Abstract
Adenoviruses are widespread in human population as well as in great apes, although the data about the naturally occurring adenovirus infections remain rare. We conducted the surveillance of adenovirus infection in wild western lowland gorillas in Moukalaba-Doudou National Park (Gabon), in order to investigate naturally occurring adenovirus in target gorillas and tested specifically a possible zoonotic transmission with local people inhabiting the vicinity of the park. Fecal samples were collected from western lowland gorillas and humans, and analyzed by PCR. We detected adenoviral genes in samples from both gorillas and the local people living around the national park, respectively: the overall prevalence rates of adenovirus were 24.1 and 35.0 % in gorillas and humans, respectively. Sequencing revealed that the adenoviruses detected in the gorillas were members of Human mastadenovirus B (HAdV-B), HAdV-C, or HAdV-E, and those in the humans belonged to HAdV-C or HAdV-D. Although HAdV-C members were detected in both gorillas and humans, phylogenetic analysis revealed that the virus detected in gorillas are genetically distinct from those detected in humans. The HAdV-C constitutes a single host lineage which is compatible with the host-pathogen divergence. However, HAdV-B and HAdV-E are constituted by multiple host lineages. Moreover, there is no evidence of zoonotic transmission thus far. Since the gorilla-to-human transmission of adenovirus has been shown before, the current monitoring should be continued in a broader scale for getting more insights in the natural history of naturally occurring adenoviruses and for the safe management of gorillas’ populations.
Electronic supplementary material
The online version of this article (doi:10.1007/s11262-016-1360-8) contains supplementary material, which is available to authorized users.
Keywords: Adenoviridae, Gorillas, Gabon, Phylogeny
Introduction
Adenoviruses (AdVs) are non-enveloped icosahedral double-stranded DNA viruses. They belong to the family of Adenoviridae, which is divided into five genera: Mastadenovirus, Atadenovirus, Aviadenovirus, Siadenovirus, and Ichtadenovirus. Members of species belonging to genera Mastadenovirus and Atadenovirus are known to infect mammalian hosts [1, 2]. Mastadenoviruses infecting primates encompass seven Human mastadenovirus species (HAdV-A to G), the accepted species Simian mastadenovirus A and candidate species SAdV-B to G, and further not yet classified mastadenoviruses [2–4]. That classification into species is based on hemagglutination features, DNA (deoxyribonucleic acid) homology, and genomic organization [5]. There are currently over 60 HAdV types with HAdV-D containing the most members [5].
Adenoviruses were first isolated from humans and identified as the causative agent of epidemic febrile respiratory disease among military recruits in the 1950s [6, 7]. It is estimated that more than 90 % of the human population is seropositive for one or more serotypes of adenoviruses [8, 9]. The molecular biology of human-derived adenoviruses has been characterized extensively for species HAdV-C, for which human adenovirus 2 (HAdV-2) and HAdV-5 serve as prototypes [10]. Adenoviruses cause a variety of non-lethal infectious diseases in humans, and lethal disseminated adenovirus infection occurs in immunosuppressed patients [10].
The first description of a simian adenovirus in the literature was of a chimpanzee AdV [11], today known as SAdV-21 within the species H. mastadenovirus B. Later, when investigating chimpanzees suffering from kuru, four novel ape AdVs were discovered [12]. Ape AdVs have been detected or isolated from African apes including chimpanzees, bonobos, and gorillas [13–18]. Gorilla adenoviruses have been proposed to be members of HAdV-B, C, E, and F [13–18]. A recent report confirmed that the species HAdV-B which includes viruses from mixed host origin [14], originated from gorillas and have switched to humans and to chimpanzees during two different host switch events [18]. Serological surveys have found that anti-AdV antibodies were prevalent in 96 % of mountain gorillas, suggesting that AdVs are circulating among these animals [19]. In addition, Hoppe et al. recently reported high prevalence of AdV in wild apes including gorillas (45−100 %) [18]. Because AdVs are shed in the feces and saliva of infected animals [13], these viruses could possibly be transmitted among host animals via the fecal–oral route and inhalation of aerosols [20].
