; Tesson et al. 2016). Experimentalist target these repair pathways, using NHEJ for gene disruption and HDR for gene correction or insertion of exogenous DNA.
Following proof of principle, the Golden Gate and FLASH assembly systems streamlined construction of this new gene editing tool set (Li et al. 2011; Miller et al. 2011; Cermak et al. 2011; Reyon et al. 2012). More recently, Ligation independent cloning (LIC), Golden Gate, and Platinum Gate assemblies reduce the time and expense and required for the TALEN construction (Christian et al. 2010; Schmid-Burgk et al. 2013; Cermak et al. 2015; Schmid-Burgk et al. 2015; Sakuma and Yamamoto 2016).
There are four main challenges in applying gene editing endonuclease (GEEN) technology to treat HIV: 1) editing efficiency; 2) delivery; 3) off target editing and cytotoxicity; and 4) immune tolerance. There are four general classes of GEEN technologies (Meganucleases, Zinc Finger Nucleases (ZFNs), TALENs, and CRISPR/Cas-9) tested on different targets to develop new HIV gene therapies (Han and Li 2016).
In this review we focus on the study of TALENs targeting both host and viral HIV genes as a promising HIV therapy. Advantages of TALENs include their minimal cytotoxicity and off-target editing (Mussolino et al. 2011; Cradick et al. 2013). However, we note that in a recent clinical trials patients tolerated treatment with ZFNs targeting host CCR5; despite the generally recognized off-target editing and cytotoxicity with ZFN technology (Cornu and Cathomen 2010). TALENs, like CRISPR/Cas-9 have high flexibility in design for specific target sites and several examples of high editing efficiency (Valton et al. 2012; Miller et al. 2015). Other advantages are the design of TALENs to target methylated HIV proviral DNA, which is relevant to latent infection, and encoding monomers with degenerate recognition to target predicted escape mutations (Bultmann et al. 2012; Valton et al. 2012; Chen et al. 2013; Strong et al. 2015). Disadvantages in using TALENs are that they take longer to build, are larger, thus more difficult to deliver, and are likely to generate an immune response. Herein, we review current progress on potential TALEN-based HIV therapies.
Host and viral targets for gene editing
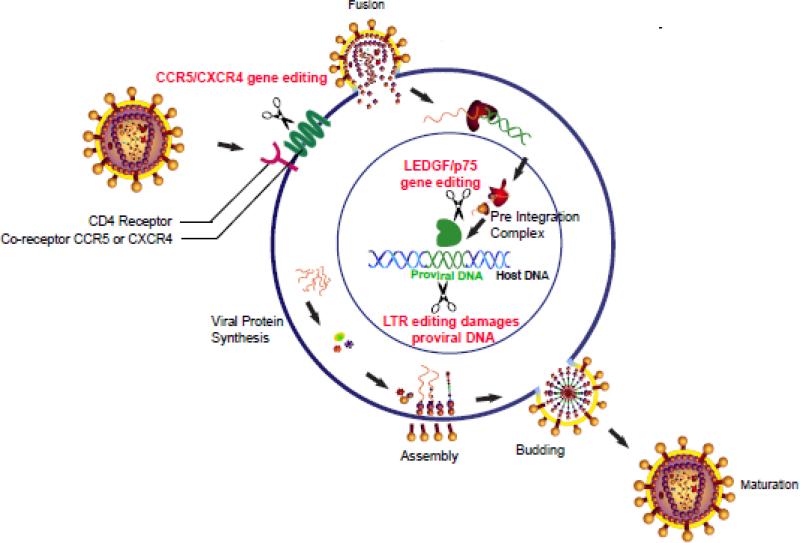
The HIV early life cycle comprises pre-transcriptional steps of virion entry into cells, uncoating of the virus envelope, reverse transcription of the viral genome, formation of the pre-integration complex, and integration of proviral DNA into the host genome (Fig. 2) (Levy 1993). Following provirus integration, HIV gene expression is precisely controlled. During the late steps of replication, the infected cell packages two copies of the HIV genome into an immature HIV particle that buds off the plasma membrane. Host or viral genes necessary for viral replication or maturation are potential therapeutic targets for gene editing.
Fig. 2. HIV and host targets of TALEN-based therapies.
Cartoon of a T-cell with cell processes labeled and TALEN targets labeled in red font.
Scientists have investigated TALEN editing of both host and viral targets. In a functional cure, HIV proliferation is controlled without eradication of the latent viral reservoir (Liu et al. 2015). The purpose of targeting host proteins is to produce a functional cure, thereby suppressing viral replication and restoring a functional immune system in the presence of infection. Thus far, the objective of targeting HIV genes is to damage the proviral DNA in infected cells, thereby eradicating the latent viral reservoir. One distinct difference in these strategies is that both copies of the host genes would need to be inactivated in the diploid genome, whereas targeting HIV genes needs to inactivate the one integrated copy of HIV provirus (Josefsson et al. 2011). This also has the advantage that error prone NHEJ is favored over homologous recombination repair of TALEN induced double strand breaks. Herein, a number of target genes selected to negatively impact HIV infection are discussed.
Target site selection
Criteria for target gene selection should include that the gene is required for viral replication or maturation, the human host should tolerate inactivation of the gene, and that the target region of the viral or host gene should not have high nucleotide sequence similarity with other regions of the host genome that could result in off-target editing.
Once a target has been identified, designing TALENs to minimize, if not completely abrogate off-target editing is important (Kim et al. 2013; Lin et al. 2014). TALENs designed to target HIV should consider that remnants of ancient retroviral infections acquired over the course of human evolution exhibit sequence homology with HIV (Schiffer et al. 2012; Ishida et al. 2015). There are two approaches to reduce off-target editing: 1) increasing the specificity of each TALENs DNA binding domain; and 2) avoiding off-target sites found elsewhere in the human genome. In target site selection, in silico tools like Tal Effector Nucleotide Targeter 2.0, Prognos, CHOPCHOP, E-TALEN, TALEN-NT, TALENoffer, and SAPTA predict potential off-target sites with good accuracy (Doyle et al. 2012; Grau et al. 2013; Heigwer et al. 2013; Fine et al. 2014; Lin et al. 2014; Montague et al. 2014).
Following TALEN construction, researchers must also consider the potential contribution of TALEN expression to the off-targeting phenomenon. Over-expression of TALENs saturate the target site, increasing the likelihood of off-target editing. This may be an issue even at nanomolar concentrations, considering the strong affinities of TALENs for their targets (Bultmann et al. 2012; Guilinger et al. 2014). However, there is a trade off with efficiency as higher expression levels increase on-target editing efficiency (Strong et al. 2015). Scientists routinely use Sanger sequencing, CEL-I, and/or T7E1 assays to evaluate off-target editing; however, whole genome sequencing and whole exome sequencing are more rigorous approaches to evaluate specificity. It is noteworthy that with proper TALEN design, off-target editing can be largely eliminated (Ding et al. 2012; Ousterout et al. 2013). Furthermore, several modifications to TALENs minimize off target editing as described later.
