Abstract
Introduction
Anaplastic lymphoma kinase (ALK) rearranged lung adenocarcinomas are responsive to the multitargeted ALK inhibitor crizotinib. One of the common mechanisms of resistance to crizotinib is the acquisition of ALK kinase domain mutations. However, the presence of ALK mutations in crizotinib-naïve tumors has not been widely reported and it is unclear if de novo ALK mutations affect the response to crizotinib.
Methods
We analyzed preclinical models of ALK rearranged lung cancers that were sensitive/resistant to ALK inhibitors, probed our institutional and other lung cancer databases for tumors with ALK kinase domain mutations, and evaluated tumor response to crizotinib.
Results
ALK rearranged cell lines with ALK kinase domain mutations were heterogeneously less inhibited by increasing concentrations of crizotinib than cells driven solely by EML4-ALK fusions. Previous ALK rearranged lung cancer cohorts did not report ALK kinase mutations in inhibitor-naïve tumors. We identified one TKI-naïve ALK rearranged tumor with an ALK kinase domain mutation: ALK-S1206F (mutations at ALK-S1206 shifted crizotinib inhibitory curves only minimally in preclinical models). The never smoker woman whose tumor harbored de novo EML4-ALK-E5;A20+ALK-S1206F achieved radiographic response to crizotinib 250mg twice daily.
Conclusions
Combining data from our and prior cohorts, ALK kinase domain mutations were uncommon events (<3% of cases) in ALK inhibitor-naïve ALK rearranged lung adenocarcinomas but their effect on intrinsic resistance to ALK inhibitors should be better evaluated.
Keywords: mutation, lung cancer, adenocarcinoma, ALK, kinase domain, crizotinib
INTRODUCTION
Anaplastic lymphoma kinase (ALK) rearrangements lead to oncogene addiction in around 5% of lung adenocarcinomas [1–3]. In preclinical models, ALK fusion proteins retain the tyrosine kinase domain (amino acid [aa] 1116 to 1383, encompassing exons 22–25) of the kinase that in turns engages proliferative and antiapoptotic downstream targets of the mitogen-activated protein kinase (MAPK)/extracellular-signal-regulated kinase (ERK) and phosphatidylinositol-3-kinases (PI3K)/protein kinase B (AKT) cascades [2]. Multitargeted ALK tyrosine kinase inhibitors (TKIs) are capable of inhibiting these intricate signaling networks to induce antiproliferative and proapoptotic effects [4, 5]. In the clinic, the multitargeted TKI crizotinib induces tumor regression in almost all cases of advanced ALK rearranged non-small-cell lung cancer (NSCLC) for a period of time before the tumor acquires resistance to monotherapy [1, 6]. The most common mechanisms of acquired resistance to crizotinib in preclinical models and clinical specimens include: ALK kinase domain mutations that shift the sensitivity profile of crizotinib [7], activation of other oncogenes (such as epidermal growth factor receptor [EGFR]) that lead to signaling bypass tracks [8], and inadequate pharmacokinetic exposure [9]. ALK kinase domain mutations - including ALK-1151Tins, L1196M, G1202R and G1269A - when present with EML4-ALK fusion proteins heterogeneously change the pattern of sensitivity to different ALK TKIs; with some resistant and others sensitive to the more potent inhibitors ceritinib, alectinib, brigatinib and lorlatinib [3, 10].
The presence of de novo ALK kinase domain mutations concurrently with ALK rearrangements in ALK TKI-naïve NSCLCs has not been reported previously by Thoracic Oncology multidisciplinary groups [7, 11, 12] or The Cancer Genome Atlas (TCGA) database [13]; and it is therefore unclear if de novo ALK mutations affect the response to evidence-based crizotinib in patients. Herein, we report the uncommon but present occurrence of de novo ALK kinase domain mutations in our institutional database of ALK rearranged NSCLCs and evaluate preclinical models to correlate with tumor response to crizotinib.
METHODS
Cell culture, proliferation assays and reagents
NCI-H3122 (H3122) cells, harboring EML4-ALK-E13;A20, were obtained as previously described [4, 5]. Variants of H3122 to model mechanisms of resistance to crizotinib, including H3122 CR1 (with ALK-L1196M provided by Dr. Jeffrey Engelman) and H3122 CR_A (with EGFR bypass track), were derived after long term exposure to crizotinib, as previously reported [7, 8]. All cells were maintained in RPMI-1640 medium (Mediatech, Manassas, VA) appended with one tenth fetal bovine serum and grown in usual conditions [14, 15].
