Abstract
Background. Epicardial adipose tissue (EAT) is identified as an atypical fat depot surrounding the heart with a putative role in the involvement of metabolic disorders, including obesity, type-2 diabetes mellitus, and atherosclerosis. We profiled miRNAs in EAT of metabolic patients with coronary artery disease (CAD) and type-2 diabetes mellitus (T2DM) versus metabolically healthy patients by microarray. Compared to metabolically healthy patients, we identified forty-two miRNAs that are differentially expressed in patients with CAD and T2DM from Xinjiang, China. Eleven miRNAs were selected as potential novel miRNAs according to P value and fold change. Then the potential novel miRNAs targeted genes were predicted via TargetScan, PicTar, and miRTarbase, and the function of the target genes was predicted via Gene Ontology (GO) analysis while the enriched KEGG pathway analyses of the miRNAs targeted genes were performed by bioinformatics software DAVID. Then protein-protein interaction networks of the targeted gene were conducted by online software STRING. Finally, using microarray, bioinformatics approaches revealed the possible molecular mechanisms pathogenesis of CAD and T2DM. A total of 11 differentially expressed miRNAs were identified and among them, hsa-miR-4687-3p drew specific attention. Bioinformatics analysis revealed that insulin signaling pathway is the central way involved in the progression of metabolic disorders. Conclusions. The current findings support the fact that miRNAs are involved in the pathogenesis of metabolic disorders in EAT of CAD patients with T2DM, and validation of the results of these miRNAs by independent and prospective study is certainly warranted.
1. Introduction
Coronary artery disease (CAD) remains one of the most common causes of morbidity and mortality in diabetic patients [1]. Thus improving the understanding of the etiology associated with CAD is highly important. Epicardial adipose tissue (EAT) is suggested to play an important role in the progression of metabolic syndrome [2]. Several findings implicate that EAT thickness may be a useful indicator for T2DM and obesity [2, 3]. Studies have shown that EAT generates several bioactive molecules, such as anti- and proinflammatory mediators and cytokines [4], which may significantly enhance paracrine effects on cardiac function or produce a variety of effects that affect many physiological processes [5]. Nowadays, beside the main factors including obesity, hypertension, and dyslipidemia, novel risk factors such as chronic low-grade inflammation, oxidative stress, and endothelial dysfunction are accepted as the decisive factors to highlight this increased cardiovascular risk in human beings [6, 7]. Experimental and clinical studies have suggested that EAT may cause CAD [8]. EAT, visceral fat depot of the heart, was found to be associated with CAD, T2DM, and other metabolic disorders [6, 9, 10]. EATs are metabolically active visceral fat deposits found around the heart, between the pericardium and myocardium [11], which are strongly associated with cardiovascular diseases (CVD) including CAD and the development of cardiac arrhythmias, predominantly due to the secretion of bioactive mediators and cytokines [12]. T2DM plays a key role in the development of CVD. EAT has great interplay with diabetic patients and has potential to influence CVD. Owing to its close proximity to the heart and coronary vasculature, EAT exerts a direct metabolic impact by secreting free fatty acids and proinflammatory factors and decreased anti-inflammatory adipokines, which promote CVD locally [9]. MicroRNAs (miRNAs) are a class of about 21–25 nucleotides in length and small noncoding RNA molecules with essential roles, of which any alteration leads to several conditions. They were first identified in 1993, and the term microRNA was created in 2001 [13]. The main functions of miRNAs are to downregulate the target gene expression in translational repression and cleavage of mRNA and in a wide range of biological processes [13]. Decisive regulatory functions exhibited by the miRNA are associated with various human diseases such as human cancer and heart disease. In addition to the link with cancer, microRNAs play a vital role in the control of cardiac-related diseases. For multiple forms of heart disease, including ventricular wall, maintenance of cardiac rhythm myocyte growth, and contractility, the misexpressions of miRNAs were shown to be necessary. However, in the literature, data regarding the relationship between miRNAs and EAT in CAD patients T2DM is scant. Hence, we aimed to investigate the role of miRNAs in EAT. We also sought to predict the targets of novel miRNAs in EAT in patients compared with the results that are obtained from control subjects [6]. The current study was carried out to assess the changes in the EAT levels of miRNAs in subjects suffering from T2DM and CAD compared to healthy control ones. In addition, bioinformatics analyses were carried out in order to find out how these possible miRNAs are associated with the incidence and pathogenesis of CAD on top of T2DM. In the present study, we depicted comparative miRNAs in “metabolically healthy” patients without metabolic disorders and in metabolic patients with CAD and T2DM. This strategy allowed us to identify a set of miRNAs characterizing EAT in health and disease as well as potential novel biological processes characterizing EAT in CAD patients with T2DM.
