Abstract
Purpose
Human papillomavirus (HPV)-associated oropharyngeal squamous cell carcinoma (OPSCC) carries a distinct clinical behavior. c-Met oncogene is an important driver for tumor progression and its relationship with HPV in OPSCC was explored in the current study.
Experimental Design
Knockdown of HPV oncogene E6 or p53 alone and in combination was performed to examine their effects on c-Met expression by western blot and qRT-PCR. The effects of c-Met inhibition on cell proliferation, migration, and colony formation were examined in HPV-positive head and neck squamous cell carcinoma (HNSCC) cells. Retrospectively collected OPSCC patient specimens (N = 78) were stained for c-Met by immunohistochemistry and the staining levels were correlated with HPV status and patient outcomes.
Results
E6 knockdown decreased c-Met protein and mRNA expression in HPV-positive HNSCC cells, which was partially abolished by the elimination of p53. Reducing c-Met decreased cell proliferation, migration, and colony formation in HPV-positive HNSCC cells. In OPSCC patient samples, high c-Met expression was associated with HPV positive status (OR = 4.11, 95%CI: 1.16–14.55, P = 0.028) and tumor stage (OR = 0.27, 95%CI: 0.08–0.93, P = 0.039) by multivariable analysis. In T3/T4 stage patients, high c-Met expression was associated with HPV positivity and low p53 levels, supporting an axis of E6-p53-c-Met regulation. Furthermore, high c-Met expression was marginally associated with poor disease-free survival in HPV-positive patients.
Conclusions
Our results suggest that c-Met may serve as a novel target for treating HPV-associated OPSCC. The data also demonstrates that HPV E6 upregulates c-Met expression partially through p53 downregulation.
Keywords: human papillomavirus, c-Met, p53, head and neck cancer
Introduction
Head and neck squamous cell carcinoma cancer (HNSCC) is the sixth most common malignancy worldwide (1). HNSCC is currently considered as two major subgroups: human papillomavirus (HPV)-related and HPV non-related. HPV-related HNSCC occurs most frequently in the oropharynx, and has demonstrated better responses to radio/chemotherapy as well as significantly improved outcomes (2–4). However, due to the heterogeneous nature of this disease, some populations are more aggressive than others. Although the incidence of HNSCC shows a decline overall, the incidence of HPV-associated disease is experiencing a steady uphill trend (5). Currently the same standard therapy applies to both HPV-related and non-related HNSCCs; although different targeted therapies have been proposed for patients with HPV-unrelated HNSCC, there remains a paucity of identified targetable pathways for HPV-related HNSCC and consequently targeted approaches are limited at present in this disease.
HPV oncoproteins, E6 and E7, contribute most to the transforming activity of high risk HPVs, maintenance of the transformed phenotype, and proliferative capability (6, 7). The tumor suppressors p53 and RB are two major targets of E6 and E7, respectively, and are inactivated by these interactions (7). Inhibition of p53 and BAX/BAK by E6 impairs apoptosis and induces chromosomal instability. Inactivation of RB and activation of cyclins A and E by E7 facilitates cell proliferation and transformation. When co-expressed, E6 and E7 can synergistically immortalize and transform various human primary cells (8). Recently we have shown that HPV E6 oncoprotein can upregulate EGFR to increase nuclear localization of β-catenin, which plays an important role in cell proliferation, migration, and invasion of HPV-associated HNSCC (9). On the other hand, little is known about whether HPV oncoproteins regulate other tyrosine kinase receptors that are frequently overexpressed in HNSCC.
c-Met is a tyrosine kinase receptor that is overexpressed in many solid tumors, including HNSCC (10). Upon binding to its sole ligand, hepatocyte growth factor (HGF), c-Met activates multiple downstream pathways, including PI3K/AKT and RAS/MAPK pathways, to promote proliferation, migration, invasion and metastasis (10). Furthermore, c-Met expression has been associated with worse outcomes in patients with locally advanced HNSCC (11) and targeting c-Met using monoclonal antibodies and specific kinase inhibitors has been proposed in clinical trials (12–14). c-Met expression and its correlation with clinical-pathological features in HNSCC have been summarized in a review article by Nisa et al (14). In spite of this, it remains unclear whether c-Met expression is associated with HPV status, especially in OPSCC in which HPV infection is prevalent.
The negative regulation of c-Met by the tumor suppressor p53 has been reported in prostate cancer (15) and primary ovarian surface epithelium cells (16). Dual mechanisms have been proposed regarding the negative regulation, including transactivation, of miR-34 that targets the MET gene and inhibition of SP1 binding to MET promoter (16). While genomic analyses of HNSCC did not reveal high mutation rate in the MET gene (17–19), c-Met overexpression (84%) and gene copy number increase (13%) have been reported (20), indicating that c-Met is a potential target in HNSCC. However, whether c-Met plays a role in HPV-associated HNSCC remains largely unknown. In the present study, we sought to explore whether c-Met expression is positively correlated with HPV status and whether its expression is correlated with worse clinical outcome in HPV-positive patients.
Materials and Methods
Cell culture
The HNSCC cell lines UD-SCC2, UM-SCC47, 93-VU-147T, JHU022, PCI-15A and UM-22B were kindly provided by Dr. Robert Ferris and the SCC090 cell line by Dr. Susanne Gollin from the University of Pittsburgh (Pittsburgh, PA). HNSCC cell line SqCCY1 was a gift of Dr. Shi-Yong Sun at Emory University (Atlanta, GA). HNSCC cell line MDA686TA (Tu686) was provided by Dr. Gary L. Clayman from University of Texas M.D. Anderson Cancer Center (Houston, TX). The cervical cancer cell line CaSki was purchased from the American Type Culture Collection. CaSki, UD-SCC2, UM-SCC47, SCC90 cells are positive for HPV with wild-type p53 (21) while 93-VU-147T cells harbor p53 mutation as confirmed by our sequencing analysis (data not shown). Most cell lines were maintained in DMEM/F12 (1:1), SCC90 in minimum essential media and CaSki and JHU022 in RPMI-1640 medium, all supplemented with 10% FBS at 37°C, 5% CO2. All cells were routinely screened for mycoplasma contamination by the MycoAlert Mycoplasma Detection Kit (Lonza Ltd., Allendale, NJ). The authenticity of UD-SCC2, UM-SCC47, SCC90, 93-VU-147T, JHU022, SqCCY1, and Tu686 cell lines was verified through the genomic short tandem repeat (STR) profile by the Research Animal Diagnostic Laboratory, University of Missouri (Columbia, MO) in September 2009, and by the Emory University Integrated Genomics Core (EIGC) in October 2013. The authenticity of PCI-15A and UM-22B was not verified by the authors, but reported by Zhao and colleagues in 2011, using the same STR profile (22).