Comprehensive studies are still needed to clarify the origin and the diversity of adenoviruses spread in human and non-human primate populations.
Thus, to fill the gap, understanding the evolution pattern of AdVs spread in non-human primates and in people frequently coming in contact with these animals is critical.
In this study, we investigated AdV infection in two habituated western lowland gorilla groups in Moukalaba-Doudou national park (MDNP). In addition, we assessed AdV infection in the local people living around the national park to evaluate potential zoonotic transmissions.
Materials and methods
Sample collection and preparation
The study site MDNP is located in the south-western part of Gabon (Fig. 1). MDNP has been reported to have a high gorilla density (more than three gorillas per square kilometer) [21], and the absence of hunting pressure from local villagers makes it a major habitat for western lowland gorillas in central Africa. From December 2010 to November 2011, during tracking, we collected 112 fresh fecal samples from 2 wild gorilla groups, which were named Group Gentil (GG) and Group 8 (G8). GG and G8 had been habituated to human observers since 2003 [21] and 2011, respectively. During the study period, GG consisted of 20−21 individuals, including 1 adult (expected age ≥13 years old) male, 6 adult (≥10 years old) females, 10 young (4–6 years old) males, and 3 young females, and all members were individually identified. In contrast, G8 was estimated to consist of 8–12 individuals, including 1 adult male, 2 adult females, and 5−8 young males and females. GG was mainly sampled near the village Doussala, in the ancient plantations, where the forest has been formerly used in various crop fields, while G8 was found far from the village in the primary forest (Fig. 1). In addition to the gorilla samples, 20 fecal samples were collected from villagers, including trackers working for the habituation of gorillas. Upon collection, each fecal specimen was immediately placed into a tube containing 2 ml of RNAlater (Ambion, Austin, TX, USA). The tubes were kept at room temperature for at most 20 days at the field camp until the samples were transported to the laboratory in Libreville, the capital city of Gabon. At the laboratory, the tubes were stored at –20 °C until DNA extraction.
Fig. 1.

Location features of the sampling area. a Map of Gabon, showing Moukalaba-Doudou National Park [21]. b The sampling area in the MDNP (blue line: rivers; black line: roads; red line: hunting area limitation; green line with black strips: national park limitation; dark green: primary forest; olive green: secondary forest; brown: savanna; spotted green: swamp; black circle: sampling points of G8 pointed by an arrow; gray circle pointed by an arrowhead: sampling points of GG; white circle: base camp; black rectangle with a black flag: village; white squares: habitations) (Color figure online)
DNA extraction and PCR
Total DNA was extracted from the sample using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. We used the following primer sets for nested PCR: (1) 4431-s/4428-as and 4428-s/4429-as (Supplementary Table 1), targeting the HAdV DNA polymerase (DPOL) gene [14] and (2) AdhexF1/AdhexR1 and AdhexF2/AdhexR2, targeting loop 1—encompassing the hypervariable region (HVR1–6)—of the hexon gene of mastadenoviruses [22]. PCR for the DPOL gene was performed in a total volume of 20 µl containing 10 µl of 2× GoTaq Green Master Mix (Promega, Madison, WI, USA), 20 pmol of each primer, and 50 ng of DNA template. The following cycling conditions, slightly modified from Wevers et al. [14], were used: 95 °C for 2 min; 35 cycles of 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min; and a 7-min final extension step at 72 °C. PCR amplification of the hexon gene (HVR1–6) was performed in a total volume of 50 µl containing 200 µM of each dNTP, 20 pmol of each primer, 1.25 U of PrimeSTAR GXL polymerase (TaKaRa, Tokyo, Japan), and 50 ng of DNA template. The cycling conditions were as follows: 98 °C for 3 min; 35 cycles of 98 °C for 10 s, 45 °C for 1 min, and 72 °C for 2 min; and a final extension of 72 °C for 7 min. For the nested reaction, 2 µl of the first PCR product was amplified as above. Amplified products were separated on 1.5 % agarose gel and purified using the QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s instructions; the amplicons were then directly sequenced with the primers for the second PCR.