2. Methods
2.1. Subjects
EAT samples were taken from 10 subjects of both genders, in the Department of Thoracic and Cardiovascular Surgery, the First Affiliated Hospital of Shihezi University School of Medicine, in which levels of miRNAs expression in EAT (using microarray) and routine parameters were measured. Subjects were divided into two groups, 5 in each group as follows: patients with T2DM and CAD and metabolically healthy control subjects without T2DM and CAD. Ten Asian patients were recruited, 5 underwent cardiac valve surgery (no evidences of CAD/T2DM/carotid atherosclerosis/metabolic syndrome), and 5 underwent coronary artery bypass graft surgery (CAD + T2DM group). The study protocol was approved by the Medical Ethics Committee of Shihezi University (School of Medicine, Xinjiang, China). Written informed consent was obtained from all subjects included in the study. This was a cross-sectional study and a review of medical records (including information on sex, age, height, weight, medications, disease duration, smoking, and history of other diseases) was undertaken. Control subjects were chosen from metabolically healthy individuals according to NCEP ATP III Metabolic syndrome criteria (two or less metabolic criteria; TG ≥ 1.7 mmol/L, blood pressure ≥ 130/85 mmHg, glucose ≥ 5.6 mmol/L, HDL-C: men < 1.03 mmol/L and women < 1.30 mmol/L, and waist circumference: men < 102 cm and women < 88 cm) as previous said [6, 14]. miRNAs microarray expression analyses were conducted on RNA extracted from perivascular EAT using the Human-MicroRNA Expression Kits.
2.2. RNA Extraction and Purification
Total RNA, including the miRNAs, was extracted from the EAT and purified using mirVana™ miRNA Isolation Kit (Cat. number AM1561, Ambion, Austin, TX, US), following the manufacturer's instructions, and checked for a RNA integrity number (RIN) to inspect RNA integration by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, US). RIN ≥ 6.0 and 28S/18S ≥ 0.7 were used for the miRNA array analysis.
2.3. RNA Labeling
miRNA molecular in total RNA was labeled by miRNA Complete Labeling and Hyb Kit (Cat. number 5190-0456, Agilent Technologies, Santa Clara, CA, US) following the manufacturer's instructions, labeling section.
2.4. Array Hybridization
MiRNA microarray assays were performed using the Agilent Human miRNA (8 ∗ 60 K) V21.0 microarray platform (design ID: 70156) at Shanghai Biotechnology Co., Ltd. (Shanghai, China). Each slide was hybridized with 100 ng Cy3-labeled RNA using miRNA Complete Labeling and Hyb Kit (Cat. number 5190-0456, Agilent Technologies, Santa Clara, CA, US) in hybridization oven (Cat. number G2545A, Agilent Technologies, Santa Clara, CA, US) at 55°C, 20 rpm for 20 hours according to the manufacturer's instructions, hybridization section. After hybridization, slides were washed in staining dishes (Cat. number 121, Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (Cat. number 5188-5327, Agilent Technologies, Santa Clara, CA, US).
2.5. Data Acquisition
Slides were scanned by Agilent Microarray Scanner (Cat. number G2565CA, Agilent Technologies, Santa Clara, CA, US) and Feature Extraction software 10.7 (Agilent Technologies, Santa Clara, CA, US) with default settings. Raw data were normalized by Quantile algorithm, Gene Spring Software 12.6 (Agilent Technologies, Santa Clara, CA, US).
2.6. Differential miRNAs Targeted Gene Prediction
The differential miRNAs targets predicted by computer-aided algorithms were obtained from TargetScan, PicTar, and miRBase targets [15]. More detailed information can be acquired from online software (http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_vertebrate.cgi; http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl; http://www.targetscan.org/).
