Abstract
Objective
Viral replication and interstitial inflammation play important roles in the pathogenesis of HIV-associated nephropathy (HIVAN). Cell-cell interactions between renal tubule epithelial cells (RTEC) and HIV-infected T cells can trigger efficient virus internalization and viral gene expression by RTEC. To understand how HIV replication initiates HIVAN, we studied the cellular response of RTECs to HIV, examining the transcriptional profiles of primary RTECs exposed to cell-free HIV or HIV-infected T cells.
Methods
HIV-induced gene expression in hRTECs was examined in vitro by Illumina RNA deep sequencing and revealed an innate response to HIV, which was subclassified by Gene Ontology (GO) biological process terms. Chemokine responses were examined by CD4+ T cell chemotaxis assays.
Results
As compared to cell-free virus infection, exposure to HIV-infected T cells elicited a stronger up-regulation of inflammatory and immune response genes. A major category of up-regulated genes are chemokine/cytokine families involved in inflammation and immune response, including inflammatory cytokines CCL20, IL6 and IL8-related chemokines: IL8, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6. Supernatants from virus-exposed RTECs contained strong chemoattractant activity on primary CD4+ T cells, which was potently blocked by a CXCR2 antagonist that antagonizes IL8-related chemokines. We observed a preferential migration of CXCR2-expressing, central memory CD4+ T cells in response to HIV infection of RTECs.
Conclusion
Interactions between primary RTECs and HIV-infected T cells result in potent induction of inflammatory response genes and release of cytokines/chemokines from RTECs that can attract additional T cells.. Activation of these genes reflects an innate response to HIV by non-immune cells.
Keywords: HIV, HIV associated nephropathy, renal tubule cell, expression profile, inflammation, chemoattractant, CXCR2
INTRODUCTION
HIV infection results in progressive CD4+ T cell depletion in immune compartments, and also causes significant pathology in non-immune organs. Advanced HIV disease can induce renal pathology and impairment of renal function called HIV-associated nephropathy (HIVAN) [1]. Clinically, HIVAN is characterized as an aggressive glomerulopathy that afflicts those with genetic risk factors, occurring more frequently in patients of West African descent[2, 3] [. Prior to the advent of highly active antiretroviral therapy, HIVAN was a leading cause of end-stage renal disease (ESRD) among African Americans in the early 1990s. Histologically, it is diagnosed as a collapsing focal segmental glomerulosclerosis (FSGS) with tubulointerstial injury and inflammation[3]. Infection of renal epithelial cells is thought to play a critical factor in promoting HIVAN in HIV-infected individuals [4, 5]. In vitro studies support that HIV can infect renal epithelial cells via unconventional CD4-independent mechanisms and induce changes in multiple host cellular pathways, especially in renal tubular cells [4, 6, 7].
A role for HIV gene expression in the pathogenesis of HIVAN is supported by an HIV-transgenic mouse (Tg26) model [8] in which HIV transgene expression within kidney induces pathological changes that resemble HIVAN. In vitro infection of renal epithelial cells with HIV results in up-regulation of cellular pathways involved in cell cycle arrest and apoptosis [5, 9]. Expression of single HIV accessory proteins such as nef and vpr are sufficient to cause cell dedifferentiation and proliferation in podocytes and cell hypertrophy in tubular epithelial cells [9, 10].
The host response to HIV infection plays a crucial role in the development of HIVAN. The pathological changes of HIVAN feature collapsed focal segmental glomerulosclerosis (FSGS), tubular dilation, interstitial inflammation and fibrosis [11]. To understand the mechanism of cellular response to HIV infection, most efforts have focused on immune cells such as monocytes or T cells [12, 13]. Comparatively, the cellular response to HIV infection in epithelial cells is less well understood as in vitro infection of renal epithelial cells by HIV is inefficient [14, 15]. To study the effects of HIV in renal cells, prior studies have infected epithelial cells with replication incompetent HIV deleted in Gag and Pol genes (pNL4-3 ΔG/P-EGFP) and pseudotyped with the vesicular stomatitis virus (VSV) glycoprotein. Transduction with pseudotyped HIV induced upregulation of inflammatory genes in renal epithelial cells, which could be related to the prominent tubulo-interstitial inflammation in the transgenic mouse model for HIVAN [16]. Infiltration of leukocytes into the tubulo-interstitial compartment is a hallmark of many renal inflammatory diseases and is an important mediator of tubular injury leading to progressive renal failure in HIVAN [17, 18]. Rather than being a passive target, resident proximal tubular epithelial cells are thought to play an active role in the inflammatory process through the elaboration of cytokines/chemokines that communicate with interstitial immune cells [19]. Activation of chemokines may explain immune cell entry into the kidney during HIV infection that underlies the high prevalence of interstitial nephritis found in kidney biopsies of HIV patients[20] even in the absence of HIVAN. The recruitment of HIV infected cells to the kidney may also play a role in reduced survival of kidney allografts after transplantation despite undetectable viremia [21].