OPSCC specimen collection
Retrospectively collected specimens from 78 primary OPSCC patients at Emory University Hospital from 1996–2007 were used for this study. The research protocol was approved by the Institutional Review Board at Emory University. Information on patient characteristics was retrieved from the medical record and the treating physician in compliance with the Health Insurance Portability and Accountability Act (HIPAA).
HPV status determination
Formalin-fixed and paraffin-embedded specimens were tested for HPV DNA by the DNA in situ hybridization (ISH) method in the Emory Hospital Pathology Department. ISH was performed using the GenPoint Catalyzed Signal Amplification System (DAKO, Carpinteria, CA) according to the manufacturer’s protocols, which can detect as few as 1–2 copies of HPV DNA, as described previously (23, 24). Two previously defined HPV-positive OPSCC tissues were adopted as positive controls.
Western blot analysis
Cell lysates were collected and quantified as total protein content. Twenty to thirty micrograms of total protein for each sample were separated by 10% SDS-PAGE and transferred to Westran S membrane (Whatman Inc.), followed by incubation with primary and secondary antibodies. Primary antibodies were mouse anti-p53 and anti-E7 from Santa Cruz Biotech; rabbit anti-c-Met, rabbit anti-EGFR, and mouse anti-HER3, rabbit anti-Akt, rabbit anti-phospho Akt, rabbit anti-Erk, and rabbit anti-phospho Erk from Cell Signaling; and mouse anti-β-actin from Sigma Aldrich. Secondary antibodies were purchased from Santa Cruz Biotech.
Small interfering RNA (siRNA) transfection
Cells were seeded into 6-cm or 6-well plates and reached 30–50% confluency before transfection. siRNAs for HPV16 E6, p53, and c-Met (Santa Cruz Biotechnology, Santa Cruz, CA) were complexed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and applied to each plate, respectively. The transfection medium was removed and replaced with complete medium after 6 hours.
Quantitative reverse transcription-PCR (qPCR)
Total cellular RNA was collected by TRIzol extraction (Invitrogen) and 2 µg RNA was used for reverse-transcription using SuperScriptIII reverse transcriptase. Two microliters of prepared cDNA sample was used for qPCR reaction using SYBR Green according to the manufacturer’s instructions (Applied Biosystems). The primers used in the study are listed below:
-
HPV E6:
Forward: 5'-TTGCTTTTCGGGATTTATGC-3',
Reverse: 5'-CAGGACACAGTGGCTTTTGA-3';
-
c-Met:
Forward: 5'-CAGGCAGTGCAGCATGTAGT-3',
Reverse: 5'-GATGATTCCCTCGGTCAGAA-3';
-
p53:
Forward: 5'-GCGCACAGAGGAAGAGAATC-3',
Reverse: 5'-CAAGGCCTCATTCAGCTCTC-3';
-
GAPDH:
Forward: 5'-TGCACCACCAACTGCTTA-3',
Reverse: 5'-GGATGCAGGGATGATGTTC-3'.
The mRNA levels of E6 and c-Met were normalized to GAPDH levels within the same sample. Each analysis was conducted in triplicate.
Sulforhodamine B (SRB) colorimetric assay
The inhibition of cell proliferation by c-Met siRNA or c-Met inhibitor SU11274 was analyzed by SRB assay as previously described (25). Briefly, 2×105 cells were seeded in 6-well plates and incubated overnight. Cells were then treated with scrambled siRNA or c-Met siRNA for 24 hour and then 2×103 cells were replated in 96-well plates and allowed to grow for 1, 2, or 3 days before harvesting for colorimetric measurements. For inhibitor treatment, 2×103 cells were plated in 96-well plates and allowed to grow overnight, then cells were exposed to different concentrations of SU11274 (0, 0.05, 0.25, 1.25, 6.25 and 31.25 µM) and allowed to grow for 1, 2, and 3 days, respectively, before harvesting for colorimetric measurements. The dose selection was based on previous studies (26, 27). All the experiments were performed in triplicate.
Wound healing assay
Wound healing assay was conducted using CytoSelect™ 24-Well Wound Healing Assay Kit (Cell BioLabs, Inc, San Diego, CA) following the manufacturer’s instructions. Briefly, 2 × 105 cells (treated with scrambled or target siRNA for 24 hours) were digested with trypsin and replated on each side of the insert in a 24-well plate. Cell were allowed to form a monolayer and migrate for approximately 48 hours before being fixed and stained with 0.2% crystal violet containing buffered formalin. Cells were then washed, air dried and photos were taken using a dissection microscope for migration measurement.
Colony formation assay
Cells were seeded into 6-well culture plates at a concentration of 250 per well. After 24 hr incubation, cells were treated with fresh medium with 0, 0.5, 1, 2, 4, and 8 µM SU11274 in DMSO (DMSO < 0.03%, v:v) for 7–12 days to allow colony formation, as previously described (25). Medium was refreshed every 3 days containing the corresponding concentrations of SU11274. The colonies were then briefly washed with PBS and stained with 0.2% crystal violet with buffered formalin (Sigma). Colony numbers were manually counted using ImageJ software. A colony is defined as ≥ 50 cells.
Immunohistochemistry (IHC) staining
Formalin-fixed, paraffin-embedded tissue sections were used for IHC staining according to the ABC method as described previously (28). In brief, the sections were incubated with a primary antibody of anti-c-Met (1:100 dilution; Cell Signaling) or anti-p53 (1:100 dilution; DAKO), followed by secondary antibody and diaminobenzidine (DAB, Vector Laboratories, Burlingame, CA) staining. Nuclei were counterstained with Hematoxylin OS (Vector Laboratories). Immunoglobulin G was used as a negative control. The cytoplasmic expression of c-Met was scored as a weighted index (WI). WI = Intensity × % of positive staining. The intensity of c-Met staining was scored as negative (0), weak (1+), intermediate (2+), and strong (3+), respectively. Two surgical pathologists (K.M. and Z.H.) evaluated the tumor sections independently.