2.7. The Interaction Network and Signaling Pathway Analysis of Differential microRNA and mRNA
DAVID [16–18], a bioinformatics analysis software, is used for the analysis of the enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) signaling pathway analysis for the interactions between microRNAs and mRNAs (http://david.abcc.ncifcrf.gov/). Online software Gene Ontology (http://geneontology.org/) was employed to perform GO enrichment analysis [19, 20]. The microRNA and mRNA of differential expression in patients were uploaded to DAVID and Gene Ontology for analysis.
2.8. Protein-Protein Interactions (PPI) Network Analysis
A number of abnormal mRNAs were found in the interaction analysis between miRNAs and mRNA. Therefore, to further understand the function of microRNA in the network, the PPI analysis was performed in the protein products of mRNAs to find out the key proteins. The selected targeted genes were put into the STRING (Search Tool for the Retrieval of Interacting Genes) database (http://string-db.org/), a metaresource that collects most of the available information on protein-protein associations and scores and weights it and augments it with predicted interactions and with the results of automatic opuses-mining searches to match the interactions of proteins [21, 22].
2.9. Statistical Analysis
Descriptive statistics for each variable were determined. Results for continuous variables were demonstrated as mean ± standard deviation. Statistical significant difference between the groups was determined by the chi-square test for categorical variables and unpaired Student's t-test for continuous variables. Differentially expressed targeted genes were studied using bioinformatics analysis and statistical analysis allowed algorithm of the selected software.
3. Results
3.1. Baseline Characteristics of Patients
The baseline characteristics of 5 patients and 5 control subjects were shown in Table 1. There were no differences with respect to the following variables between patients and control subjects, age, gender, waist circumference (WC), systolic blood pressure (SBP), diastolic blood pressure (DBP), and body mass index (BMI). Compared to “controls,” CAD patients with T2DM were characterized by significantly increased waist-hip ratio, LDL-C, and systemic inflammation, while HDL-C was decreased.
Table 1.
Baseline characteristics of patients.
| Non-CAD + T2DM (n = 5) | CAD + T2DM (n = 5) | P value | |
|---|---|---|---|
| Age (years) | 61.8 ± 5.2 | 54.6 ± 7.0 | 0.392 |
| BMI (kg/m2) | 29.47 ± 5.83 | 28.63 ± 4.26 | 0.213 |
| WC (cm) | 99.64 ± 10.67 | 101.23 ± 10.34 | 0.492 |
| Waist-hip ratio | 0.96 ± 0.14 | 0.99 ± 0.09 | <0.001 |
| FPG (mg/dL) | 106.5 (98.5–115.5) | 116.0 (100.5–138.0) | 0.103 |
| Total cholesterol | 166.48 ± 37.10 | 159.55 ± 39.41 | 0.335 |
| LDL-C (mg/dL) | 93.51 ± 26.59 | 98.44 ± 23.14 | <0.01 |
| HDL-C (mg/dL) | 40.66 ± 10.08 | 34.39 ± 9.64 | <0.01 |
| Triglyceride (mg/dL) | 139 (105.5–223.5) | 154 (106.0–234.0) | 0.276 |
| hsCRP (mg/dL) | 1.03 ± 2.31 | 3.15 ± 5.22 | <0.001 |
| Adiponectin (μg/mL) | 12.37 ± 6.55 | 9.16 ± 4.78 | <0.05 |
| Fibrinogen (mg/dL) | 523.0 (453.0–638.0) | 603.5 (510.5–766.5) | <0.01 |
FPG: fasting plasma glucose, LDL-C: low density lipoprotein cholesterol, HDL-C: high density lipoprotein cholesterol, and hsCRP: hypersensitive C reactive protein. Adiponectin, hsCRP, and fibrinogen indicate systemic inflammation.