Reagents
Crizotinib and afatinib were purchased from LC Laboratories (Woburn, MA). Ceritinib was purchased from Active Biochemicals (Hong Kong). All reagents were dissolved in dimethyl sulfoxide (DMSO) and stored at −80°C.
Proliferation assay, western blot and antibodies
Selected cells were overlaid in 96-well plates, allowed to attach and then treated with or without TKIs (crizotinib, certinib or afatinib) for prespecified time points. Cell viability was determined by CellTiter 96 Aqueous One solution proliferation assay (Promega, Madison, WI). Experiments were performed in triplicate. Inhibitory proliferation curves and the 50% inhibitory concentration (IC50) were made using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). To highlight in an isogenic system different ALK kinase domain mutants’ effects on EML4-ALK, we recycled previously published IC50 values obtained using modified Ba/F3 cells from Friboulet et al [10]. H3122, H3122 CR1 and H3122 CR_A were lysed after exposure to TKIs or DMSO, lysates were separated, transferred to membranes and analyzed as previously described [8, 16]. AKT, phospho-AKT (pS473), phospho-ERK1/2 (pT202/pY204), phospho-ALK (pY1604) and ALK were purchased from Cell Signaling Technology (Beverly, MA). EGFR was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). ERK1/2 antibody was purchased from BD Transduction Laboratories (Lexington, KY). Primary antibodies were diluted 1:1000 and their recommended secondary antibodies diluted 1:10000.
Tumor and data collection
Patient-tumor pairs followed at the Thoracic Oncology multidisciplinary clinic at Beth Israel Deaconess Medical Center (BIDMC) with a diagnosis of NSCLC were registered through an ongoing Institutional Review Board-approved study [17–19]. Pathologic data, tumor genotype and radiographic images were assembled from retrospective chart extraction. Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 was utilized. Data was collected and managed using the REDCap electronic data capture held at BIDMC. Additional published ALK rearranged NSCLC cohorts were identified by literature review [7, 11, 12]. In addition, the 2014 TCGA lung adenocarcinoma mutation database [13] was reviewed and collated for ALK genotypes and co-existing mutations using cBioPortal (http://www.cbioportal.org/index.do).
ALK fusion partner and kinase domain analysis
ALK fluorescence in situ hybridization (FISH) using the Vysis break apart probe set, as detailed previously [18], was our institution’s screening test for ALK rearrangements. RNA was isolated from tumor tissue for evaluation of fusion partners using PCR-based or next generation sequencing assays, while DNA was isolated to sequence exons corresponding to the kinase domain of ALK (exons 21 to 27), as previously reported [16, 17, 20, 21].
RESULTS
Preclinical models of ALK rearranged NSCLC and resistance to crizotinib
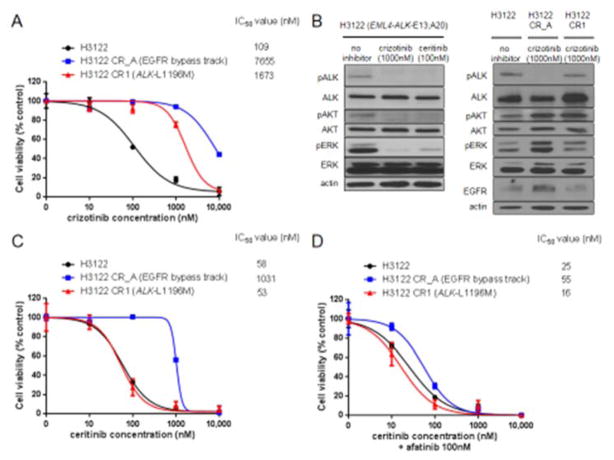
We selected one cell line (H3122 with EML4-ALK-E13;A20) to mimic sensitivity to ALK TKIs and two representative cell lines (H3122 CR1 and H3122 CR_A) to mirror crizotinib resistance. H3122 had the lowest nanomolar IC50 values of dose-dependent proliferation experiments for crizotinib; while H3122 CR1 (with the ALK-L1196M mutation as the resistance mechanism) and H3122 CR_A (with EGFR bypass as the resistance mechanism) were not inhibited by clinically achievable levels [22] of crizotinib (Fig. 1A). Intracellular signaling analysis indicated that ALK TKIs suppressed phosphorylation of ALK, AKT and ERK in H3122 (Fig. 1B). Crizotinib was unable to inhibit phosphorylation of ALK, AKT or ERK in H3122 CR1, whereas it inhibited ALK in H3122 CR_A but enhanced EGFR levels continued to drive AKT and ERK (Fig. 1B).