Statistical Analysis
The univariate association of c-Met expression with covariates was examined with analysis of variance or Wilcoxon rank-sum test for numerical covariates and chi-square test or Fisher’s exact test for categorical covariates, where appropriate. Multivariable analysis of c-Met expression was carried out by entering all covariates into a logistic regression model and using a backward variable selection method with an alpha level of removal of 0.2. Survival functions were estimated by the Kaplan-Meier method and a log-rank test was used to assess the difference in disease-free survival (DFS) and overall survival (OS) between patients with high (≥ median) and low (< median) c-Met and p53 expressions (29). A Cox proportional hazards model (30) was employed to examine the effect of c-Met and p53 expressions as well as covariates on DFS or OS. Multivariable survival analysis was carried out by entering all covariates into a Cox proportional hazards model and using a backward variable selection method with an alpha level of removal of 0.2. c-Met expression was forced in the model. The proportional hazards assumption was evaluated with Schoenfeld residuals (31) and a Kolmogorov-type supremum test. Analyses were performed using SAS 9.3 (SAS Institute, Inc., Cary, North Carolina) with two-sided tests and a significant level of 0.05.
Results
c-Met protein expression is regulated by E6 in HPV-positive HNSCC cell lines
We initially screened nine HNSCC cell lines and one cervical cancer cell line as the HPV-positive control (CaSki). The results showed that there was no clear association between c-Met status and HPV-positivity in these cell lines (Fig. S1).
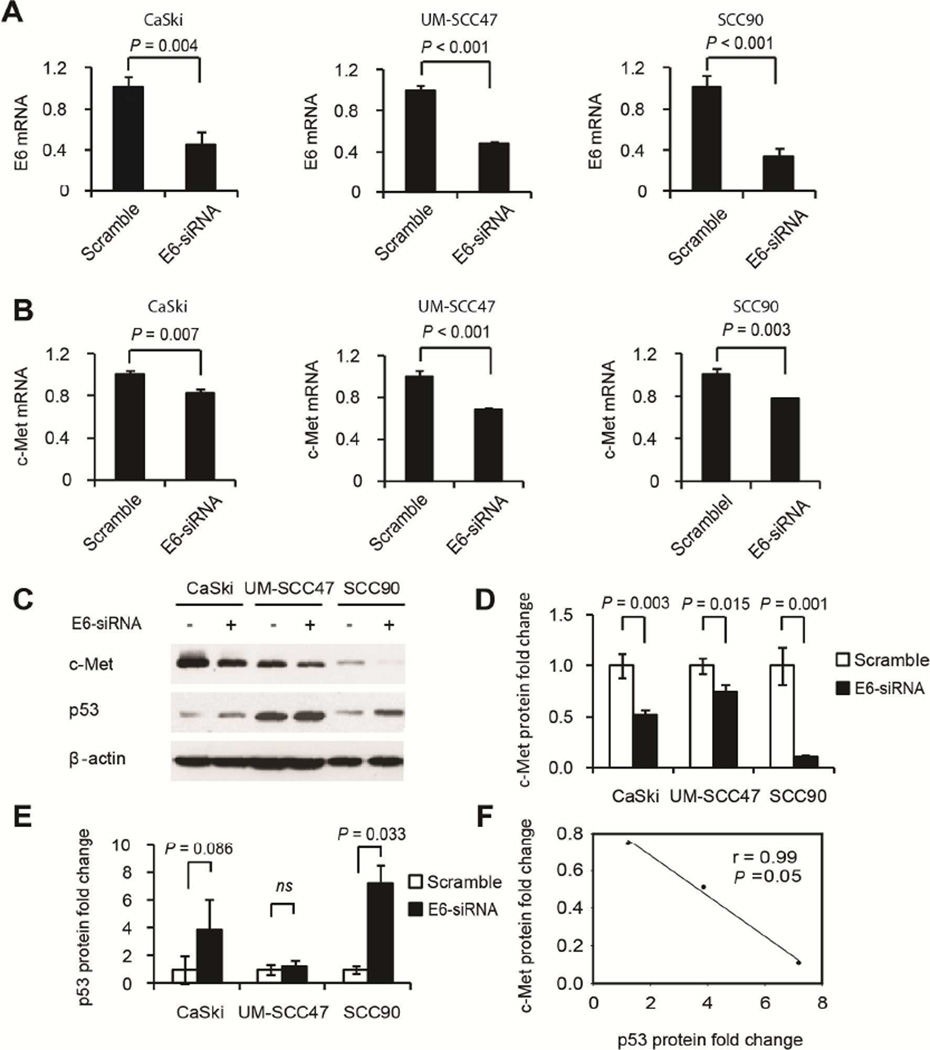
To test whether there is transcriptional regulation of c-Met by E6, we conducted qPCR analysis of c-Met mRNA change upon HPV E6 knockdown by a pool of 3–5 siRNAs specifically against E6 in CaSki, UM-SCC47, and SCC90 cells. The results showed that E6 knockdown significantly decreased c-Met mRNA levels in the three HPV-positive cancer cell lines, indicating a transcriptional regulation of c-Met by E6 (Fig. 1A and 1B). Furthermore, knockdown of E6 by siRNA significantly decreased c-Met protein levels in all of the HPV-positive cancer cell lines tested (Fig. 1C) without affecting other receptor tyrosine kinases such as EGFR or HER3 (Fig. S2). Quantification of c-Met and p53 protein level changes is shown in Fig. 1D and 1E, respectively.
Figure 1.
c-Met expression was decreased upon HPV E6 knockdown and correlated with upregulation of p53 in several HPV-positive cancer cell lines. E6 knockdown efficiency was shown in (A), which correlated with c-Met mRNA (B) and protein (C) inhibition in CaSki, UM-SCC47, and SCC90 cells. GAPDH was used as the internal control for mRNA expression. Quantification of the changes in c-Met and p53 expression in C is shown in D and E. Furthermore, a linear correlation existed between changes in protein levels of c-Met and p53 (r = −0.99, P = 0.05, F). ns, no significance. Error bars represent mean ± SD. Experiments were repeated three times.
Meanwhile, we also confirmed that E6 knockdown upregulated p53 protein levels, which correlated with decreased c-Met protein levels in these HPV-positive CaSki, UM-SCC47 and SCC90 cell lines. Statistical analysis showed that there is a significant negative correlation between changes in c-Met and p53 expression (r = −0.99, P = 0.05, Fig. 1F).