3.2. Data of Microarray
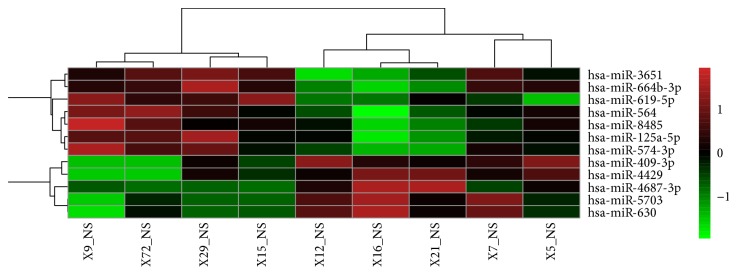
In order to measure the miRNAs expression patterns that characterize EAT in CAD patients with T2DM from EAT in control group, we used the whole-genome miRNAs microarrays. Overall, the resulting signal intensity of miRNAs genes is statistically different at the Wilcoxon signed-rank test in EAT in both groups, thus underscoring the profound diversity of EAT. Unsupervised hierarchical clustering was presented in Figure 1. Compared to metabolically healthy patients, we identified forty-two miRNAs that are differentially expressed in patients with CAD and T2DM (26 downregulated; 16 upregulated, data was not shown). Eleven miRNAs were selected as potential novel miRNAs according to P value and fold change (Table 2: 6 were significantly downregulated; 5 were significantly upregulated in both subgroups of patients with P < 0.05; fold change > 2 times).
Figure 1.
Unsupervised hierarchical clustering (heat map). Heat map generated by hierarchical clustering for differentially expressed miRNAs in the EAT from CAD with T2DM patients versus control subjects. Hierarchical clustering for differentially expressed miRNAs in CAD with T2DM (n = 4) versus control (n = 5) (P < 0.05 and fold change > 2 times). Columns display the clustering of EAT samples; rows show the clustering of genes. The expression intensity of each miRNA in each sample varies from red to green, which indicates relative high or low expression, respectively. Expression clusters representing different patterns of upregulation to downregulation are depicted on the side of figure.
Table 2.
Disregulated miRNAs (CAD + T2DM versus control).
| miRNA | P value | Fold change |
|---|---|---|
| hsa-miR-4429 | 0.04437405 | 0.16922769 |
| hsa-miR-409-3p | 0.012379586 | 0.184890055 |
| hsa-miR-6802-5p | 0.047496135 | 0.305662149 |
| hsa-miR-5703 | 0.008804377 | 0.40973895 |
| hsa-miR-630 | 0.020813173 | 0.43854972 |
| hsa-miR-4687-3p | 0.034377945 | 0.44430727 |
| hsa-miR-3651 | 0.04317499 | 2.063333894 |
| hsa-miR-574-3p | 0.025198143 | 2.156938357 |
| hsa-miR-619-5p | 0.00179165 | 2.179715733 |
| hsa-miR-664b-3p | 0.04815202 | 2.325766896 |
| hsa-miR-146b-5p | 0.044983658 | 2.450112684 |
Lists of the deregulated miRNAs between CAD + T2DM and control. Fold change < 0.5 indicated downregulation significantly and fold change > 2 indicated upregulation significantly.
3.3. Results of Bioinformatics Analyses
To depict the possible role of miRNAs in EAT, we selected these miRNAs as potential novel biomarkers that were significantly different (patients versus controls) in the overall population (P < 0.05; fold change > 2). To provide a framework for interpretation of our results, we then functionally clustered significant biological pathways using the bioinformatics analyses.