Entry co-receptors CCR5 and CXCR4
HIV mainly uses the host CCR5 or CXCR4 as co-receptors to CD4 for cell entry. Individuals homozygous for a CCR5Δ32 deletion display resistance to HIV infection (Dean et al. 1996; Huang et al. 1996; Zimmerman et al. 1997; Benkirane et al. 1997). CCR5 silencing, anti-CCR5 antibodies, and small molecular inhibitors inhibit HIV infection, confirming the strategy of blocking CCR5 function (Lopalco 2010; Mock et al. 2015). The HIV resistance of CCR5 depleted cells gained acceptance when the “Berlin patient”, who received HLA compatible stem cell transplantation from an individual homozygous for CCR5-Δ32, remained free of virus without HAART therapy (Yukl et al. 2013). However the “Essen patient”, who received very similar treatment, relapsed with a X4 tropic HIV, indicating that targeting of CCR5 alone may not prove a comprehensive approach (Kordelas et al. 2014).
These observations laid the groundwork for a therapeutic gene editing approach initially targeting the CCR5 co-receptor. Studies of CCR5 gene disruption with ZFNs have been reviewed (Drake and Bates 2015; Gu 2015; Allers and Schneider 2015). More recently, editing of this target has progressed to the stage of testing in humans (Reardon 2014; Tebas et al. 2014). A phase I clinical trial including 12 patients treated with ZFNs to inactivate CCR5 was tolerated for almost a year and showed efficacy for treatment in one patient (Tebas et al. 2014). While promising, a major concern is that ZFNs produce off-target editing and cytotoxicity, and this was not examined in these patients (Händel and Cathomen 2011).
One advantage of TALENs is their minimal cytotoxicity and off-target editing compared to other GEENs (Mussolino et al. 2011; Cradick et al. 2013). TALENs are also among the gene editing techniques targeting CCR5 (Manjunath et al. 2013). The CCR5 target sites of TALENs include DNA sequences encoding transmembrane domains (Mussolino et al. 2011; Ru et al. 2013; Liu et al. 2014; Mock et al. 2015). A noteworthy caveat is that HIV switches its preference from CCR5 to the CXCR4 co-receptor in the later stages of infection (Hütter et al. 2015). Hou et al. disrupted the CXCR4 gene with CRISPR/Cas-9, increasing resistance to X4 tropic HIV in cell culture. However, using CXCR4 as a gene editing target is debatable, considering its role in thymic differentiation, progenitor cell migration and homing, and hematopoietic cells (Hou et al. 2015).
TALEN editing to treat HIV infection has limitations like sequence variability in the divergent strains of HIV, high mutation rates in TALE binding sites that could produce escape mutations, and inefficient delivery of TALENs to latent cells that are less than <1% of the total T cells. To address escape mutations, TALENs are unique among GEENs, where monomers with NS RVDs bind and cleave, targets with degenerate positions (Boch et al. 2009). For example, Strong et al. showed that TALENs engineered to recognize degeneracy edited mutated targets at poorly conserved positions in the HIV long terminal repeat (Strong et al. 2015). Despite this advantage, CRISPR/Cas-9 editing of the HIV provirus produced some escape indels, raising the possibility that some TALEN editing may also produce indel escape mutations (Wang et al. 2016). There may also be limitations in accessing the HIV genome packaged in heterochromatinized DNA in latent cells, which would be a problem for all GEEN therapies. However, TALEN design modification that recognize methylated cytosine make it possible for targeting the compactly packaged DNA including silenced LTRs in latent cells (Valton et al. 2012).
LEDGF/p75
Several observations warrant the investigation of host Lens epithelium-derived growth factor (LEDGF/p75) as a target for gene editing. LEDGF/p75 binds integrase and facilitates HIV proviral DNA integration into the host genome (Cherepanov et al. 2005; Shun et al. 2007). Additionally, LEDGF/p75 complexes with Iws1 and Spt6 promoting latency (Gérard et al. 2015). Inhibitors or the LEDGF/p75-Integrase interaction reduce HIV replication (Christ et al. 2012). Fadel et al. knocked out the exon coding regions for the integrase binding domain, as well as the complete LEDGF/p75 gene using TALENs (Fadel et al. 2014). The knockout severely affected HIV replication in Jurkat cells. Interestingly, LEDGF/P75 is also a therapeutic target for mixed lineage leukemia (MLL)(Cermáková et al. 2014). However, the majority of LEDGF/P75 knock out mice die prenatally and the surviving develop phenotypic abnormalities, raising concerns for LEDGF/P75 inactivation in human cells (Sutherland et al. 2006).
Long Terminal Repeat
Resting memory T cells can serve as a reservoir of latent HIV infection, persisting while under HAART therapy and rebounding after HAART cessation (Chun et al. 2010; Le et al. 2011). GEENs, like TALENs, can edit targets in the integrated proviral genome. Targeting HIV genes or regulatory regions has the advantage that it does not interfere with the host genes. One of the two non-coding Long Terminal Repeats (LTR) contains the HIV promoter, which drives expression of viral proteins. The trans-activation response element (TAR) region of the LTR is critical for viral replication, thus is a viable target.
Strong et al targeted this highly conserved region that occurs in both the 5’ and 3’ LTR of the viral genome using TALENs. A dose dependent increase in TALEN expression enhanced editing efficiency with maximal 55-60% editing efficiency. The linear dose-response suggests that even higher expression would produce higher editing efficiency. Furthermore, TALEN-mediated LTR editing abolished virion production in cells harboring an integrated complete HIV-1 genome (Strong et al. 2015). A side-by-side comparison of TALENs and CRISPR/Cas-9 technologies showed that TALEN-mediated editing of the LTR was significantly more efficient than editing by CRISPR/ Cas-9 in a T cell line (Ebina et al. 2015).