Figure 1. Preclinical models of acquired resistance to crizotinib in ALK rearranged lung cancers.
A. Dose-inhibition curves for crizotinib using H3122, H3122 CR_A and H3122 CR1, with 50% inhibitory concentration (IC50) using nanomolar (nM) concentrations indicated. B. Western blot results showing the intracellular signaling effects of crizotinib 1000nM and certinib 100nM after 6 hours of exposure to H3122 cells, with inhibition of phosphorylated (p) levels of each protein indicating drug activity. The same intracellular signaling is shown for H3122 CR_A and H3122 CR1 cells grown in the presence of continuous 1000nM of crizotinib. C. Dose-inhibition curves for ceritinib using H3122, H3122 CR_A and H3122 CR1, with IC50 concentrations indicated. D. Dose-inhibition curves for ceritinib in the presence of afatinib 100nM using H3122, H3122 CR_A and H3122 CR1, with IC50 concentrations indicated.
To further confirm the aforementioned mechanisms of resistance to crizotinib, we were able to show that the more potent ALK TKI ceritinib was able to inhibit the proliferation of H3122 CR1 but not of H3122 CR_A (Fig. 1C). Co-inhibition of ALK and EGFR with ceritinib and afatinib, respectively, led to antiprofilerative effects at low ceritinib doses in H3122 CR_A (Fig. 1D).
These preclinical models confirmed that ALK kinase domain mutations (such as ALK-L1196M) can induce resistance to crizotinib and led us to question if de novo ALK mutations could be present in TKI-naïve ALK rearranged NSCLCs and/or could lead to primary insensitivity to crizotinib.
Tumors with ALK rearrangements and de novo ALK kinase domain mutations
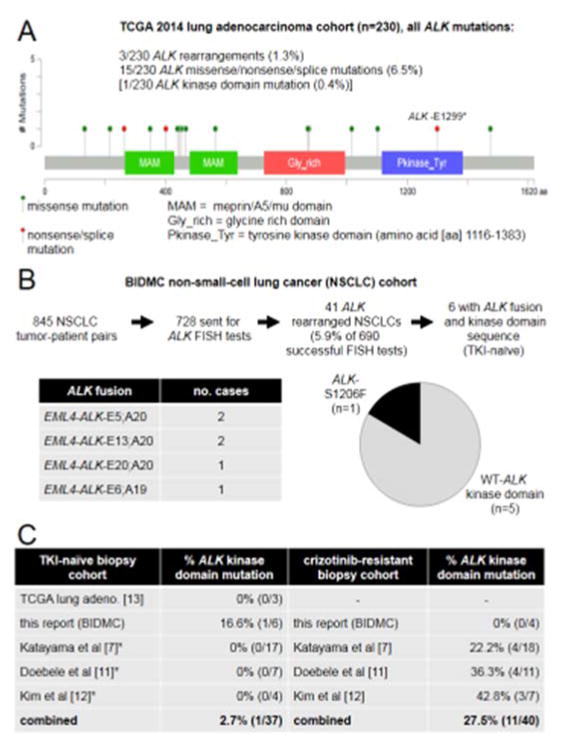
To evaluate the spectrum of genomic ALK changes in NSCLC, we first turned to the TCGA 2014 cohort of 230 surgically resected lung adenocarcinomas [13]. The frequency of ALK rearrangements was 1.3% (none co-occurring with ALK mutations) and of any type of ALK exon mutation 6.5% (Fig. 2A). Interestingly, most mutations seems to be “passenger events” (e.g., outside the catalytic kinase domain) and only one sample (without co-occurring ALK rearrangement) had an ALK kinase domain mutation, and the mutation reported was an ALK-E1299* that would be considered inactivating/non-functional (Fig. 2B). Our BIDMC NSCLC cohort identified ALK rearrangements in 5.9% (41/690) of TKI-naïve cases screened by ALK FISH, and detailed information on fusion partners plus ALK kinase domain sequence was available for 6 tumors (Fig. 2B). One case with EML4-ALK-E5;A20 also harbored a de novo ALK-S1206F mutation.
Figure 2. ALK kinase domain mutations in different NSCLC cohorts.