3.3.1. Bioinformatics Analyses of miRNAs Targeted Genes
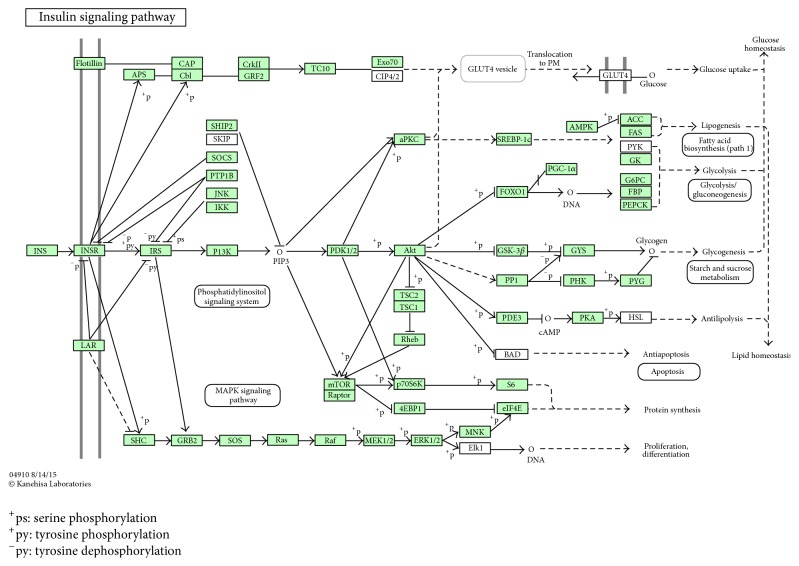
In order to investigate the possible regulation mechanisms of miRNAs in the process of CAD complicated with T2DM, we first utilized three online bioinformatics databases (TargetScan, PicTar, and miRBase targets) to select plausible targets and validated targets of miRNAs and finally obtained target genes for the following analysis; then we analyzed biological processes, molecular functions, and cellular components through Gene Ontology and enriched KEGG pathways by DAVID. The results showed that the predicted target genes mainly are enriched in the following biological processes: detection of chemical stimulus involved in sensory perception of smell, G-protein coupled receptor signaling pathway, translation, axon guidance, protein phosphorylation, and so forth (P < 0.001), they mainly are enriched in the following molecular functions: G-protein coupled receptor activity, olfactory receptor activity, protein binding, structural constituent of ribosome, and so forth (P < 0.001), and significant cellular components were extracellular region and Golgi apparatus (P < 0.001). Enriched KEGG pathways by DAVID displayed in KEGG pathway database showed that the predicted target genes of miRNAs were significantly enriched in the insulin signaling pathway (Figure 2), adipocytokine signaling pathway, MAPK signaling pathway, FoxO signaling pathway, and other signaling pathways (P < 0.05).
Figure 2.
Insulin signaling pathway. ERK1/2(MAPK1), GRB2, PKA (PRKAR), P13K (PIK3), and PDK1/2 (PDPK1) are the main target genes of differential miRNAs.
3.3.2. Protein-Protein Interaction (PPI) Network
We analyzed the protein-protein interaction network of selecting miRNAs target genes using STRING 10 and removed the target genes which were linked to lax isolated nodes through data analysis; the result showed that the interaction existed in total 148 proteins targeted by the predicted genes, which together formed the target gene interaction network. The network consists of 148 nodes which represent 148 proteins and many lines with different colors that represent the types of evidence for the association. From the result we can see that PDPK1, PIK3R3, PPP1R3B, PRKAR1A, SOCS3, SREBF1, PPARGC1A, SHC4, MAPK1, GRB2, and MKNK2 played key roles in maintaining stability in the network, especially PIK3R3, MAPK1, and GRB2 whose connections were very close, so the protein encoded by them may be important downstream target proteins (Figure 3).
Figure 3.
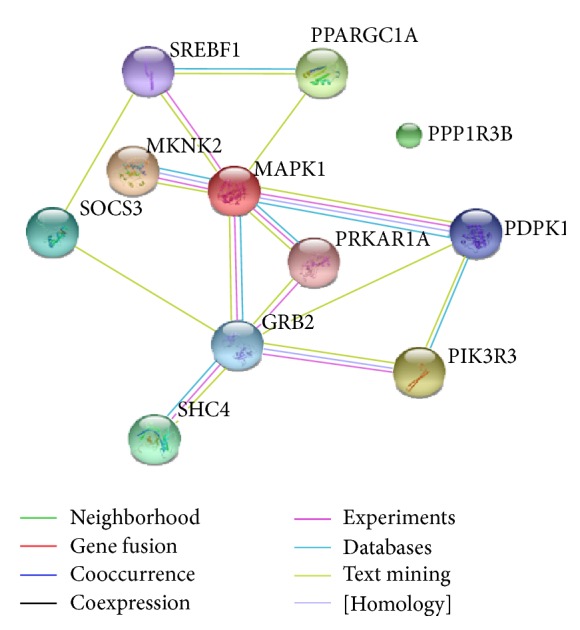
Protein-protein interaction (PPI) network of target proteins of the selecting miRNAs.