Refining TALENs gene editing
Since the introduction of the TALEN technology, several modifications improved editing efficiency (sensitivity) and reduced off-target editing (specificity). Modifications of FokI and scaffolds enhance TALEN editing efficiency (Fig. 3). The Sharkey mutations and obligate heterodimer mutations in the FokI nuclease domain of ZFNs increased editing efficiency and have been transferred to TALENs (Guo et al. 2010; Doyon et al. 2011). Directed evolution experiments in bacteria identified the Sharkey mutations in FokI (S148P and K441E) that increase editing 3-6 fold, over wild type (Guo et al. 2010). Structure-guided engineering of the FokI nuclease dimerization interface identified a set of obligate heterodimer mutations (ELD: Q486E, I449L, N496D with KKR: E490K, I153K, and H537K) (Doyon et al. 2011). Accordingly, a FokI nuclease domain with the ELD mutation will only dimerize with its partner domain containing the KKR mutation, which enhances cleavage site specificity, as dimerization is necessary for endonuclease activity. Fortuitously, the Sharkey and heterodimer mutations cooperatively improve TALENs editing efficiency, but not yet tested in targeting HIV targets (Nakajima and Yaoita 2013; Guilinger et al. 2014).
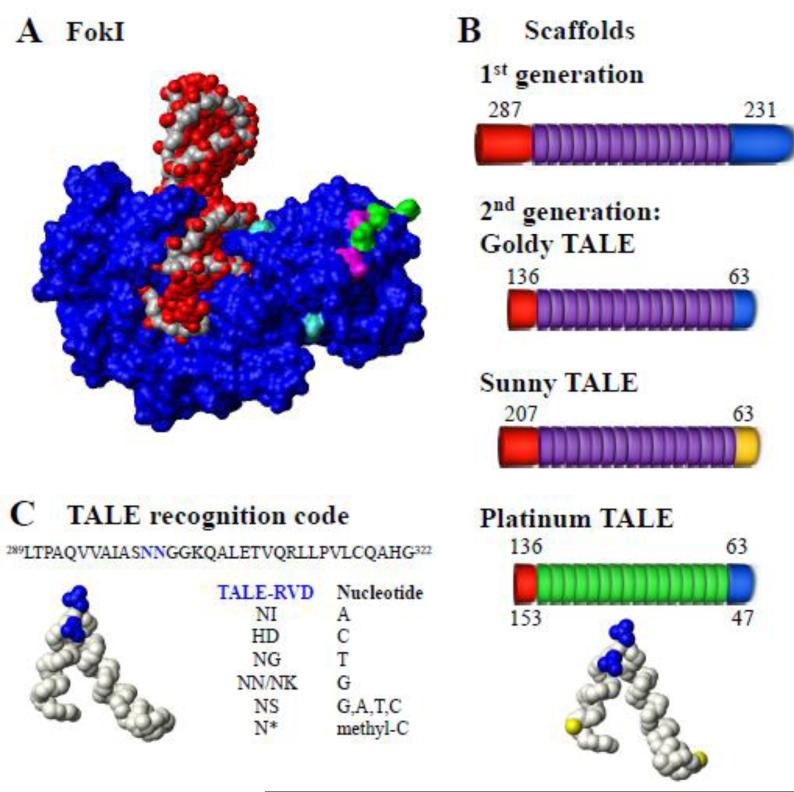
Fig. 3. Location of Sharkey and obligate heterodimer mutations in the FokI endonuclease.
A. Structure of complex (Protein Data Bank [PDB] accession = 1FOK) of wild type Flavobacterium okeanokoites FokI protein (Blue) with DNA (red/grey). Mutations are colored: magenta and green indicate the ELD and KKR obligate heterodimers mutations of the FokI, respectively [3]; Turquoise indicates the Sharkey mutations [3]. B. TALE recognition code. Protein sequence and structure of PthXo1 TALE (PDB accession = 3UGM, amino acids 289-322) with positions of the RVD colored blue(Mak et al. 2012). Nucleotide recognition code for substituted RVDs is shown; * indicated deletion. C. Scaffolds used to build TALENs. Lengths of N- and C-terminal domains are shown with TALE repeats. Red indicates the C-terminus; purple indicates 1st generation TALE repeats and green indicates Platinum TALE repeats; blue indicates 1st generation C-terminal domain; and yellow indicates mutated Sunny TALE C-terminal domain. PthXo1TALE structure (amino acids 289-322) with positions of the RVD (blue) and substitutions in the repeat scaffold at positions 4 and 35 (yellow) shown for Platinum TALEs (PDB accession = 3UGM). Structure figures were created with MOLMOL 2K.1 (Koradi et al. 1996).
The TALEN scaffold determines the overall architecture of the TALEN protein. Improvements in TALEN scaffolds include the length of N- and C-terminal domains, sequences of non-RVD residues in TALE repeats, and substitution of non-classical RVD monomers. First generation TALENs scaffolds were typical of the TAL effector architecture from Xanthomonas spp, with an N-terminal domain of 287 residues, C-terminal domain of 231 residues, and 13-30 bp between the target binding sites (Christian et al. 2010). However, a series of TALEN constructs helped elucidate that truncated N-terminal and C- terminal domain with 136 and 63 residues, respectively (+136/+63 scaffold), more efficiently edit several human genes when compared to other scaffolds (Miller et al. 2011).
The Miller et al. study laid the foundation for second-generation TALEN scaffold modifications (Goldy, Platinum, and Sunny TALENs). Goldy TALENs, have a +158/+63 scaffold (N-terminus / C-terminus lengths), exhibit greater editing efficiency when compared to first generation TALENs, and are built with Golden Gate cloning (Cermak et al. 2011; Bedell et al. 2012; Ma et al. 2013). Platinum TALENs have a +136/+63 or +153/+47 scaffold with non-RVD variants in the 4th and 32nd residues, compared with classic TALE monomers whose residues are similar except at the RVD. These non-RVD variants can be D-D, D-A, E-A, A-D (Sakuma et al. 2013; Miller et al. 2011). Unlike Goldy and Platinum TALENs, Sunny TALENs have a relatively longer scaffold of +207/+63 with a point mutation (P11H) in the C-terminal domain (Sun et al. 2012a). These second generation TALENs increased the editing efficiency across many cell lines and organisms (Sun et al. 2012b; Nakajima and Yaoita 2013; Sakuma et al. 2013).
In addition, other RVD mutations broaden the TALE cipher (AA binds T, HN binds G, CI binds A, and RD binds C) (Miller et. al, 2015) or target epigenetic modifications that may impair TALEN accessibility. DNA targets containing 5-methylated cytosines, generally found in CpG islands, can prevent TALENs from binding their designated targets (Bultmann et al. 2012; Valton et al. 2012; Chen et al. 2013). Fortunately, substitution of the HD monomer that recognizes cytosine with a unique monomer (N*) recognizing methylated cytosine is a viable work-around in select instances (Valton et al. 2014). Several TALEs with mutated lysines or arginines in the C-terminal domain produced enhanced editing efficiency (Guilinger et al. 2014). Mutation of three (Q3) or seven (Q7) cationic amino acids to glutamine enhanced the DNA binding specificity (Guilinger et al. 2014). When targeting CCR5, the Q3 and Q7 TALENs produced approximately 24 and 120 fold high specificity, respectively, for the CCR5 target site (Guilinger et al. 2014).