p53 knockdown blocks the upregulation of c-Met by E6 in HNSCC cells
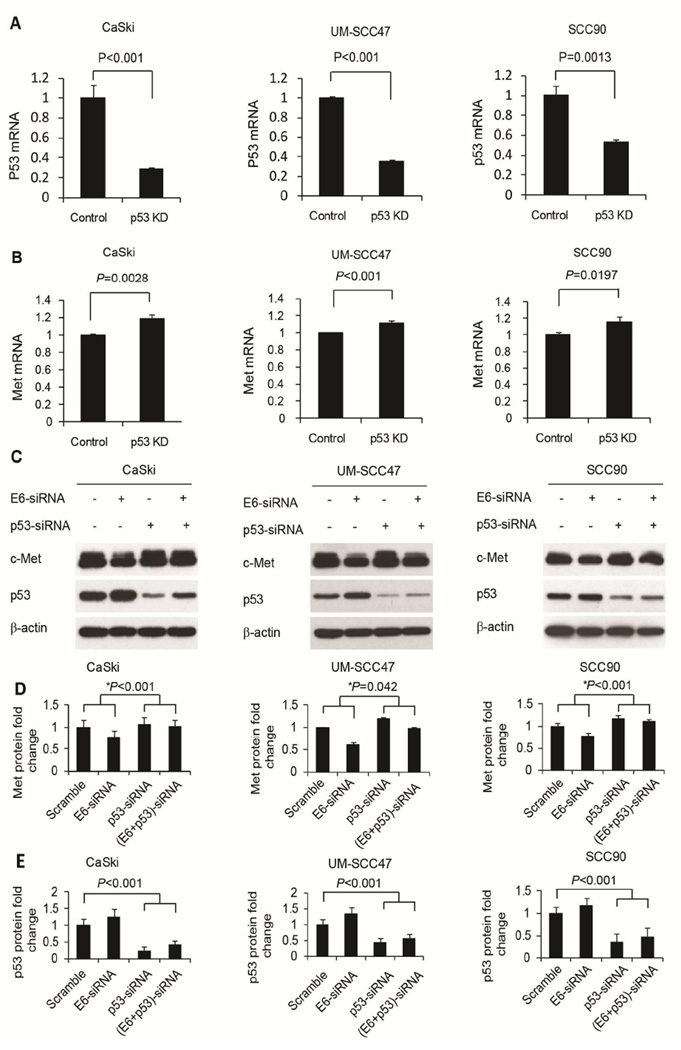
Based on recent evidence showing that p53 inhibits c-Met expression (15, 16), we tested whether the regulation of c-Met by E6 is mediated by p53. First, we found that p53 siRNA knockdown increased the mRNA level of c-Met in CaSki cells and two HNSCC cell lines (Fig. 2A and 2B). Consistent with these mRNA changes, upregulation of c-Met protein was also found as a result of p53 siRNA knockdown (Fig. 2C).
Figure 2.
E6 upregulated c-Met through p53. p53 knockdown increased c-Met mRNA expression in several HPV-positive cancer cell lines. p53 knockdown efficiency in CaSki, UM-SCC47, and SCC90 cells is shown in A. Corresponding increases in c-Met mRNA are shown in B. Error bars represent mean ± SD. Experiments were repeated three times. E6 knockdown also decreased c-Met protein levels and this regulation was mitigated when p53 was knocked down in CaSki, UM-SCC47, and SCC90 cells (C). Quantifications of c-Met and p53 levels are shown in D and E, respectively. * indicates comparison of c-Met changes due to E6 knockdown in the absence or presence of p53 knockdown. Error bars represent mean ± SD from three independent experiments.
To test the hypothesis that E6 upregulation of c-Met is mediated by p53, we further knocked down both p53 and E6 by siRNA to assess whether the regulation of c-Met by E6 can be alleviated by p53 knockdown. The results showed that while E6 knockdown alone decreased c-Met expression, simultaneous knockdown of p53 reduced this inhibitory effect in CaSki, UM-SCC47, and SCC90 cells. Quantification of protein levels of c-Met and p53 is shown in Fig. 2D and 2E, respectively.
c-Met inhibition decreases cell proliferation, migration, and colony formation of HPV-positive HNSCC cells
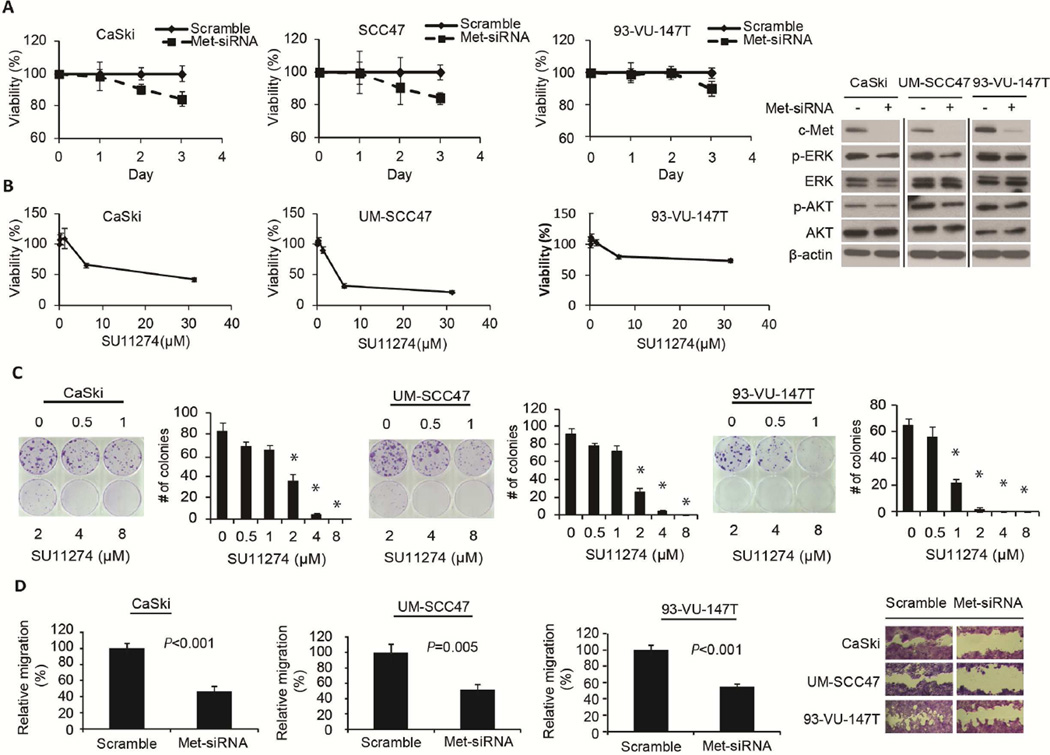
To test whether c-Met is important in the proliferation of HPV-positive cancer cells, we knocked down c-Met by siRNA in CaSki, UM-SCC47, and 93-VU-147T cells and examined cell proliferation using SRB assay. We found that cell proliferation was inhibited by c-Met knockdown in a time-dependent manner in CaSki, UM-SCC47, and 93-VU-147T cells and the downstream signaling of c-Met was also inhibited (Fig. 3A). To confirm this observation, treatment with a c-Met kinase inhibitor, SU11274, also inhibited cell proliferation dose-dependently at 72h (Fig. 3A). Similarly, a dose-dependent decrease in colony formation was found at concentrations of SU11274 ≥ 1 µM in CaSki, UM-SCC47, and 93-VU-147T cells (Fig. 3B). In addition, the downstream signaling of c-Met was also inhibited after treatment with 10 µM SU11274 for 24h and 48h, respectively (Fig. S3).