4. Discussion
Better understanding of the biological characteristics can provide vital theoretical basis for the prevention and treatment of disease. The aim of this study was to investigate the profiles of miRNAs and the interaction network of novel microRNA and mRNA as well as related signaling pathway in EAT through analyzing the expression profile of microRNA and mRNA to provide novel insights in the biological characteristics in CAD patients with T2DM. According to newly added studies, it is clear that miRNAs are involved in the regulation of metabolic and inflammation functions, and the alterations in CAD and T2DM diseases have been analyzed through the identification and evaluation of miRNAs profiles in patients as in animal models [13, 23, 24]. microRNAs are key components of many cellular processes. Different studies have demonstrated that miRNA expression is tissue-specific, tightly regulated during embryogenesis, and overexpressed/underexpressed in many diseases, including CAD and T2DM pathologies [25]. They are easily detected in a quantitative way by real-time polymerase chain reaction (qRT-PCR) or microarrays and by other less frequently used identification methods, such as PCR-based restriction fragment length polymorphisms (PCR-RLFP), traditional northern blotting, direct sequencing using next generation sequencing (NGS), and platforms ligation based measurement [26]. Risk prediction for T2DM and CVD remains suboptimal even after the introduction of global risk assessment by various methods. This has prompted the search for additional biomarkers [27]. EAT is a source of several inflammatory mediators in high-risk cardiac patients [4]. Adipose tissue may function as an endocrine organ that contributes to an inflammatory burden in patients at risk of CVD. Obesity, adiposopathy, and insulin resistance induce EAT enlargement, inflammation, and dysfunction and trigger CAD [10]. EAT is an atypical fat depot surrounding the heart with a putative role in the development of atherosclerosis [28]. Given the close anatomic relationship between perivascular EAT and coronary arteries and the positive correlation between EAT and the presence of coronary atherosclerosis, several results point to EAT as a putative actor in CAD and/or T2DM [3, 10]. EAT thickness is an independent risk factor for CAD. EAT could thus be able to modulate heart and coronary artery pathophysiology, and mounting evidences point to EAT as a candidate player in pathophysiology of CAD. miRNAs act on multiple targets and complex pathogenesis, thus representing candidate regulators of adipocyte differentiation, metabolic homeostasis, and inflammation. miRNAs have also been described as differentially modulated in adipose tissue during metabolic disease, thus being considered candidate biomarkers for metabolic disorders, CAD, T2DM, and putative targets for therapy [28]. We present evidence that a profile of miRNAs was dysregulated in EAT in CAD patients with T2DM compared to metabolic health patients, supporting the concept that miRNAs in EAT are involved in pathogenesis of CAD and/or T2DM. The current study identified a total of 11 differentially expressed miRNAs, and among them, hsa-miR-4687-3p drew specific attention for the largest targeted genes which were identified. Particularly, bioinformatics analysis revealed that insulin signaling pathway is the central way involved in the progression of metabolic disorders. Previous studies indicated that the circulating levels of miR-133a can be used as a predictor for diagnosing CAD, since increased miR-133a level may be used to predict both the presence and severity of coronary lesions in CAD patients [29]. Evidence from a recent study had showed that miR-370 was significantly increased in patients with T2DM and CAD and CAD only and patients with T2DM [23]. This was also concomitant with another study that showed the use of certain miRNA imprints including miR-370, in the screening of patients at risk for developing CAD [30]. Because of the highly metabolic paracrine and endocrine functions of EAT, it has been proposed to play a role in the pathogenesis of CVD by releasing proinflammatory and proatherogenic factors. EAT plays important roles in CAD, not only in its location, but also by its blood supply. EAT derives its blood supply from coronary circulation. There is a functional and anatomic relationship between EAT and muscular components of the heart as these components share the same coronary blood supply. The release of proinflammatory and proatherogenic factors into the circulation advancing CVD is more significantly linked to the progression of CAD [10]. It is the close anatomical relationship between EAT and the coronary arteries combined with its biologically active properties that participates in the pathogenesis of diabetic coronary atherosclerosis [31]. EAT-specific miRNAs are miR-196b-5p, miR-196a-5p (a promoter of brown adipogenesis), miR-18a-3p (a member of the miR-17/92 cluster that promotes adipocyte differentiation), and miR-10a-3p (an anti-inflammatory agent) which were analyzed by one article that also supported our study. In EAT of CAD patients, miR-135b-3p (a direct target of inflammatory pathways) was found upregulated, while miR-455-3p (a driver of during brown adipocyte differentiation), miR-193b-3p (promoting adiponectin secretion in human adipocytes), and Let-7a-3p and miR-127-3p (negative modulators of inflammatory pathways) were found downregulated [32, 33]. These findings are in accordance with our results that miRNAs are involved in the pathogenesis of metabolic disorders in EAT of CAD patients with T2DM. Overall, we were able to depict a novel miRNA signature of EAT in CAD patients with T2DM characterized by dysregulated miRNAs profile which are probably involved in pathogenesis of CAD. The results of the current study support the hypothesis that miRNA expression is deregulated in epicardial adipose tissue in patients suffering from T2DM and CAD disease.