BLAST search
BLAST searches were carried out in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the determined nucleotide sequence as a query in the BLASTN program. The queries with at least 90 % identity with the deposited adenovirus gene sequences were considered for AdV species identification.
Sequencing and phylogenetic analysis
Twenty-four of the 27 positive samples (DNA quantity ≥5 ng/µl) were subjected to direct sequencing of DPOL gene fragments. Six samples were selected randomly for cloning and sequencing of DPOL and hexon HVR1–6 gene fragments. The PCR products were cloned into plasmid vector pCR-Blunt II-TOPO using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Plasmid extraction was carried out using the Wizard Miniprep Kit (Promega), and the extracted plasmids were sequenced by Big Dye terminator cycle sequencing (Applied Biosystems, Foster City, CA, USA).
The hexon HVR1–6 and DPOL gene sequences were edited and aligned using GENETYX software version 12.0 (Genetyx Co., Tokyo, Japan) and MEGA software version 5.05 [23]. The nucleotide sequences of DPOL (528-bp, corresponding to the position 29,200–29,727 in the reference simian adenovirus 21) and 782-bp fragments of the hexon gene (corresponding to the position 18,867–19,635 in the reference simian adenovirus 21) were aligned using MUSCLE, with the default parameters for gap opening and gap extension. These alignments were used for phylogenetic analyses. Phylogenetic trees were constructed using the neighbor-joining method in MEGA 5.05 [23]. A statistical test for the phylogeny was computed by means of bootstrapping. Percentages of 100 bootstrap replicates at the node were calculated to ensure the reliability of the trees.
Nucleotide sequence accession numbers
Preliminary names were given to candidate novel HAdVs following the method used by Wevers et al. [14]. The gorilla adenoviruses detected in this study were named as follows: Gorilla gorilla AdV B11-B23 (KM886307-KM886309, KM886311, KM886325-KM886328, KM886331-KM886335), Gorilla gorilla AdV C10-C18 (KM886310, KM886320-KM886324, KM886329), and Gorilla gorilla AdV E1 (KM886330). The sequences used as references for phylogenetic analysis are presented in Supplementary Table 2.
Results
Detection of AdV genes in western lowland gorillas in MDNP
To survey AdV infection in gorillas in MDNP, we collected fecal samples from two gorilla groups (GG: well-habituated group, G8: newly habituated group) and analyzed them by nested PCR targeting the DPOL and hexon genes. The DPOL and hexon genes were detected in both groups (Table 1). The overall prevalence of AdV in the gorilla population was 24.1 % (27/112): of the 86 samples from GG, 21 were positive for both genes, 4 were positive only for the DPOL gene, and 1 sample was positive only for the hexon gene. In contrast, only 1 of the 26 samples was positive for both tested genes in G8 (Table 1). These data suggest that AdVs are naturally circulating among gorillas in MDNP. To confirm the detected AdV species, we further determined the nucleotide sequences of the amplicons and determined the species of the detected AdVs by BLAST searches. Of the tested samples, 16 belonged to HAdV-B; 10 to HAdV-C; and 1 to HAdV-E.
Table 1.
Detection of adenovirus DPOL and hexon genes in samples from gorilla groups in MDNP
| Gorilla groups | No. of tested samples | No. of positive samples in PCR (%) | HAdV Species | ||
|---|---|---|---|---|---|
| No. of samples | |||||
| B | C | E | |||
| GG | 86 | 26 (30.2 %) | 16 | 9 | 1 |
| G8 | 26 | 1 (3.8 %) | 0 | 1 | 0 |
| Total | 112 | 27 (24.1 %) | 16 | 10 | 1 |
Detection of AdV genes in local people living around the national park
The prevalence of AdVs in well-habituated gorillas (30.2 % in GG group) was higher than that of newly habituated ones (3.8 % in G8 group), raising two possibilities either the AdVs in gorillas are derived from humans during the habituation process or AdVs are ubiquitous in the environment in and around the areas of human habitation. Therefore, we screened the local people (village Doussala in Fig. 1) for AdV infection. The prevalence in the local people was 35.0 % (7/20): two samples were positive for both DPOL and hexon genes, and five were positive only for the hexon gene (Table 2). These results revealed that the local people including trackers were also infected with AdVs. We sequenced the detected virus genes and identified the species of AdVs: one sample was infected with a HAdV-C type, and the others harbored HAdV-D members.