Figure 3.
c-Met inhibition decreased cell proliferation, colony formation, and migration of HPV-positive cancer cell lines. c-Met siRNA and inhibitor SU11274 inhibited cell proliferation of CaSki, UM-SCC47, and 93-VU-147T cells and downstream signaling of c-Met (A). SU11274 treatment showed dose-dependent inhibition of colony formation in CaSki, UM-SCC47, and 93-VU-147T cells (B). Relative cell migration of CaSki, UM-SCC47, and 93-VU-147T cells was significantly decreased upon c-Met siRNA knockdown, after adjusting for the influence of cell proliferation (divided by percentage of viability from SRB assay under identical treatment conditions) (C). Error bars represent mean ± SD. Experiments were repeated three times.
Furthermore, we tested the effects of c-Met knockdown on cell migration and calculated relative migration after adjusting for the influence of cell proliferation. The data showed that cell migration was significantly decreased after c-Met knockdown in CaSki (P < 0.001), UM-SCC47 (P = 0.005), and 93-VU-147T (P < 0.001) cells, as shown in a representative image of cell migration in Fig. 3C.
Association of c-Met and p53 with HPV status in OPSCC patients
Among all patients with primary OPSCC, approximately 74% were HPV-positive. The majority of patients were Caucasian (88%), 8% were African American, and two patients were Hispanic. Given the limited numbers of non-Caucasian cases these were combined as non-Caucasians in the analysis. Patient characteristics are shown in Table S1 and detailed information for each patient is shown in Table S2.
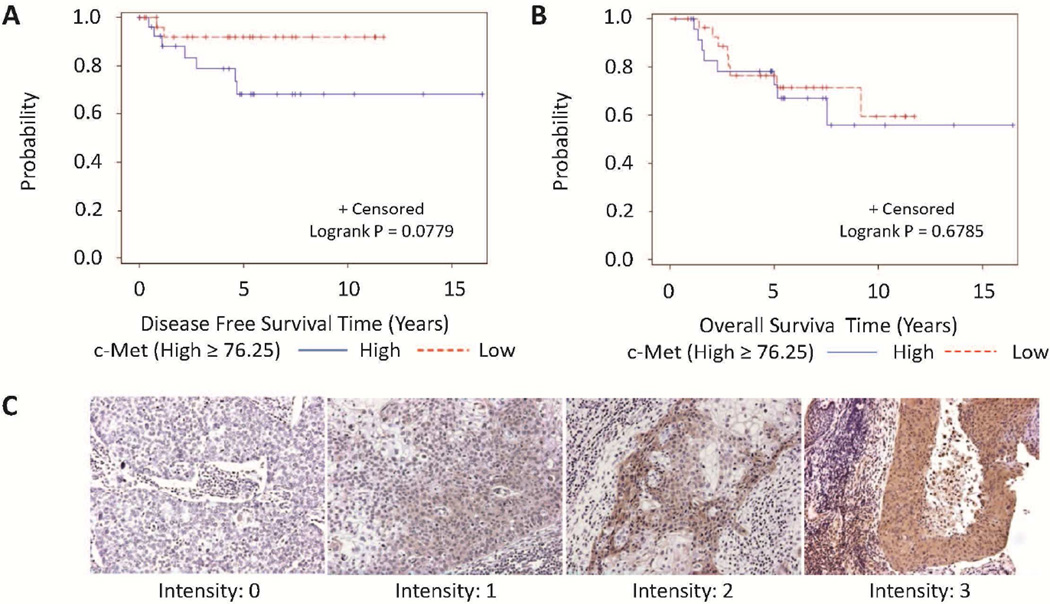
IHC staining of c-Met and p53 are shown in Fig. 4C and Fig. S4, respectively. c-Met expression was found in the majority of the OPSCC patient samples (94%). Samples from four HPV-positive patients (7%) and one HPV-negative patient (5%) did not show c-Met expression. p53 expression was observed in 29% of HPV-positive OPSCC compared to 50% in HPV-negative samples, and chi-square test showed a marginal significance of P = 0.09. Univariate analysis showed that c-Met was marginally related with tumor stage but not HPV status in the overall group of OPSCC patients (data not shown). However, multivariable analysis showed that c-Met protein levels were significantly related with HPV status and tumor stage after adjusting for race. High c-Met expression (≥ median) was found to be significantly associated with HPV positivity (HR = 4.11, 95%CI: 1.16–14.55, P = 0.028, Table 1) and advanced tumor stage (T1/T2 vs. T3/T4, HR = 0.27, 95% CI: 0.08–0.93, P = 0.039, Table 1). Further univariate analysis stratifying the OPSCC patients by tumor stage showed that only in T3/T4 patients, high c-Met levels (≥ median) was significantly associated with HPV positivity (P < 0.001) and marginally associated with low p53 levels (< median) (P = 0.066) and a younger age (P = 0.078, Table 2).
Figure 4.
Kaplan–Meier estimates of overall survival (OS) and disease-free survival (DFS) using c-Met and p53 levels in patients with HPV-positive OPSCC. High c-Met and p53 levels marginally correlated with worse DFS (A) while in contrast, neither c-Met nor p53 showed prognostic significance for OS (B). Representative images of c-Met are shown in C.
Table 1.
Multivariate association of c-Met (categorical) with covariates.
| Covariate | Level | c-Met high (High ≥ 70.416) |
P-value | |
|---|---|---|---|---|
| Odds Ratio (OR) | (95% CI) | |||
| HPV status | Positive | 4.11 | 1.16–14.55 | 0.028 |
| Negative | 1 (Ref) | |||
| Race | White | 0.31 | (0.05–1.86) | 0.199 |
| Non-White | 1 (Ref) | |||
| Tumor Stage | T1/T2 | 0.27 | 0.08–0.93 | 0.039 |
| T3/T4 | 1 (Ref) | |||
Note: logistic regression model was used. Backward selection with an alpha level of removal of 0.2 was used. Age, Gender, Node Metastasis, Stage, Differentiation, p53, and Smoking were removed from the model.
Table 2.