We provided a comprehensive potential novel miRNAs expression signature of EAT in CAD patients with T2DM, and we showed that the targets of the miRNAs are necessarily associated with the pathogenesis of CAD and T2DM.
Acknowledgments
The authors would like to acknowledge the help from the First Hospital Affiliated to Medical College of Shihezi University. The work was supported in part by NFSC 81460059 from the National Science Foundation of China and doctor/scientific Grant 2014BB019 from Xinjiang Production and Construction Crops.
Disclosure
Yang Liu and Wenbo Fu should be considered co-first authors.
Competing Interests
The authors declare that they have no competing interests regarding the publication of this paper.
Authors' Contributions
Yang Liu and Wenbo Fu contributed equally to this work.
References
- 1.Coutinho M., Gerstein H. C., Wang Y., Yusuf S. The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care. 1999;22(2):233–240. doi: 10.2337/diacare.22.2.233. [DOI] [PubMed] [Google Scholar]
- 2.Song D. K., Hong Y. S., Lee H., Oh J.-Y., Sung Y.-A., Kim Y. Increased epicardial adipose tissue thickness in type 2 diabetes mellitus and Obesity. Diabetes and Metabolism Journal. 2015;39(5):405–413. doi: 10.4093/dmj.2015.39.5.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohar D. S., Salcedo J., Hoang K. C., et al. Epicardial adipose tissue volume as a marker of coronary artery disease severity in patients with diabetes independent of coronary artery calcium: Findings from the CTRAD Study. Diabetes Research and Clinical Practice. 2014;106(2):228–235. doi: 10.1016/j.diabres.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazurek T., Zhang L., Zalewski A., et al. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation. 2003;108(20):2460–2466. doi: 10.1161/01.cir.0000099542.57313.c5. [DOI] [PubMed] [Google Scholar]
- 5.Iacobellis G., Leonetti F. Epicardial adipose tissue and insulin resistance in obese subjects. Journal of Clinical Endocrinology and Metabolism. 2005;90(11):6300–6302. doi: 10.1210/jc.2005-1087. [DOI] [PubMed] [Google Scholar]
- 6.Akbas E. M., Hamur H., Demirtas L., et al. Predictors of epicardial adipose tissue in patients with type 2 diabetes mellitus. Diabetology and Metabolic Syndrome. 2014;6(1, article 55) doi: 10.1186/1758-5996-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker A. R., Silva N. F. D., Quinn D. W., et al. Human epicardial adipose tissue expresses a pathogenic profile of adipocytokines in patients with cardiovascular disease. Cardiovascular Diabetology. 2006;5, article 1 doi: 10.1186/1475-2840-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y., Cheng X., Hong K., Huang C., Wan L. How to interpret epicardial adipose tissue as a cause of coronary artery disease: a meta-analysis. Coronary Artery Disease. 2012;23(4):227–233. doi: 10.1097/mca.0b013e328351ab2c. [DOI] [PubMed] [Google Scholar]
- 9.Noyes A. M., Dua K., Devadoss R., Chhabra L. Cardiac adipose tissue and its relationship to diabetes mellitus and cardiovascular disease. World Journal of Diabetes. 2014;5(6):868–876. doi: 10.4239/wjd.v5.i6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iacobellis G., Bianco A. C. Epicardial adipose tissue: emerging physiological, pathophysiological and clinical features. Trends in Endocrinology and Metabolism. 2011;22(11):450–457. doi: 10.1016/j.tem.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacobellis G., Corradi D., Sharma A. M. Epicardial adipose tissue: anatomic, biomolecular and clinical relationships with the heart. Nature Clinical Practice Cardiovascular Medicine. 2005;2(10):536–543. doi: 10.1038/ncpcardio0319. [DOI] [PubMed] [Google Scholar]
- 12.Wang C.-P., Hsu H.-L., Hung W.-C., et al. Increased Epicardial Adipose Tissue (EAT) volume in type 2 diabetes mellitus and association with metabolic syndrome and severity of coronary atherosclerosis. Clinical Endocrinology. 2009;70(6):876–882. doi: 10.1111/j.1365-2265.2008.03411.x. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan S., Selvan S. T., Archunan G., Gulyas B., Padmanabhan P. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics. 2013;3(12):930–942. doi: 10.7150/thno.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy S. M., Cleeman J. I., Merz C. N. B., et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. Journal of the American College of Cardiology. 2004;44(3):720–732. doi: 10.1016/j.jacc.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Fu H. L., Wu D. P., Wang X. F., et al. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochemistry and Biophysics. 2013;67(2):657–668. doi: 10.1007/s12013-013-9554-3. [DOI] [PubMed] [Google Scholar]