Delivery
Criteria for selecting the optimal delivery method for TALENs must consider: the target cell, target cell proliferation status, size of the TALEN genes, desired gene expression level, toxicity, delivery efficiency, ease of delivery system construction, and immune response, if delivered in vivo. To translate TALEN-based HIV therapy from the bench to bedside, scientists must carefully balance these often competing criteria. While the preferred delivery method for TALENs is in vitro transfection, this approach generally does not work well in differentiated cells and in vivo. Instead, scientists most often select recombinant viral vectors for in vivo experimentation. Adenoviral, adeno-associated vectors, and lentiviral vectors are among the most frequent vectors used in clinical trials testing gene therapy. Consequently, this section focuses on these vectors.
Adenoviral vector
Adenoviruses, non-enveloped doubled stranded DNA viruses, interact with host cell receptors through viral fiber proteins extending from the viral capsid. More than 50 human adenoviruses have been characterized, and adenoviral vector constructs based upon serotype 5 (Ad5) are in common use (Holkers et al. 2014). Ad5 adenovirus infects cells through binding the coxsackievirus-adenovirus receptor (CAR). First generation adenoviral vectors replaced genes required for replication (E1) with exogenous gene(s) of interest. These vectors, with limited packaging capacity accommodate small inserts and require complement cell lines for propagation. Second generation adenoviral vectors lack genes for replication and viral packaging (such as E1, E3, and E4), and can accommodate up to a 10 kb transgene. Correspondingly, “empty” adenoviruses have a packaging capacity of about 35kb. The empty virus lacks most viral genes excluding the Inverted Terminal Repeat (ITR) and packing signal, thus requires a helper vector and complement cell line to produce virus (Holkers et al. 2014).
While adenoviruses with a TALEN transgene targeting HIV do not yet exists, Ad5 has been pseudotyped for T cells, and adenoviral vectors that express TALEN for gene editing were tested. Adenoviral vectors infect both proliferating and non-proliferating cells with relatively high transduction efficiency, thus could target HIV-infected T cells with active and latent infection. Adenoviruses cannot infect T cells or macrophages because of low CAR expression. As such, chimeric pseudotyped Ad5/F35 adenoviral vectors with displaying the F35 fiber bind host CD46 and transduce hematopoietic cells via CD46 (Schroers et al. 2004). Pseudotyped Ad5/F35 could be engineered to express TALENs. The viral DNA does not integrate into the host genome and transiently expresses transgenes. Infection with a set of recombinant Ad5 adenoviral vector delivered monocistronic TALEN pairs producing editing of the Ddx3y gene (Zhang et al. 2013). Optimally, it would be better to use a bicistronic vector with the Ad5, but packaging both large TALEN genes may prove difficult.
Lentiviral vector
Lentiviral vectors, derivatives of HIV, produce virions with an icosahedral nucleocapsid and a genome size of 3-9kb. Generation of lentiviral vectors involves various plasmids including: a packaging plasmid with rev-response element, gag, pol, tat, and rev sequences, an expression plasmid consisting of a packaging signal, gene of interest, and LTR and a pseudotyping plasmid consisting of a VSV envelope glycoprotein sequence. These plasmids are propagated and packaged in complement cell lines (Zheng et al. 2015). The VSV-G pseudotyping of lentiviral vectors confers broad tissue tropism resulting in infection of cells in vitro and in vivo, as used in clinical trials for metachromatic leukodystrophy indication (Cronin et al. 2005). Lentiviral vectors have relatively high transduction efficiency observed in primary hematopoietic cells (~60%) and stably express integrated genes (Li et al. 2005). However, VSV-G psuedotyped lentiviral vectors cannot specifically infect resting CD4+ T cells. In such cases, lentiviral vectors psuedotyped with Vpx-VLP (Viral protein x and Virus-like particle) and CXCR4-tropic HIV-1 envelope can be used to mediate delivery (Agosto et al. 2009; Geng et al. 2014). Non-targeted integration into tumor suppressor genes loci is a major cause for concern when using lentiviral vectors (Maldarelli et al. 2014). Furthermore, an 8 kb packaging capacity is inadequate for some bicistronic TALEN pairs, and stable gene expression may not be appropriate for editing HIV entry receptors or proviral DNA long term. However, Ebina et al. demonstrated that bicistronic lentiviral vectors effectively mediate delivery of TALENs to T cells, with editing efficiencies at about 80% in various T cell lines (Ebina et al. 2015). In any case, lentiviral vectors may become a more attractive gene therapy tool if the potential of integrase-defective lentiviral vectors is realized (Hanoun et al. 2016).
Adeno-associated viral vector (rAAV)
The AAV genome, approximately 5 kb in length, presents with a packaging capacity of about 4.5 kb. This packaging capacity is a limiting factor with regard to bicistronic construction of TALEN pairs. The ability of AAV to integrate into the host genome, typically into the AAVS1 site in the human chromosome 19 (Ayuso 2016) may be a double-edged sword depending on therapeutic context. However, the beauty of AAV is its non-pathogenicity, negligible immunogenicity, and broad tissue tropism in human host. Taken together, these characteristics make AAV an ideal delivery candidate for gene therapy. In fact, AAV-mediated delivery of TALENs and megaTAL (fusion of TALE binding domain with a meganuclease cleavage domain) can edit the CCR5 gene in primary human T cells (Sather et al. 2015).
Non-viral delivery
Several methods for non-viral delivery of TALENs to edit genes are therapeutically relevant for treating HIV infection. For example, the Pseudomonas aeruginosa type III secretion system (injectosome) delivery of TALEN proteins into the nuclei of HeLa cells produces editing (Jia et al. 2014). Likewise, liposomes and nanoparticles have been used to deliver anti-oncogenic drugs and may also show promise for delivering TALEN proteins to nuclei of cells (Malam et al. 2009). Because HIV-infected cells can exist in anatomical reservoirs such as the brain or gastrointestinal tract. An all-hands-on-deck approach to exploring delivery systems that can safely cargo TALENs to T cells, macrophages, and dendritic cells, should be pursued.