Univariate association of c-Met with covariates stratified by Tumor Stage.
| Tumor Stage | Covariate | Level | All patients (N=78) |
c-Met (High ≥ 70.416) |
P-value* | |
|---|---|---|---|---|---|---|
| High (N=39) | Low (N=39) | |||||
| T1/T2 | HPV status | Negative | 20 (25.64) | 7 (50) | 7 (50) | 0.692 |
| Positive | 58 (74.36) | 18 (43.9) | 23 (56.1) | |||
| Age | Mean (± SD) | 55.13 (± 9.2) | 55.68 (± 8.14) | 53.9 (± 9.92) | 0.476 | |
| Gender | Female | 10 (12.82) | 3 (60) | 2 (40) | 0.65 | |
| Male | 68 (87.18) | 22 (44) | 28 (56) | |||
| Race | Non-White | 8 (11.76) | 3 (50) | 3 (50) | 1 | |
| White | 60 (88.24) | 18 (41.86) | 25 (58.14) | |||
| Node Metastasis | Negative | 7 (9.21) | 3 (50) | 3 (50) | 1 | |
| Positive | 69 (90.79) | 20 (42.55) | 27 (57.45) | |||
| Differentiation | MD | 21 (26.92) | 9 (56.25) | 7 (43.75) | 0.432 | |
| NK | 50 (64.1) | 16 (43.24) | 21 (56.76) | |||
| WD | 7 (8.97) | 0 (0) | 2 (100) | |||
| Stage | I/II | 9 (11.69) | 5 (55.56) | 4 (44.44) | 0.716 | |
| III/IV | 68 (88.31) | 20 (43.48) | 26 (56.52) | |||
| Smoking | Current | 26 (35.14) | 7 (50) | 7 (50) | 0.556 | |
| Former | 30 (40.54) | 12 (46.15) | 14 (53.85) | |||
| Never | 18 (24.32) | 4 (30.77) | 9 (69.23) | |||
| p53 | Median (Range) | 0 (0 – 285) | 0 (0 – 240) | 0 (0 – 240) | 0.316 | |
| T3/T4 | HPV status | Negative | 20 (25.64) | 0 (0) | 6 (100) | <0.001 |
| Positive | 58 (74.36) | 14 (93.33) | 1 (6.67) | |||
| Age | Mean (± SD) | 55.13 (± 9.2) | 54 (± 7.61) | 61.86 (± 11.7) | 0.078 | |
| Gender | Female | 10 (12.82) | 2 (50) | 2 (50) | 0.574 | |
| Male | 68 (87.18) | 12 (70.59) | 5 (29.41) | |||
| Race | Non-White | 8 (11.76) | 2 (100) | 0 (0) | 1 | |
| White | 60 (88.24) | 11 (64.71) | 6 (35.29) | |||
| Node Metastasis | Negative | 7 (9.21) | 0 (0) | 1 (100) | 0.333 | |
| Positive | 69 (90.79) | 14 (70) | 6 (30) | |||
| Differentiation | MD | 21 (26.92) | 4 (80) | 1 (20) | 0.475 | |
| NK | 50 (64.1) | 8 (72.73) | 3 (27.27) | |||
| WD | 7 (8.97) | 2 (40) | 3 (60) | |||
| Smoking | Current | 26 (35.14) | 6 (50) | 6 (50) | 0.164 | |
| Former | 30 (40.54) | 2 (66.67) | 1 (33.33) | |||
| Never | 18 (24.32) | 5 (100) | 0 (0) | |||
| p53 | Median (Range) | 0 (0 – 285) | 0 (0 – 27) | 100 (0 – 240) | 0.066 | |
Note: Data are presented as number of patients (%), mean (± SD) or median (range).
The P-value is calculated by ANOVA or Wilcoxon rank-sum test for numerical covariates and chi-square test or Fisher's exact test for categorical covariates, where appropriate.
Prognostic values of c-Met in OPSCC patients
We found that c-Met expression was not associated with DFS while high p53 levels was associated with worse DFS on both univariate (data not shown) and multivariable analysis (Table S3) in OPSCC patients overall. Neither c-Met nor p53 was associated with OS in OPSCC patients overall (data not shown). We further examined the prognostic value of c-Met and p53 stratified by HPV status. We found that high c-Met and p53 was marginally associated with worse DFS (P = 0.078 and 0.065, respectively, Fig. 4A; Table S4), but not OS in HPV-positive patients upon univariate analysis. Non-Caucasian patients and those with T3/T4 disease also tended to have a worse DFS (Table S4). Multivariable analysis showed that non-White, T3/T4 stage and high p53 expression level were significant predictors of worse DFS (Table S5).
Clinical covariates such as tumor stage and smoking status were associated with OS on univariate analysis in HPV-positive patients (Table S6). Multivariable analysis showed that current smoking, older age, and high p53 levels were associated with worse OS in HPV-positive patients (Table S7). In HPV-negative patients, c-Met and p53 levels were not associated with either DFS or OS (data not shown).
Discussion
HPV-associated HNSCC has been recognized as a distinct clinical identity with the biology largely depending upon inactivation of the p53 and pRB tumor suppressors. Detailed molecular mechanisms underlying the particular phenotypes of HPV-associated OPSCC, however, remain poorly understood. We found that HPV E6 upregulates c-Met expression partially through p53 downregulation. This finding provides novel insights into the biology of HPV-driven HNSCC and may suggest a novel therapeutic target for this specific disease.
In this study we found that c-Met expression was significantly higher in HPV-positive OPSCC patient tumor samples and more prominently in patients with higher T stage (T3/T4) (Table 2). To our knowledge, this is the first study demonstrating such a positive link between c-Met and HPV in OPSCC. Previous studies showed that overexpression of HGF/c-Met was strongly correlated with HPV positivity in cervical cancer (32) and that c-Met overexpression significantly increased during cancer progression associated with HPV in anal cancer (33), supporting our novel findings in OPSCC. Recent TCGA data from 279 cases of HNSCC revealed that HPV-positive tumors have significant somatic mutations in the oncogene PIK3CA, amplification of E2F1, and loss of TRAF3 but no significant genomic alterations in receptor tyrosine kinases (e.g., EGFR, HER3, or c-Met) (34), supporting that these receptor tyrosine kinases are most likely genetically intact in HPV-positive tumors. It would be interesting to define the association between HPV status and transcriptional/translational regulation of c-Met. Moreover, high c-Met expression marginally correlated with worse DFS only in HPV-positive patients (Fig. 4 and Table S4). Controversial findings exist regarding the prognostic significance of c-Met in HNSCC (11, 35, 36). These discrepancies are likely due to differences in the tumor sites investigated, analytical methods used (whether HPV status is stratified, e.g.), and sample sizes. Future large-scale, prospective studies stratifying HPV status are necessary to dissect the prognostic significance of c-Met in HNSCC.