Table 2.
Adenovirus infection in humans
| Sample ID | PCR DPOL | PCR hexon |
|---|---|---|
| H1 | HAdV-C | HAdV-C |
| H2 | HAdV-D | |
| H3 | ||
| H4 | ||
| H5 | ||
| H6 | HAdV-D | |
| H7 | ||
| H8 | HAdV-D | |
| H9 | ||
| H10 | ||
| H11 | HAdV-D | |
| H12 | HAdV-D | |
| H13 | ||
| H14 | ||
| H15 | ||
| H16 | ||
| H17 | HAdV-D | HAdV-D |
| H18 | ||
| H19 | ||
| H20 |
Phylogenetic analysis
HAdV-C genes were detected in both gorillas and humans in MDNP, suggesting zoonotic transmission of AdV between the human and gorilla populations. To investigate this possibility, as well as to gain insights into the genetic diversity of adenoviruses in MDNP, we performed phylogenetic analyses.
In gorillas, on the tree based on the DPOL gene, 14 AdV genes identified in this study were divided into two groups; they clustered with SAdV-28.2, SAdV-46, SAdV-47, and gorilla AdV strains 6588 and 6575, which are representative strains of HAdV-B in gorillas, and unidentified simian adenoviruses recently described [18] (Fig. 2 and Supplementary Fig. 1). Nine AdV genes were clustered with simian AdV-45 and simian AdV-43, which are representative strains of HAdV-C in gorilla and new unidentified simian adenoviruses [18] (Fig. 2 and Supplementary Fig. 1). In contrast, one AdV gene clustered with SAdV-26 and chimpanzee AdV strain Y25, which are chimpanzee-specific strains belonging to HAdV-E (Fig. 2 and Supplementary Fig. 1). On the hexon gene-based trees, five HAdV-B (Supplementary Fig. 2a) and HAdV-E (Fig. 3) strains were identified among those isolated from gorillas. HADV-E is divided into four groups (Fig. 3): two groups of human origin and two of simian origin. The HAdV-E detected in gorillas in this study belonged to the H mastadenovirus E of simian origin (Fig. 3).
Fig. 2.
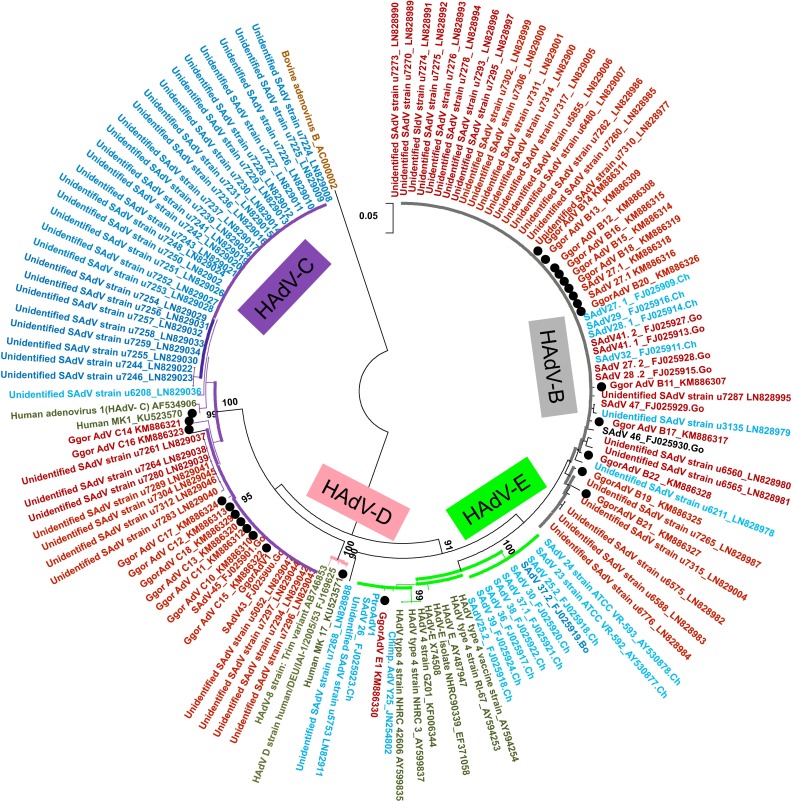
Phylogenetic tree of adenovirus (AdV) DPOL. The tree was constructed based on the alignment of AdV DPOL (539 bp) by using the neighbor-joining bootstrap-confirmed method in MEGA 5.05 software with 100 replicates. The names of simian isolates include the serotype nomenclature and the animal species of isolation (Ch chimpanzee, Go gorilla, Bo bonobo). Names of novel sequences obtained in this study are indicated with black dots. Bootstrap values <90 % are omitted. Scale bar, nucleotide substitutions per site
Fig. 3.

Phylogenetic tree of the adenovirus hexon gene loop 1 of HAdV-E. The tree was constructed based on the alignment of a 792-bp sequence of the hexon gene by using the neighbor-joining bootstrap-confirmed method in MEGA 5.05 software with 100 replicates. The names of simian isolates include the serotype nomenclature and the animal species of isolation (Ch chimpanzee, Go gorilla Bo bonobo). Names of novel sequences obtained in this study are indicated with black dots