Editing stem cell and transplantation
The biggest advantages of introducing a patients own stem cells as a therapy for HIV is that these cells can be edited ex vivo, selected based on a resistance marker, expanded, and then the cells can be used to deliver the edited genome; furthermore, they are expected to be well tolerated by the immune system. This allows broader options in using other delivery systems for in vitro editing and then screening for edited cells prior to transplantation. Since the stems cells are edited before being introduced in the systemic circulation, they overcome the uncertainty of delivery and editing under in vivo conditions. Furthermore, cryopreserved cells can be saved for treatment of viremia relapse. Gene editing in the human pluripotent CD34+ stem cells, followed by transplantation into humanized mice (HM) have proven successful for the CCR5 loci (Cornu et al. 2015). Harvested somatic cells can be reprogrammed to develop induced pluripotent stem cells (iPSCs). iPSCs with homozygous CCR5Δ32 were generated with TALENs or CRISPR/Cas-9 and Piggybag technology (Ye et al. 2014).
Animal Models for HIV gene editing
An increase in gene delivery and genome editing efficiency is one of the current aims in the humanized mice (HM) field, because success in these areas should translate to new therapies in humans. Research groups have been able to establish a delivery efficiency between 30-40% in cells in which the editing efficiency resulted in 10 to 25% depending on the analyzed tissue (Holt et al. 2010; Hauber et al. 2013).
The HM class of models offer many well-known advantages that make it a suitable option to test gene editing therapies in living organisms. Mice reproduce quickly, are relatively inexpensive for research, and are amenable to genetic manipulation, thus allowing the production of highly immunodeficient strains. The generation of HM is produced through the engraftment of human hematopoietic stem cells (HSCs) which go on to produce a wide variety of human white blood cells, of which T cells and monocytes/macrophages are the main targets of HIV-1 infection (Berges et al. 2006; Berges et al. 2008; Akkina et al. 2011; Berges and Rowan 2011; Sanchez and Berges 2013). HM infected with HIV-1 develop persistent infections and AIDS. Moreover, long-term HIV-1 infection has been shown in Rag2−/−γc−/− which was not possible in prior generations of HM models (Berges et al. 2010). Due to the generation of the hematopoietic lineage of the stem cells, AIDS disease development is remarkably similar to human pathology (Berges et al. 2006). Novel techniques are also arising with the aim of transducing stem cells prior to engraftment, thus allowing the HM to contain desirable genes in all progeny white blood cells (Holt et al. 2010; Hauber et al. 2013).
Despite the great advances accomplished with the HM, some limitations must be considered. Even though engraftment is a successful method for accomplishing human hematopoiesis in mice, each animal must be engrafted individually, and the engraftment process requires about two months to reach a mature human immune system. In addition, a source of human umbilical cord blood or fetal liver is required to obtain the primary human cells required for engrafting. Although HM are always made following the same protocol, human adaptive immune responses can be inconsistent. Stem cells extracted from umbilical cord blood come from multiple donors. Hence, scientists experiment with both homogeneous (single donor) and heterogeneous populations (multiple donors) of HM.
On the other hand, some researchers use non-human primates (NHPs) for their animal models. NHPs are more similar in size to humans and their genetics are more similar; however there are significant inconveniences that can disqualify this organism to be used in research: (i) Even though it is feasible to infect some types of NHPs with HIV-1, no AIDS-like pathogenesis has been documented, and therefore it is not possible to use NHPs to study a therapy intended for use in humans (McClure et al. 2000). (ii) NHP maintenance is considerably more expensive than mouse maintenance. (iii) NHPs require specialized facilities and expert veterinarians to ensure the welfare of the animals. Due to these facts many scientific institutions are unable to provide an appropriate environment.
Original HM were unable to develop a wide hematopoietic cell repertoire required to study HIV-1 pathogenesis, and in some models infection only lasted a few months (Mosier et al. 1991; Aldrovandi et al. 1993; Mosier 1996; Jamieson et al. 1996). Nevertheless, advances in this field over the last few decades produced a new generation of mice with different mutations and cell types engrafted that make them more suitable to perform these kinds of studies. Common immunosuppressive mutations are combinations of Rag1 or Rag2 mutations to prevent T and B lymphocyte development, paired with a mutation in the gene encoding the common gamma chain receptor that prevents natural killer cell and T cell development. Mutations causing the severe combined immunodeficiency (SCID) phenotype or in combination with the non-obese diabetic (NOD) phenotype have also been extremely useful (Berges and Rowan 2011). The resulting strains and the use of HSCs have proven to be the most effective to date for generating a primary human adaptive immune response showing high levels of CD4+ T-cells, monocytes and macrophages, and dendritic cells, which are the main HIV-1 targets (Shultz et al. 2007). Furthermore, HR serves as a model for many different types of pathogenesis because infections can last more than a year (Berges et al. 2010; Berges and Rowan 2011).
The ability of HIV-1 to develop resistance to drug treatment and to escape from immune surveillance has been widely studied. Studies with HM advanced our understanding of HIV-1 evolution by: 1) Mutations in the pol gene render HIV-1 resistant to ART (Choudhary et al. 2009); 2) In addition, mutations in the env gene can affect glycosylation patterns and promote escape from neutralizing antibodies (Ince et al. 2010); and 3) Resistance to anti-HIV-1 drugs can also result from sub-lethal A3-mediated G-to-A mutations (Sato et al. 2010); this results from the action of APOBEC3 proteins, specifically APOBEC3D, APOBEC3F, and APOBEC3G which induce G-to-A hypermutations in the HIV-1 genome. When modified, APOBEC3D and APOBEC3F preferred sites of actions are related to anti-HIV-1 drug resistance (Sato et al. 2014). Cellular double-stranded RNA adenosine deaminase, modifies the viral RNA sequence, so that the HIV-1 genome is not recognized by anti-sense RNA encoded by VRX494 (Mukherjee et al. 2010). Nevertheless, these modifications usually result in defective replication (Mukherjee et al. 2010).
Ethical issues concerning the use of the HM in the laboratory includes the justification for the use of the animal; it specially focuses on the number of mice studied and their welfare; including potential pain or distress that they may suffer during the procedures (Brown and Murray 2006). The use of the mice in the laboratory must always take place under directions and instructions from the Institutional Animal Care and Use Committee. Some researchers use fetal liver as a source of human HSCs, but it can be challenging to obtain human fetal tissues due to governmental regulations in some states or countries.
Table 1.
Characteristics of viral vectors targeting T cells, macrophages, dendritic cells, hematopoietic stem cells for HIV-1 gene therapy1.
| Gene therapy characteristics | Adenoviral vector | Lentiviral vector | Adeno-associated viral vector |
|---|---|---|---|
| Tissue tropism | proliferating and non-proliferating cells | proliferating and non-proliferating cells | proliferating and non-proliferating cells |
| Packaging capacity | 7-35 kb | 8 kb | 4.5 kb |
| Relative delivery efficiency | high | very high | high |
| Gene expression level | transient | stable | stable or transient |
| Integration site | 2N/A | random site | AAVS1 site in chromosome 19 |
| Immunogenicity | high | low | negligible |
| Pseudotyped with | Ad5/35 fiber | 3VSV- glycoprotein | 2N/A |
| Receptors | CD46 | 4LDL receptor | Heparin sulfate proteoglycan (serotype 2) |
Table adapted from (Schroers et al. 2004; Zhang et al. 2013; Ayuso 2016; Summerford and Samulski 1998; Finkelshtein et al. 2013).