Considering the better response and more favorable outcomes of HPV-positive HNSCC patients, de-intensification of therapeutic regimens has been proposed and several clinical trials are ongoing to explore this approach (37, 38). However, our data suggest that HPV-positive patients have heterogeneous outcomes in terms of DFS and therefore caution should be given with regard to de-intensification of therapy. A subgroup of HPV-positive patients seems to have a worse prognosis than others and identifying those patients is clinically important. A recent review paper concludes that discriminating between HPV-positive tumors that are related to smoking and alcohol consumption and those are not related to these risk factors is critical for prognosis analysis (39). Our current data also show that co-existing conditions such as smoking may affect patient outcomes (Table S6 and Table S7). The identification of specific tumor-related biomarkers may help better direct treatment options. In the present study we showed that c-Met may serve as a prognostic marker in HPV-positive OPSCC, which warrants further validation in large-scale studies.
c-Met and its sole ligand HGF, play a crucial role in the oncogenic process, including the regulation of cell proliferation, migration, invasion and angiogenesis (40). In the present study, we found that inhibition of c-Met expression by siRNA and a c-Met tyrosine kinase inhibitor, SU11274, remarkably decreased cell proliferation, migration, and colony formation of CaSki, UM-SCC47, and 93-VU-147T cancer cells, indicating an important role of c-Met in maintaining the aggressive phenotypes of HPV-positive cancer cells. These findings are similar to those of previous studies including our recent paper showing that HPV E6 knockdown results in decrease/elimination of oncogenic behaviors, such as cell proliferation and invasion of HPV-positive cancer cells (28, 41).
It is speculated that high-risk HPVs may have evolved to adopt a stem cell-like state of the host cells in order to establish a persistent infection (42). Since c-Met expression has proven important to promote self-renewal and maintain the stem cell features of HNSCC cells (43, 44), it is possible that c-Met overexpression is favored by HPV E6 oncoprotein to facilitate its oncogenic process. We confirmed this hypothesis by showing that knockdown of HPV E6 significantly decreased c-Met expression at both the protein and mRNA level in diverse HPV-positive cancer cells (Fig. 1A-1C) without affecting other receptor tyrosine kinases such as EGFR and HER3 (Fig. S2).
Mechanistically, we hypothesize that p53 downregulation by E6 may mediate the upregulation of c-Met in HPV-positive cancer cells. Our results showed that HPV E6 knockdown increased wild-type p53 protein levels and not surprisingly, a significant negative correlation between the change in c-Met and p53 expression was observed (Fig. 1), consistent with previous findings (15, 16). Overexpression of wild-type p53 in cells often leads to cell cycle arrest and apoptosis (45, 46), which adds to the complexity of elucidating of the relationship between p53 and c-Met. Instead we performed wild-type p53 knockdown in several HPV-positive cancer cells and showed that p53 knockdown upregulated c-Met mRNA and protein levels (Fig. 2). We further showed that c-Met downregulation by E6 knockdown was obviously mitigated when p53 knockdown was performed simultaneously, demonstrating the interdependence of E6 and c-Met on p53 in HPV-positive disease. These in vitro data were also consistent with the clinical observation of the positive association between HPV status and c-Met expression. The limitation of our current study is that we did not distinguish wild-type p53 from mutant p53 in our OPSCC tissue IHC staining. It is possible that in HPV-positive population, there may be certain percentage of mutant p53 proteins which contribute to poor DFS. Taken together, our data strongly support that the regulation of c-Met expression by E6 is mediated through wild-type p53 downregulation and that c-Met may be a valid molecular target in HPV-positive HNSCC. The function of mutant p53 in regulation of c-Met expression remains currently unknown.
In summary, our data reveal a novel regulation of c-Met by HPV E6 oncoprotein that is mediated by p53 downregulation. In tumor samples from OPSCC patients, we also demonstrated that HPV-positive OPSCC samples have higher c-Met protein expression than HPV-negative tumors and that c-Met expression is prognostic of worse DFS in HPV-positive OPSCC. These findings together support the clinical investigation of c-Met as a prognostic factor as well as a molecular target in HPV-positive OPSCC patients. Future studies are warranted to validate these findings in different clinical settings.
Supplementary Material
Highlights.
Reducing HPV oncogene E6 decreased both mRNA and protein expressions of c-Met.
The upregulation of c-Met by HPV E6 is mediated by p53.
High c-Met protein levels correlated with HPV positivity in OPSCC specimens.
High c-Met protein levels correlated with worse DFS in HPV-positive OPSCC patients.
Acknowledgments
The authors thank Dr. Anthea Hammond for her editing of the manuscript. The present study was supported by grants from HPV Small Business Innovation Research (SBIR) program Phase I (HHSN261201000125C) and II Award (HHSN261201200097C, PI: Wang AY), National Institutes of Health (R33 CA161873, PI: Chen ZG), National Institute of Health (R21 CA182662, PIs Saba NF and Chen ZG), and Georgia Cancer Coalition Distinguished Scholar Award to Dr. Chen ZG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
All authors declared no conflicts of interest.