In the case of humans, the tree based on the DPOL gene showed one AdV gene clustered with HAdV-1 (HAdV-C), which is genetically different from the strains detected in gorillas (Fig. 2 and Supplementary Fig. 2b), and one clustered with the human AdV type 44 and human AdV type 47, which belong to the HAdV-D (Supplementary Fig. 1). The HAdV-D seems to be exclusively limited to the human population as reported earlier [18].
Discussion
In this study, we detected several species of AdVs in western lowland gorillas in MDNP as well as in local people residing nearby. Interestingly, the positive rate in the well-habituated group (30.2 %) was higher than that of the newly habituated group (3.8 %). In addition, members of HAdV-C were detected in both gorillas and humans. However, the phylogenetic analyses revealed that the AdVs detected from gorillas are genetically distinct from those from local people living around the national park. Therefore, gorilla viruses and human viruses may have been separately circulating in each population in this region, and transmission between human and animals does not seem to happen easily in either direction, although we cannot exclude the possibility that we just missed zoonotically transmitted AdVs in this study. The difference in the prevalence between groups GG and G8 may be attributed to the quality of samples, because samples from GG might have been fresher than the ones from G8; GG was sampled while following animals, but G8 was sampled on trails, sometimes without observing the animals. In contrast, AdVs were reported to be transmitted between humans and non-human primates, indicating that AdVs have zoonotic potential [15, 18], despite the belief that AdVs have co-evolved with their hosts and are usually not transmitted to other species.
Adenovirus infections have been reported in high prevalence in wild gorillas’ populations as well as in other great apes [15, 17, 18]. In this study, the overall prevalence of AdV infection in gorillas was 24.1 %, which is lower than the previously reported figure of 44.9 % in free-ranging gorillas in Congo Republic [17] or of 48 % in free-ranging gorillas in Loango National Park (Gabon) [18]. These differences might be due to the quality of the samples and/or sensitivity of the PCR. In addition, the PCR systems used in this study targeted the conserved DNA polymerase gene of mastadenovirus or the hypervariable region of the hexon gene, but in some samples, only one of the two genes was amplified. This shows that our PCR system might not be able to amplify all gene variants or that the samples could have been partially degraded [18]. Alternatively, the DNA amount of AdV in the gorillas included in this study was lower than detection limit. Further systematic studies are needed to assess these possibilities.