Not applicable.
Vesicular stomatitis virus.
Low density lipoprotein.
Acknowledgements
Grants from the National Institutes of Health (R56 AI109156) and the Nevada Governors Office of Economic Development supported this work.
Footnotes
Conflict of Interest
Martin R. Schiller and Christy L. Strong have a patent pending for using TALENs to treat HIV, thus have a potential conflict of interest.
References
- Agosto LM, Yu JJ, Liszewski MK, et al. The CXCR4-Tropic Human Immunodeficiency Virus Envelope Promotes More-Efficient Gene Delivery to Resting CD4+ T Cells than the Vesicular Stomatitis Virus Glycoprotein G Envelope. J Virol. 2009;83:8153–8162. doi: 10.1128/JVI.00220-09. doi: 10.1128/JVI.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkina R, Berges BK, Palmer BE, et al. Humanized Rag1−/− γc−/− mice support multilineage hematopoiesis and are susceptible to HIV-1 infection via systemic and vaginal routes. PLoS ONE. 2011;6:e20169. doi: 10.1371/journal.pone.0020169. doi: 10.1371/journal.pone.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrovandi GM, Feuer G, Gao L, et al. The SCID-hu mouse as a model for HIV-1 infection. Nature. 1993;363:732–736. doi: 10.1038/363732a0. doi: 10.1038/363732a0. [DOI] [PubMed] [Google Scholar]
- Allers K, Schneider T. CCR5Δ32 mutation and HIV infection: basis for curative HIV therapy. Current Opinion in Virology. 2015;14:24–29. doi: 10.1016/j.coviro.2015.06.007. doi: 10.1016/j.coviro.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Ayuso E. Manufacturing of recombinant adeno-associated viral vectors: new technologies are welcome. Molecular Therapy — Methods & Clinical Development. 2016;3:15049. doi: 10.1038/mtm.2015.49. doi: 10.1038/mtm.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, et al. In vivo Genome Editing Using High Efficiency TALENs. Nature. 2012;491:114–118. doi: 10.1038/nature11537. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Jin DY, Chun RF, et al. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- Berges BK, Akkina SR, Folkvord JM, et al. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2−/− gammac −/− (RAG-hu) mice. Virology. 2008;373:342–351. doi: 10.1016/j.virol.2007.11.020. doi: 10.1016/j.virol.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Akkina SR, Remling L, Akkina R. Humanized Rag2(−/−)gammac(−/−) (RAG-hu) mice can sustain long-term chronic HIV-1 infection lasting more than a year. Virology. 2010;397:100–103. doi: 10.1016/j.virol.2009.10.034. doi: 10.1016/j.virol.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Rowan MR. The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment. Retrovirology. 2011;8:65. doi: 10.1186/1742-4690-8-65. doi: 10.1186/1742-4690-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Wheat WH, Palmer BE, et al. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−gamma c−/− (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Brown MJ, Murray KA. Phenotyping of genetically engineered mice: humane, ethical, environmental, and husbandry issues. ILAR J. 2006;47:118–123. doi: 10.1093/ilar.47.2.118. [DOI] [PubMed] [Google Scholar]
- Budhagatapalli N, Rutten T, Gurushidze M, et al. Targeted Modification of Gene Function Exploiting Homology-Directed Repair of TALEN-Mediated Double-Strand Breaks in Barley. G3 (Bethesda) 2015;5:1857–1863. doi: 10.1534/g3.115.018762. doi: 10.1534/g3.115.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultmann S, Morbitzer R, Schmidt CS, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermáková K, Tesina P, Demeulemeester J, et al. Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res. 2014;74:5139–5151. doi: 10.1158/0008-5472.CAN-13-3602. doi: 10.1158/0008-5472.CAN-13-3602. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Starker CG, Voytas DF. Efficient design and assembly of custom TALENs using the Golden Gate platform. Methods Mol Biol. 2015;1239:133–159. doi: 10.1007/978-1-4939-1862-1_7. doi: 10.1007/978-1-4939-1862-1_7. [DOI] [PubMed] [Google Scholar]
- Chen S, Oikonomou G, Chiu CN, et al. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio ALB, Rahman S, et al. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary SK, Rezk NL, Ince WL, et al. Suppression of human immunodeficiency virus type 1 (HIV- 1) viremia with reverse transcriptase and integrase inhibitors, CD4+ T-cell recovery, and viral rebound upon interruption of therapy in a new model for HIV treatment in the humanized Rag2−/−{gamma}c−/− mouse. J Virol. 2009;83:8254–8258. doi: 10.1128/JVI.00580-09. doi: 10.1128/JVI.00580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ F, Shaw S, Demeulemeester J, et al. Small-Molecule Inhibitors of the LEDGF/p75 Binding Site of Integrase Block HIV Replication and Modulate Integrase Multimerization. Antimicrob Agents Chemother. 2012;56:4365–4374. doi: 10.1128/AAC.00717-12. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, et al. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T-W, Justement JS, Murray D, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24:2803–2808. doi: 10.1097/QAD.0b013e328340a239. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI, Cathomen T. Quantification of zinc finger nuclease-associated toxicity. Methods Mol Biol. 2010;649:237–245. doi: 10.1007/978-1-60761-753-2_14. doi: 10.1007/978-1-60761-753-2_14. [DOI] [PubMed] [Google Scholar]
- Cornu TI, Mussolino C, Bloom K, Cathomen T. Editing CCR5: a novel approach to HIV gene therapy. Adv Exp Med Biol. 2015;848:117–130. doi: 10.1007/978-1-4939-2432-5_6. doi: 10.1007/978-1-4939-2432-5_6. [DOI] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J, Zhang X-Y, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, et al. Genetic Restriction of HIV-1 Infection and Progression to AIDS by a Deletion Allele of the CKR5 Structural Gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee Y-K, Schaefer EAK, et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.11.011. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–122. doi: 10.1093/nar/gks608. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Drake MJ, Bates P. Application of gene-editing technologies to HIV-1. Curr Opin HIV AIDS. 2015;10:123–127. doi: 10.1097/COH.0000000000000139. doi: 10.1097/COH.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H, Kanemura Y, Misawa N, et al. A High Excision Potential of TALENs for Integrated DNA of HIV-Based Lentiviral Vector. PLoS One. 2015 doi: 10.1371/journal.pone.0120047. doi: 10.1371/journal.pone.0120047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Morrison JH, Saenz DT, et al. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014;88:9704–9717. doi: 10.1128/JVI.01397-14. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine EJ, Cradick TJ, Zhao CL, et al. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42:e42. doi: 10.1093/nar/gkt1326. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelshtein D, Werman A, Novick D, et al. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Doitsh G, Yang Z, et al. Efficient delivery of lentiviral vectors into resting human CD4 T cells. Gene Ther. 2014;21:444–449. doi: 10.1038/gt.2014.5. doi: 10.1038/gt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard A, Ségéral E, Naughtin M, et al. The Integrase Cofactor LEDGF/p75 Associates with Iws1 and Spt6 for Postintegration Silencing of HIV-1 Gene Expression in Latently Infected Cells. Cell Host & Microbe. 2015;17:107–117. doi: 10.1016/j.chom.2014.12.002. doi: 10.1016/j.chom.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Grau J, Wolf A, Reschke M, et al. Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol. 2013;9:e1002962. doi: 10.1371/journal.pcbi.1002962. doi: 10.1371/journal.pcbi.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Meth. 2014;11:429–435. doi: 10.1038/nmeth.2845. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W-G. Genome editing-based HIV therapies. Trends Biotechnol. 2015;33:172–179. doi: 10.1016/j.tibtech.2014.12.006. doi: 10.1016/j.tibtech.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Händel E-M, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- Hanoun N, Gayral M, Pointreau A, et al. Initial Characterization of Integrase-Defective Lentiviral Vectors for Pancreatic Cancer Gene Therapy. Human Gene Therapy. 2016;27:184–192. doi: 10.1089/hum.2015.151. doi: 10.1089/hum.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Li Q. [Application progress of CRISPR/Cas9 genome editing technology in the treatment of HIV-1 infection]. Yi Chuan. 2016;38:9–16. doi: 10.16288/j.yczz.15-284. doi: 10.16288/j.yczz.15-284. [DOI] [PubMed] [Google Scholar]
- Hauber I, Hofmann-Sieber H, Chemnitz J, et al. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog. 2013;9:e1003587. doi: 10.1371/journal.ppat.1003587. doi: 10.1371/journal.ppat.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Walther N, et al. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt789. doi: 10.1093/nar/gkt789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Cathomen T, Gonçalves MAFV. Construction and characterization of adenoviral vectors for the delivery of TALENs into human cells. Methods. 2014;69:179–187. doi: 10.1016/j.ymeth.2014.02.017. doi: 10.1016/j.ymeth.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Chen S, Wang S, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep. 2015;5:15577. doi: 10.1038/srep15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YX, Paxton WA, Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- Hütter G, Bodor J, Ledger S, et al. CCR5 Targeted Cell Therapy for HIV and Prevention of Viral Escape. Viruses. 2015;7:4186–4203. doi: 10.3390/v7082816. doi: 10.3390/v7082816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince WL, Zhang L, Jiang Q, et al. Evolution of the HIV-1 env gene in the Rag2−/− gammaC−/− humanized mouse model. J Virol. 2010;84:2740–2752. doi: 10.1128/JVI.02180-09. doi: 10.1128/JVI.02180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Gee P, Hotta A. Minimizing off-Target Mutagenesis Risks Caused by Programmable Nucleases. Int J Mol Sci. 2015;16:24751–24771. doi: 10.3390/ijms161024751. doi: 10.3390/ijms161024751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson BD, Aldrovandi GM, Zack JA. The SCID-hu mouse: an in-vivo model for HIV-1 pathogenesis and stem cell gene therapy for AIDS. Semin Immunol. 1996;8:215–221. doi: 10.1006/smim.1996.0027. doi: 10.1006/smim.1996.0027. [DOI] [PubMed] [Google Scholar]
- Jia J, Jin Y, Bian T, et al. Bacterial Delivery of TALEN Proteins for Human Genome Editing. PLoS One. 2014 doi: 10.1371/journal.pone.0091547. doi: 10.1371/journal.pone.0091547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, King MS, Makitalo B, et al. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proceedings of the National Academy of Sciences. 2011;108:11199–11204. doi: 10.1073/pnas.1107729108. doi: 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kordelas L, Verheyen J, Esser S. Shift of HIV Tropism in Stem-Cell Transplantation with CCR5 Delta32 Mutation. New England Journal of Medicine. 2014;371:880–882. doi: 10.1056/NEJMc1405805. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- Le T, Farrar J, Shikuma C. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2011;25:871–872. doi: 10.1097/QAD.0b013e32834490b1. author reply 872–873. doi: 10.1097/QAD.0b013e32834490b1. [DOI] [PubMed] [Google Scholar]
- Levy JA. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-J, Kim J, Li S, et al. Long-Term Inhibition of HIV-1 Infection in Primary Hematopoietic Cells by Lentiviral Vector Delivery of a Triple Combination of Anti-HIV shRNA, Anti-CCR5 Ribozyme, and a Nucleolar-Localizing TAR Decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Lin Y, Fine EJ, Zheng Z, et al. SAPTA: a new design tool for improving TALE nuclease activity. Nucleic Acids Res. 2014;42:e47. doi: 10.1093/nar/gkt1363. doi: 10.1093/nar/gkt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang W, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ma X, Liu B, et al. HIV-1 functional cure: will the dream come true? BMC Medicine. 2015 doi: 10.1186/s12916-015-0517-y. doi: 10.1186/s12916-015-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Patterson JT, et al. Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE. 2014;9:e85755. doi: 10.1371/journal.pone.0085755. doi: 10.1371/journal.pone.0085755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopalco L. CCR5: From Natural Resistance to a New Anti-HIV Strategy. Viruses. 2010;2:574–600. doi: 10.3390/v2020574. doi: 10.3390/v2020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AC, Lee HB, Clark KJ, Ekker SC. High Efficiency In Vivo Genome Engineering with a Simplified 15-RVD GoldyTALEN Design. PLoS One. 2013 doi: 10.1371/journal.pone.0065259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz M, Li L, Shamimuzzaman M, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AN-S, Bradley P, Cernadas RA, et al. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malam Y, Loizidou M, Seifalian AM. Liposomes and nanoparticles: nanosized vehicles for drug delivery in cancer. Trends in Pharmacological Sciences. 2009;30:592–599. doi: 10.1016/j.tips.2009.08.004. doi: 10.1016/j.tips.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Yi G, Dang Y, Shankar P. Newer Gene Editing Technologies toward HIV Gene Therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure J, Schmidt AM, Rey-Cuille MA, et al. Derivation and characterization of a highly pathogenic isolate of human immunodeficiency virus type 2 that causes rapid CD4+ cell depletion in Macaca nemestrina. J Med Primatol. 2000;29:114–126. doi: 10.1034/j.1600-0684.2000.290304.x. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotech. 2011;29:143–148. doi: 10.1038/nbt.1755. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Miller JC, Zhang L, Xia DF, et al. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods. 2015;12:465–471. doi: 10.1038/nmeth.3330. doi: 10.1038/nmeth.3330. [DOI] [PubMed] [Google Scholar]