References
- 1.Duvvuri U, Myers JN. Cancer of the head and neck is the sixth most common cancer worldwide. Curr Probl Surg. 2009;46(2):114–117. doi: 10.1067/j.cpsurg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia A, Burtness B. Human Papillomavirus-Associated Oropharyngeal Cancer: Defining Risk Groups and Clinical Trials. J Clin Oncol. 2015;33(29):3243–3250. doi: 10.1200/JCO.2015.61.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110(5):525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 7.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10(8):550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 8.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8(12):3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Muller S, Qian G, Xu J, Kim S, Chen Z, et al. Human papillomavirus 16 oncoprotein regulates the translocation of beta-catenin via the activation of epidermal growth factor receptor. Cancer. 2015;121(2):214–225. doi: 10.1002/cncr.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006;12(12):3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 11.Baschnagel AM, Williams L, Hanna A, Chen PY, Krauss DJ, Pruetz BL, et al. c-Met expression is a marker of poor prognosis in patients with locally advanced head and neck squamous cell carcinoma treated with chemoradiation. Int J Radiat Oncol Biol Phys. 2014;88(3):701–707. doi: 10.1016/j.ijrobp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Blumenschein GR, Jr, Mills GB, Gonzalez-Angulo AM. Targeting the hepatocyte growth factor-cMET axis in cancer therapy. J Clin Oncol. 2012;30(26):3287–3296. doi: 10.1200/JCO.2011.40.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9(6):314–326. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- 14.Nisa L, Aebersold DM, Giger R, Zimmer Y, Medova M. Biological, diagnostic and therapeutic relevance of the MET receptor signaling in head and neck cancer. Pharmacol Ther. 2014;143(3):337–349. doi: 10.1016/j.pharmthera.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Cheng CY, Hwang CI, Corney DC, Flesken-Nikitin A, Jiang L, Oner GM, et al. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep. 2014;6(6):1000–1007. doi: 10.1016/j.celrep.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci U S A. 2011;108(34):14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21(3):632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaykalova DA, Mambo E, Choudhary A, Houghton J, Buddavarapu K, Sanford T, et al. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PLoS One. 2014;9(3):e93102. doi: 10.1371/journal.pone.0093102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiwert TY, Jagadeeswaran R, Faoro L, Janamanchi V, Nallasura V, El Dinali M, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009;69(7):3021–3031. doi: 10.1158/0008-5472.CAN-08-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wald AI, Hoskins EE, Wells SI, Ferris RL, Khan SA. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck. 2011;33(4):504–512. doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17(23):7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans MF, Aliesky HA, Cooper K. Optimization of biotinyl-tyramide-based in situ hybridization for sensitive background-free applications on formalin-fixed, paraffin-embedded tissue specimens. BMC Clin Pathol. 2003;3(1):2. doi: 10.1186/1472-6890-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiedorn KH, Kuhl H, Galle J, Caselitz J, Vollmer E. Comparison of in-situ hybridization, direct and indirect in-situ PCR as well as tyramide signal amplification for the detection of HPV. Histochem Cell Biol. 1999;111(2):89–95. doi: 10.1007/s004180050338. [DOI] [PubMed] [Google Scholar]
- 25.Jiang N, Wang D, Hu Z, Shin HJ, Qian G, Rahman MA, et al. Combination of anti-HER3 antibody MM-121/SAR256212 and cetuximab inhibits tumor growth in preclinical models of head and neck squamous cell carcinoma. Mol Cancer Ther. 2014;13(7):1826–1836. doi: 10.1158/1535-7163.MCT-13-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18(7):1118–1122. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolle CE, Kanteti R, Surati M, Nandi S, Dhanasingh I, Yala S, et al. Combined MET inhibition and topoisomerase I inhibition block cell growth of small cell lung cancer. Mol Cancer Ther. 2014;13(3):576–584. doi: 10.1158/1535-7163.MCT-13-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Muller S, Qian G, Xu J, Kim S, Chen Z, et al. Human papillomavirus 16 oncoprotein regulates the translocation of beta-catenin via the activation of epidermal growth factor receptor. Cancer. 2014 doi: 10.1002/cncr.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D. KJ, L. PR. The Statistical Analysis of Failure Time Data. Second. Hoboken, NJ: John Wiley & Sons, Inc; 2002. [Google Scholar]
- 30.Cox DR. Regression Models and Life Tables. J Royal Stat Society. 1972;B34(2):187–220. [Google Scholar]
- 31.Schoenfeld DA. Partial residuals for the proportional hazards regression model. Biometrika. 1982;(69):239–241. [Google Scholar]
- 32.Walker F, Kermorgant S, Darai E, Madelenat P, Cremieux AC, Henin D, et al. Hepatocyte growth factor and c-Met in cervical intraepithelial neoplasia: overexpression of proteins associated with oncogenic human papillomavirus and human immunodeficiency virus. Clin Cancer Res. 2003;9(1):273–284. [PubMed] [Google Scholar]
- 33.Walker F, Abramowitz L, Benabderrahmane D, Duval X, Descatoire V, Henin D, et al. Growth factor receptor expression in anal squamous lesions: modifications associated with oncogenic human papillomavirus and human immunodeficiency virus. Hum Pathol. 2009;40(11):1517–1527. doi: 10.1016/j.humpath.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chau NG, Perez-Ordonez B, Zhang K, Pham NA, Ho J, Zhang T, et al. The association between EGFR variant III, HPV, p16, c-MET, EGFR gene copy number and response to EGFR inhibitors in patients with recurrent or metastatic squamous cell carcinoma of the head and neck. Head Neck Oncol. 2011;3:11. doi: 10.1186/1758-3284-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon MJ, Kim DH, Park HR, Shin HS, Kwon JH, Lee DJ, et al. Frequent hepatocyte growth factor overexpression and low frequency of c-Met gene amplification in human papillomavirus-negative tonsillar squamous cell carcinoma and their prognostic significances. Hum Pathol. 2014;45(7):1327–1338. doi: 10.1016/j.humpath.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Masterson L, Moualed D, Liu ZW, Howard JE, Dwivedi RC, Tysome JR, et al. De-escalation treatment protocols for human papillomavirus-associated oropharyngeal squamous cell carcinoma: a systematic review and meta-analysis of current clinical trials. Eur J Cancer. 2014;50(15):2636–2648. doi: 10.1016/j.ejca.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Bhatia A, Burtness B. Human Papillomavirus-Associated Oropharyngeal Cancer: Defining Risk Groups and Clinical Trials. J Clin Oncol. 2015 doi: 10.1200/JCO.2015.61.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duray ALD, Demoulin S, Delvenne P, Saussez S. Prognosis of HPV-positive head and neck cancers: implication of smoking and immunosuppression. Advances in Cellular and Molecular Otolaryngology. 2014;(2):25717. [Google Scholar]
- 40.Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther. 2014;142(3):316–338. doi: 10.1016/j.pharmthera.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22(38):5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- 42.Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78(21):11451–1160. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun S, Wang Z. Head neck squamous cell carcinoma c-Met(+) cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer. 2011;129(10):2337–2348. doi: 10.1002/ijc.25927. [DOI] [PubMed] [Google Scholar]
- 44.Lim YC, Kang HJ, Moon JH. C-Met pathway promotes self-renewal and tumorigenecity of head and neck squamous cell carcinoma stem-like cell. Oral Oncol. 2014;50(7):633–639. doi: 10.1016/j.oraloncology.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Seth P, Katayose D, Li Z, Kim M, Wersto R, Craig C, et al. A recombinant adenovirus expressing wild type p53 induces apoptosis in drug-resistant human breast cancer cells: a gene therapy approach for drug-resistant cancers. Cancer Gene Ther. 1997;4(6):383–390. [PubMed] [Google Scholar]
- 46.Li P, Bui T, Gray D, Klamut HJ. Therapeutic potential of recombinant p53 overexpression in breast cancer cells expressing endogenous wild-type p53. Breast Cancer Res Treat. 1998;48(3):273–286. doi: 10.1023/a:1005961705860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.