A. Frequencies of ALK genomic changes and graphic representation of the ALK protein with ALK mutations identified in the TCGA 2014 lung adenocarcinoma cohort indicated. B. ALK rearrangements and ALK kinase domain mutations identified in the BIDMC NSCLC tumor-pair cohort. C. Tabulated ALK rearranged NSCLC cohort and percentage (%) of ALK kinase domain mutations in TKI-naïve and crizotinib-resistant biopsies. References to the original publication of the cohort are indicated in parenthesis. * We assumed that crizotinib-resistant samples with wild-type (WT) ALK kinase domain would also have lacked a mutation in the TKI-naïve setting and we excluded cases that had ALK kinase mutations in the crizotinib-resistant setting but were not analyzed in the TKI-naïve biopsy.
To further expand the aforementioned results, we collated data from three published cohorts of advanced ALK rearranged NSCLCs in which information was available in crizotinib-naïve and –resistant biopsy specimens. None of these studies reported de novo ALK kinase domain mutations, and combining all cohorts one can come up with a frequency of only 2.7% (1/37) for ALK kinase mutations (Fig. 2C) in TKI-naïve ALK rearranged NSCLC (Fig. 2C). The latter numbers contrast with the combined frequency of 27.5% of activating ALK kinase domain mutations in crizotinib-resistant rebiopsy samples (Fig. 2D; p=0.0035 compared to TKI-naïve, Fisher’s exact test).
Preclinical evaluation of ALK kinase domain mutations
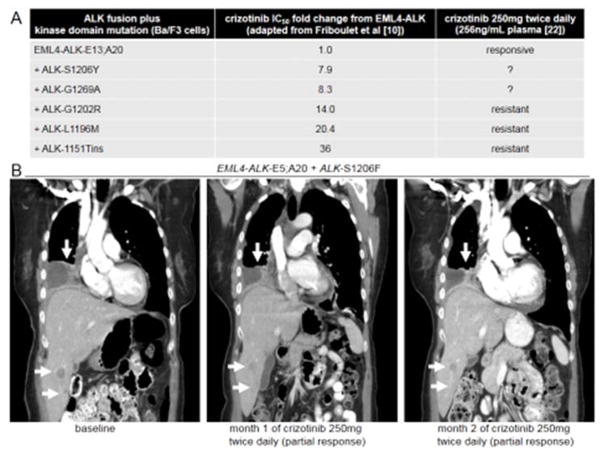
We were curious to understand the extent of heterogeneity between different types of ALK kinase domain mutant proteins against crizotinib in the background of EML4-ALK. We identified a previously published report, from Friboulet et al [10], with a comprehensive set of Ba/F3 cell lines driven by EML4-ALK-E13;A20 and a variety of patient-identified ALK kinase domain mutants treated with crizotinib (Fig. 3A).
Figure 3. ALK kinase domain mutants and EML4-ALK+ALK-S1206F.
A. Crizotinib 50% inhibitory concentration (IC50) fold changes induced by different ALK kinase mutants in the background of EML4-ALK-E13;A20, as reported by Friboulet et al (reference 10), tabulated with possible clinical response to achievable concentrations of crizotinib 250mg twice daily. B. Computed tomography (CT) to exemplify response to crizotinib in the patient whose lung adenocarcinoma harbored de novo EML4-ALK-E5;A20+ALK-S1206F. Shown are pre-crizotinib (baseline), month 1 and month 2 of crizotinib 250mg twice daily representative images. The white arrows highlight sites of tumor burden in the thorax and liver.
Of interest, the ALK mutant at position S1206 (ALK-S1206Y) showed the least fold inhibition of crizotinib IC50 when compared to EML4-ALK (Fig. 3A), raising the question if this mutant coexisting with EML4-ALK would or would not lead to clinical resistance in a patient in which crizotinib was given at the initial prescribed dose of 250mg twice daily [3, 9]. Whereas other ALK mutants (such as ALK-G1202R, L1196M, 1151Tins) had IC50s >10 fold higher than EML4-ALK-E13;A20, and would be expected to lead to resistance to achievable concentrations of crizotinib (Fig. 3A).