- 16.Huang D. W., Sherman B. T., Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D. W., Sherman B. T., Tan Q., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research. 2007;35(2):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D. W., Sherman B. T., Tan Q., et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8(9, article R183) doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle E. I., Weng S., Gollub J., et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J., Sun S., Tracy A., et al. NetAffix gene ontology mining tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20(9):1462–1463. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 21.Jensen L. J., Kuhn M., Stark M., et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research. 2009;37(1):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 2011;39(1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motawae T. M., Ismail M. F., Shabayek M. I., Seleem M. M. MicroRNAs 9 and 370 association with biochemical markers in T2D and CAD complication of T2D. PLoS ONE. 2015;10(5, article e0126957) doi: 10.1371/journal.pone.0126957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karolina D. S., Tavintharan S., Armugam A., et al. Circulating miRNA profiles in patients with metabolic syndrome. Journal of Clinical Endocrinology and Metabolism. 2012;97(12):E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 25.Bronze-Da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. BioMed Research International. 2014;2014:23. doi: 10.1155/2014/985408.985408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etheridge A., Lee I., Hood L., Galas D., Wang K. Extracellular microRNA: a new source of biomarkers. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesi. 2011;717(1-2):85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herder C., Karakas M., Koenig W. Biomarkers for the prediction of type 2 diabetes and cardiovascular disease. Clinical Pharmacology and Therapeutics. 2011;90(1):52–66. doi: 10.1038/clpt.2011.93. [DOI] [PubMed] [Google Scholar]
- 28.Vacca M., Di Eusanio M., Cariello M., et al. Integrative miRNA and whole-genome analyses of epicardial adipose tissue in patients with coronary atherosclerosis. Cardiovascular Research. 2016;109(2):228–239. doi: 10.1093/cvr/cvv266. [DOI] [PubMed] [Google Scholar]
- 29.Wang F., Long G., Zhao C., et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. Journal of Translational Medicine. 2013;11(1, article 222) doi: 10.1186/1479-5876-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoekstra M., van der Lans C. A. C., Halvorsen B., et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochemical and Biophysical Research Communications. 2010;394(3):792–797. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 31.Kremen J., Dolinkova M., Krajickova J., et al. Increased subcutaneous and epicardial adipose tissue production of proinflammatory cytokines in cardiac surgery patients: possible role in postoperative insulin resistance. The Journal of Clinical Endocrinology & Metabolism. 2006;91(11):4620–4627. doi: 10.1210/jc.2006-1044. [DOI] [PubMed] [Google Scholar]
- 32.Marchington J. M., Mattacks C. A., Pond C. M. Adipose tissue in the mammalian heart and pericardium: structure, foetal development and biochemical properties. Comparative Biochemistry and Physiology—Part B. 1989;94(2):225–232. doi: 10.1016/0305-0491(89)90337-4. [DOI] [PubMed] [Google Scholar]
- 33.Gaborit B., Venteclef N., Ancel P., et al. Human epicardial adipose tissue has a specific transcriptomic signature depending on its anatomical peri-atrial, peri-ventricular, or peri-coronary location. Cardiovascular Research. 2015;108(1):62–73. doi: 10.1093/cvr/cvv208. [DOI] [PubMed] [Google Scholar]