We detected members of three species: HAdV-B, HAdV-C, and HAdV-E in western lowland gorillas in MDNP; these AdV species have been reported earlier [15–18] in western lowland gorillas as well as in other gorilla sub-species in sub-Saharan Africa. The gorilla adenoviruses of this study mainly belong to the HAdV-B (59 %). This confirms the gorilla as the major host of HAdV-B in sub-Saharan Africa. Based on the hexon tree (Supplementary Fig. 2a), the new virus named Gorilla gorilla adenovirus B19, together with the Human mastadenovirus B isolates 6560 and 6674 constitutes a single clade. The pattern observed within the species Human mastadenovirus C (Supplementary Fig. 1) is compatible with the host-pathogen divergence as previously reported [13, 15, 18]. All the lineages in HAdV-C are host specific [18]. The only member of HAdV-E detected in this study clusters with chimpanzee strains (Fig. 3). This finding supports previous report describing the non-human primate AdVs members of the HAdV-E to originate from chimpanzees [18]. We can suspect the Gorilla gorilla adenovirus E1 of this study to be the result of chimpanzee-to-gorilla transmission, as chimpanzees and gorillas are living sympatrically in MNDP. Broader screening would clarify the evolution of viruses belonging to HAdV-E.
On the other hand, the adenoviruses detected in the human population around MDNP are mainly members of the HAdV-D (85.71 %) which confirms that the species HAdV-D originated in humans [18] and so far has been exclusively human specific. Four different serotypes were detected in this study, highlighting the diversity of adenoviruses circulating in the target human population. Further systematic studies should clarify the circulation of AdVs in human population.
Taken together, our results show that AdVs are naturally present among gorillas and humans in MDNP in Gabon. Although there is no evidence of zoonotic transmission of AdVs in this region, our data show the feasibility of monitoring viral agents in wild habituated gorillas [24] and in local people living nearby for the safe management of wild gorilla populations and human health, as well as for understanding the evolution of virus. Since the zoonotic transmission of adenovirus already occurred during hominin evolution, assessing the zoonotic transmission of that virus in the context of habituation sites such as MNDP is recommended.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Dr. Bernhard Ehlers for providing the sequence alignment data of the last published AdV sequences in non-human primates, and to Professor Toshihiro Ito for his valuable comments on this paper. This study was part of a program supported by the Japanese Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA) (PROCOBHA: Principal Investigator: JY). We would like to thank Centre National de la Recherche Scientifique et Technologique (CENAREST, Gabon), Institut de Recherche en Ecologie Tropicale (IRET/CENAREST, Gabon), and Agence Nationale des Parcs Nationaux (ANPN, Gabon) for permission to conduct study in Moukalaba-Doudou National Park. We also acknowledge the field assistants for help with data and sample collection, as well as the local people in Doussala, Konzi, and Mboungou for their full support during the field study.
Author contributions
CNN and SF designed the research. MH, YK, NO, YK, KM, TM, SE, AM, TM, MO, OY, AN, JY, and KTK helped with the experiments, data analysis, and discussion. CNN, MH, SF, and KTK have written the manuscript.