Radiographic response to crizotinib in lung adenocarcinoma harboring de novo EML4-ALK-E5;A20+ALK-S1206F
The patient with de novo EML4-ALK-E5;A20+ALK-S1206F mutated lung adenocarcinoma was a 69 year-old never smoker woman who presented with pulmonary, pleural, nodal, osseous and hepatic metastases (Fig. 3B). The diagnostic specimen analyzed was a pleural effusion cellblock with 60% tumor content. 47% of cells were ALK FISH positive [nuc_ish (3′ALKx2~4,5′ALKx1~2)(3′ALKcon5′ALKx1~2)-(94/200)], the allele frequency of ALK-S1206F was 28% (i.e., one mutant allele per tumor cell) and TP53-R175C was additionally identified. The patient was prescribed crizotinib at 250mg twice daily and tolerated therapy without significant gastro-intestinal, visual or laboratorial adverse events. Within one week of therapy her clinical condition improved and imaging studies at the one month mark of crizotinib therapy disclosed remarkable decrease in tumor burden that was maintained at the two month mark (Fig. 3B), compatible with partial response by RECISTv1.1 with 30.3% decrease in target lesions. The patient continues on therapy at time of this report (month four of therapy).
The clinical/radiographic response suggests that the ALK-S1206F mutant, similar to ALK-S1206Y (Fig. 3A), may be fully inhibited in the background of EML4-ALK by initial plasma and tumor concentrations achieved by crizotinib 250mg twice daily in vivo [22].
DISCUSSION
The use of crizotinib and other multitargeted ALK TKIs has revolutionized the care of ALK rearranged NSCLCs; however, acquired resistance usually ensues after a period of 6–24 months [3]. ALK kinase domain mutations have been recognized in a portion of rebiopsy specimens upon progression on crizotinib, ceritinib, alectinib and lorlatinib [7, 10–12, 23]. Different mutations have divergent patterns of sensitivity to ALK TKIs in preclinical models. As examples, EML4-ALK with ALK-G1202R leads to inhibitory curves that suggest resistance to crizotinib, ceritinib, alectinib and brigatinib, but sensitivity to lorlatinib [10, 23]; while EML4-ALK with ALK-L1196M leads to resistance only to crizotinib and sensitivity to other aforementioned ALK TKIs [7, 10]. Approximately one third, as confirmed in this report (Fig. 1C), of ALK-driven tumors at time of acquired resistance to crizotinib display ALK kinase domain mutations while the same frequencies are unclear for other ALK TKIs. NSCLC cells can modulate p-glycoprotein expression as a mechanisms of systemic pharmacokinetic resistance to crizotinib [24], and this observation raises concern that crizotinib concentrations may decrease linearly over time within the cancer cell to a level where even “less resistant” ALK kinase mutants can exert inhibitory effects.
Our group questioned if de novo ALK kinase domain mutations co-occur with EML4-ALK and if these compound genomic changes explain some of the rare cases of primary insensitivity of ALK-driven NSCLCs to crizotinib. The data presented here suggest that de novo ALK kinase domain mutations are uncommon in tumors with ALK rearrangements (<3% of cases) and even less common in NSCLCs in general (<1% of cases). Nonetheless, the only TKI-naïve case with co-occurring EML4-ALK-E5;A20 plus ALK-S1206F was quite instructive and shows how clinical-level data can complement preclinical systems. The patient received crizotinib 250mg twice daily and had a rapid response to TKI monotherapy (Fig. 3B), suggesting that the minimal fold change of crizotinib by ALK-S1206 mutations [10, 25] in preclinical systems (Fig. 3A) is insufficient to offset the inhibition by initial achievable intratumor levels of crizotinib at its recommended starting dose [6, 22]. The presence of different de novo ALK kinase mutations (such as ALK-L1196M or G1202R that in vitro lead to many fold levels of resistance to crizotinib [10, 23]) could have led to a scenario where crizotinib at 250mg twice daily would have been incapable of overcoming the inhibitory hurdles of these mutants in ALK rearranged NSCLC and another ALK TKI should have been selected to best match the sensitivity pattern of the compound EML4-ALK plus ALK kinase domain mutant. Additional reports of these rare cases of ALK rearrangements co-occurring with ALK kinase mutations prior to starting an ALK TKI may shed light in these intriguing clinical decisions that can further improve precision oncology choices for this important cohort of NSCLCs and help explain hitherto unexplained cases of primary insensitivity to crizotinib in clinical practice.
In summary, our institutional database combined with prior cohorts demonstrates that ALK kinase domain mutations seem to be uncommon events in ALK inhibitor-naïve ALK rearranged lung adenocarcinomas but their effect on intrinsic resistance to achievable doses of clinically-available ALK inhibitors should be better evaluated in preclinical models and clinical cases.
HIGHLIGHTS.