- Mock U, Machowicz R, Hauber I, et al. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43:5560–5571. doi: 10.1093/nar/gkv469. doi: 10.1093/nar/gkv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucl Acids Res gku410. 2014 doi: 10.1093/nar/gku410. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mosier DE. Human immunodeficiency virus infection of human cells transplanted to severe combined immunodeficient mice. Adv Immunol. 1996;63:79–125. doi: 10.1016/s0065-2776(08)60855-x. [DOI] [PubMed] [Google Scholar]
- Mosier DE, Gulizia RJ, Baird SM, et al. Human immunodeficiency virus infection of human-PBLSCID mice. Science. 1991;251:791–794. doi: 10.1126/science.1990441. [DOI] [PubMed] [Google Scholar]
- Mukherjee R, Plesa G, Sherrill-Mix S, et al. HIV sequence variation associated with env antisense adoptive T-cell therapy in the hNSG mouse model. Mol Ther. 2010;18:803–811. doi: 10.1038/mt.2009.316. doi: 10.1038/mt.2009.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucl Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Comparison of TALEN scaffolds in Xenopus tropicalis. Biol Open. 2013;2:1364–1370. doi: 10.1242/bio.20136676. doi: 10.1242/bio.20136676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousterout DG, Perez-Pinera P, Thakore PI, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon S. Gene-editing method tackles HIV in first clinical test. Nature. 2014 doi: 10.1038/nature.2014.143. [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru R, Yao Y, Yu S, et al. Targeted genome engineering in human induced pluripotent stem cells by penetrating TALENs. Cell Regen (Lond) 2013 doi: 10.1186/2045-9769-2-5. doi: 10.1186/2045-9769-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Ochiai H, Kaneko T, et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Scientific Reports. 2013 doi: 10.1038/srep03379. doi: 10.1038/srep03379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Yamamoto T. Engineering Customized TALENs Using the Platinum Gate TALEN Kit. Methods Mol Biol. 2016;1338:61–70. doi: 10.1007/978-1-4939-2932-0_6. doi: 10.1007/978-1-4939-2932-0_6. [DOI] [PubMed] [Google Scholar]
- Sanchez FM, Berges BK. Characterization of HIV-1 infection in the humanized Rag2−/−γc−/− mouse model. Methods Mol Biol. 2013;1031:215–222. doi: 10.1007/978-1-62703-481-4_24. doi: 10.1007/978-1-62703-481-4_24. [DOI] [PubMed] [Google Scholar]
- Sather BD, Ibarra GSR, Sommer K, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Science Translational Medicine. 2015;7:307ra156–307ra156. doi: 10.1126/scitranslmed.aac5530. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Izumi T, Misawa N, et al. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J Virol. 2010;84:9546–9556. doi: 10.1128/JVI.00823-10. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Takeuchi JS, Misawa N, et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer JT, Aubert M, Weber ND, et al. Targeted DNA Mutagenesis for the Cure of Chronic Viral Infections. J Virol. 2012;86:8920–8936. doi: 10.1128/JVI.00052-12. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Hornung V. Ligation-independent cloning (LIC) assembly of TALEN genes. Methods Mol Biol. 2015;1239:161–169. doi: 10.1007/978-1-4939-1862-1_8. doi: 10.1007/978-1-4939-1862-1_8. [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Kaiser V, et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroers R, Hildebrandt Y, Hasenkamp J, et al. Gene transfer into human T lymphocytes and natural killer cells by Ad5/F35 chimeric adenoviral vectors. Exp Hematol. 2004;32:536–546. doi: 10.1016/j.exphem.2004.03.010. doi: 10.1016/j.exphem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- Shun M-C, Raghavendra NK, Vandegraaff N, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong CL, Guerra HP, Mathew KR, et al. Damaging the Integrated HIV Proviral DNA with TALENs. PLoS One. 2015 doi: 10.1371/journal.pone.0125652. doi: 10.1371/journal.pone.0125652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol BioSyst. 2012a;8:1255–1263. doi: 10.1039/c2mb05461b. doi: 10.1039/C2MB05461B. [DOI] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol BioSyst. 2012b;8:1255–1263. doi: 10.1039/c2mb05461b. doi: 10.1039/C2MB05461B. [DOI] [PubMed] [Google Scholar]
- Sutherland HG, Newton K, Brownstein DG, et al. Disruption of Ledgf/Psip1 Results in Perinatal Mortality and Homeotic Skeletal Transformations. Mol Cell Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Remy S, Ménoret S, et al. Genome Editing in Rats Using TALE Nucleases. Methods Mol Biol. 2016;1338:245–259. doi: 10.1007/978-1-4939-2932-0_18. doi: 10.1007/978-1-4939-2932-0_18. [DOI] [PubMed] [Google Scholar]
- Valton J, Cabaniols J-P, Galetto R, et al. Efficient strategies for TALEN-mediated genome editing in mammalian cell lines. Methods. 2014;69:151–170. doi: 10.1016/j.ymeth.2014.06.013. doi: 10.1016/j.ymeth.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Valton J, Dupuy A, Daboussi F, et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pan Q, Gendron P, et al. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Reports. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Ye L, Wang J, Beyer AI, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl SA, Boritz E, Busch M, et al. Challenges in Detecting HIV Persistence during Potentially Curative Interventions: A Study of the Berlin Patient. PLoS Pathog. 2013;9:e1003347. doi: 10.1371/journal.ppat.1003347. doi: 10.1371/journal.ppat.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang S, Huang X, et al. Rapid Assembly of Customized TALENs into Multiple Delivery Systems. PLoS ONE. 2013;8:e80281. doi: 10.1371/journal.pone.0080281. doi: 10.1371/journal.pone.0080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Yu F, Wu Y, et al. Broadening the versatility of lentiviral vectors as a tool in nucleic acid research via genetic code expansion. Nucl Acids Res. 2015;43:e73–e73. doi: 10.1093/nar/gkv202. doi: 10.1093/nar/gkv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman PA, BucklerWhite A, Alkhatib G, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]