References
- 1.Lehmkuhl HD, Hobbs LA. Serologic and hexon phylogenetic analysis of ruminant adenoviruses. Arch. Virol. 2008;153:891–897. doi: 10.1007/s00705-008-0063-4. [DOI] [PubMed] [Google Scholar]
- 2.Pantó L, Podgorski II, Jánoska M, Márkó O, Harrach B. Taxonomy proposal for Old World monkey adenoviruses: characterisation of several non-human, non-ape primate adenovirus lineages. Arch. Virol. 2015;160:3165–3177. doi: 10.1007/s00705-015-2575-z. [DOI] [PubMed] [Google Scholar]
- 3.Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gal J, Hornyak A, Mandoki M, Bakonyi T, Balka G, Szeredi L, Marosan M, Ludanyi T, Forgach P, Sos E, Demeter Z, Farkas SL. Novel mastadenovirus infection and clinical disease in a pygmy marmoset (Callithrix [Cebuella] pygmaea) Vet. Microbiol. 2013;167:695–699. doi: 10.1016/j.vetmic.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. Molecular evolution of human adenoviruses. Sci. Rep. 2013;3:1812. doi: 10.1038/srep01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilleman MR, Werner JH. Recovery of new agents from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 1954;85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 7.Rowe WP, Huebner RJ, Gilmore LK, Parrot RH, Ward TG. Isolation of a cytopathic agent from human adenoids undergoing spontaneous degradation in tissue culture. Proc. Soc. Exp. Biol. Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 8.D’Ambrosio E, Del Grosso N, Chicca A, Midulla M. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J. Hyg. (Lond) 1982;89:155–161. doi: 10.1017/S0022172400070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadell GA, Allard A, Johansson M, Svensson L, Uhnoo I. Enteric adenoviruses. Chichester: Wiley; 1987. pp. 73–91. [DOI] [PubMed] [Google Scholar]
- 10.Fields BN, Knipe DM, Howley PM, et al. Adenoviruses. In: Fields BN, et al., editors. Fields virology. Philadelphia: Lippincott-Raven Publishers; 1996. p. 1504. [Google Scholar]
- 11.Rowe WP, Hartley JW, Huebner RJ. Additional serotypes of the APC virus group. Proc. Soc. Exp. Biol. Med. 1956;91:260–262. doi: 10.3181/00379727-91-22231. [DOI] [PubMed] [Google Scholar]
- 12.Rogers NG, Basnight M, Gibbs CJ, Gajdusek DC. Latent viruses in chimpanzees with experimental kuru. Nature. 1967;216:446–449. doi: 10.1038/216446a0. [DOI] [PubMed] [Google Scholar]
- 13.Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wevers D, Leendertz FH, Scuda N, Boesch C, Robbins MM, Head J, Ludwig C, Kuhn J, Ehlers B. A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla) Virol. J. 2010;7:303. doi: 10.1186/1743-422X-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH, Ehlers B. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J. Virol. 2011;85:10774–10784. doi: 10.1128/JVI.00810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan M, Cranfield MR, Torano H, Kuete HM, Lee GP, Glenn A, Bruder JT, Rangel D, Brough DE, Gall JG. Adenoviruses isolated from wild gorillas are closely related to human species C viruses. Virology. 2013;444:119–123. doi: 10.1016/j.virol.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 17.Seimon TA, Olson SH, Lee KJ, Rosen G, Ondzie A, Cameron K, Reed P, Anthony SJ, Joly DO, McAloose D, Lipkin WI. Adenovirus and herpesvirus diversity in free-ranging great apes in the Sangha region of the Republic Of Congo. PLoS One. 2015;10:e0118543. doi: 10.1371/journal.pone.0118543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, Karhemere S, Madinda NF, Mugisha L, Muyembe JJ, Todd A, Petrzelkova KJ, Gray M, Robbins M, Bergi RA, Wittig RM, K.Zuberbuler, Boesch C, Schubert G, Leendertz FH, Ehlers B, Calvignac-Spencer S. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 2015;32:2072–2084. doi: 10.1093/molbev/msv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C.A. Whittier, Ph.D. thesis, North Carolina State University College of Veterinary Medicine, 2009www.lib.ncsu.edu/resolver/1840.16/6215
- 20.Fong TT, Phanikumar MS, Xagoraraki I, Rose JB. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ando C. Habituation and conservation of gorillas in Moukalaba-Doudou. Gorilla J. 2009;38:28–29. [Google Scholar]
- 22.Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch. Virol. 2006;151:1587–1602. doi: 10.1007/s00705-005-0722-7. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K.V. Gilardi, T.R. Gillespie, F.H. Leendertz, E.J. Macfie, D.A. Travis, C.A. Whittier, and E.A. Williamson. Best practice guidelines for health monitoring and disease control in great ape populations. Occasional Papers of the IUCN Species Survival Commission (2015), pp. 56
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