ALK rearranged lung adenocarcinomas are responsive to crizotinib
However, ALK kinase mutations in crizotinib-naïve tumors have not been widely reported
ALK kinase mutations were uncommon (<3%) in ALK inhibitor-naïve lung adenocarcinomas
Acknowledgments
This work was funded in part through an American Cancer Society grant RSG 11-186 (DBC), and National Cancer Institute grants P50CA090578 (DBC), R01CA169259 (SSK) and R21CA17830 (SSK).
Footnotes
CONFLICT OF INTEREST STATEMENT
Daniel B. Costa has received consulting fees and honoraria from Pfizer Inc (unrelated to the current work), consulting fees from Ariad Pharmaceuticals (unrelated to the current work), and honoraria from Boehringer Ingelheim (unrelated to the current work).
Antonio R. Lucena-Araujo, Jason P. Moran, Paul A. VanderLaan, Dora Dias-Santagata, Erik Folch, Adnan Majid, Michael S. Kent, Sidharta P. Gangadharan, Deepa Rangachari, Mark S. Huberman, and Susumu S. Kobayashi have no conflicts to disclose.
No other conflict of interest is stated.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 3.Costa DB. ALK inhibitors: plateauing systemic and intracranial activity? Lancet Oncol. 2016;17:404–06. doi: 10.1016/S1470-2045(16)00025-5. [DOI] [PubMed] [Google Scholar]
- 4.Yasuda H, Figueiredo-Pontes LL, Kobayashi S, Costa DB. Preclinical Rationale for Use of the Clinically Available Multitargeted Tyrosine Kinase Inhibitor Crizotinib in ROS1-Translocated Lung Cancer. J Thorac Oncol. 2012;7:1086–90. doi: 10.1097/JTO.0b013e3182570919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorge SE, Schulman S, Freed JA, et al. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer. 2015;90:369–74. doi: 10.1016/j.lungcan.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa DB, Shaw AT, Ou SH, et al. Clinical Experience With Crizotinib in Patients With Advanced ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastases. J Clin Oncol. 2015;33:1881–8. doi: 10.1200/JCO.2014.59.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4:120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi N, Lucena-Araujo AR, Nakayama S, et al. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung Cancer. 2014;83:37–43. doi: 10.1016/j.lungcan.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa DB, Kobayashi S, Pandya SS, et al. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol. 2011;29:e443–e445. doi: 10.1200/JCO.2010.34.1313. [DOI] [PubMed] [Google Scholar]
- 10.Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–73. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–82. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, Kim TM, Kim DW, et al. Heterogeneity of Genetic Changes Associated with Acquired Crizotinib Resistance in ALK-Rearranged Lung Cancer. J Thorac Oncol. 2013;8:415–22. doi: 10.1097/JTO.0b013e318283dcc0. [DOI] [PubMed] [Google Scholar]
- 13.Collisson EA, Campbell JD, Brooks AN Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–79. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Figueiredo-Pontes LL, Wong DW, Tin VP, et al. Identification and Characterization of ALK Kinase Splicing Isoforms in Non-Small-Cell Lung Cancer. J Thorac Oncol. 2014;9:248–53. doi: 10.1097/JTO.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangachari D, VanderLaan PA, Le X, et al. Experience with targeted next generation sequencing for the care of lung cancer: insights into promises and limitations of genomic oncology in day-to-day practice. Cancer Treat Commun. 2015;4:174–81. doi: 10.1016/j.ctrc.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanderLaan PA, Yamaguchi N, Folch E, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer. 2014;84:39–44. doi: 10.1016/j.lungcan.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folch E, Yamaguchi N, VanderLaan PA, et al. Adequacy of lymph node transbronchial needle aspirates using convex probe endobronchial ultrasound for multiple tumor genotyping techniques in non-small-cell lung cancer. J Thorac Oncol. 2013;8:1438–44. doi: 10.1097/JTO.0b013e3182a471a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–58. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–84. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 22.Tan W, Wilner KD, Bang Y, et al. Pharmacokinetics (PK) of PF-02341066, a dual ALK/MET inhibitor after multiple oral doses to advanced cancer patients. J Clin Oncol. 2010;28 (suppl; abstract 2596) [Google Scholar]
- 23.Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med. 2016;374:54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama R, Sakashita T, Yanagitani N, et al. P-glycoprotein Mediates Ceritinib Resistance in Anaplastic Lymphoma Kinase-rearranged Non-small Cell Lung Cancer. EBioMedicine. 2016;3:54–66. doi: 10.1016/j.ebiom.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Wang F, Keats J, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]