Our previous studies on the interaction between HIV-infected T cells and renal epithelial cells revealed efficient cell-cell contact mediated virus uptake, as well as viral gene expression in renal epithelial cells [6]. In this study, we investigate the cellular response of primary renal epithelial cells upon exposure to either cell-free, or primary T cells infected with HIV-1 without the use of viral pseudotypes or transgene expression to enhance the delivery of HIV to cells. We find that as compared to cell-free virus, cell-associated virus infection of renal epithelial cells evoked larger number of genes with greater magnitude of changes, particularly genes related to immune/inflammatory response, and chemotaxis. The transcriptional response was surprisingly robust even without the requirement for pseudotyped virus to enhance viral transduction. Trans-well cell migration of T cells, especially central memory T cells, in response to the supernatants collected from HIV infected renal epithelial cells demonstrate that non-pseudotyped, virus-exposed primary epithelial cells can activate immune cell recruitment.
CONCISE METHODS
Cells and tissue culture
Primary Human Renal Cortical Epithelial Cells (HRCEpC) isolated from the cortex of the human kidney and stain positive for cytokeratin were obtained from a commercial source (PromoCell GmbH, Germany, #C-12660) and are of unknown racial background. HRCEpC were cultured in Renal Epithelial Cell Growth Medium 2 (PromoCell GmbH, Germany, #C-26030). Human peripheral blood CD4+ T cells were isolated from buffy coat cells obtained from New York Blood Center, using CD4+ T cell isolation kit II from Macs Miltenyi Biotec (#130-091-155) according to manufacture’s instructions. CD4+ T cell were cultured in RMPI1640 medium plus 10% fetal bovine serum (FBS) and penicillin streptomycin, or stimulated with phytohemaglutanin (PHA, 1ug/ml) for 48–72 hours and maintained with IL-2 (10 units/ml).
HIV infection of CD4+ T cells
Infectious virus (NL-GI) was produced by transient transfection of human embryonic kidney cells, 293T [22]. One million CD4+ cells were spinnoculated with 25 ng of cell-free virus. Virus expression was monitored by GFP signal using flow cytometry. At 48h after infection, infected cells were washed and resuspended in culture medium at a density of 2 × 106/ml and used as donors in all experiments described hereafter.
CD4+ T cell and RTEC coculture
Cell-associated virus infection was performed in a coculture system using HIV infected CD4+ T cell as donor and HRCEpC cell as target as previous described [6] with minor modifications. The target cells were labeled with 2 μM of CellTracker orange CMTMR fluorescent dye (Molecular Probes, USA) at 37°C for 30 min, according to manufacturer’s instructions. Dye-labeled target cells were washed with PBS, cultured overnight and washed again with PBS before coculture with donor cells. Donor cells were added to target epithelial monolayer at a ratio of 4:1 and cocultured for 3h. The coculture was terminated by removing the donor cells and performing three PBS washes. Adherent cells were detached by trypsin-EDTA treatment at 37°C for 5 min, washed once before being single-cell sorted by flow cytometry using a biohazard-contained flow sorter. Sorted cells were cultured alone for the indicated time. RNA was extracted from cell pellets and culture supernatant was collected at each time point post infection. Non-infected primary CD4+ T cells were also included as a control group (cell-mock infection). Cell-free virus infection was performed as previously described [6]
Differential gene expression analysis
RNA sequencing was performed at Duke sequencing facility using 50bp single read at 6 multiplexing per lane resulted in total reads per lane at 1.18 – 1.6 Gb. The RNAseq data was analyzed by following the procedure described below. Briefly, after sequence quality filtering at a cutoff of a minimum quality score Q20 in at least 90% bases, the good quality reads were aligned to several human reference databases including mm9 genome, RefSeq exons, splicing junction sequences as well as contamination sequences of human ribosomes, mitochondria and phix genome using BWA (Burrows-Wheeler Alignment) algorithm[23]. The alignments with greater than 2 mismatches were discarded. After filtering out the reads that aligned to contaminating sequences, the reads that are uniquely aligned to the exon and splicing-junction sites for each transcript were combined to calculate an expression level for a corresponding transcript and further normalized based on reads per kilobase per million reads (RPKM) to compare transcription levels amongst samples. Gene expression values were transformed to the log 2 base scale. The differentially expressed genes in HIV-infected vs. control cell line at 12, 24 or 72 hours were identified by the R package DEGseq and the common differentially-expressed genes from duplicated experiments were subjected to Gene Ontology enrichment by Fisher-exact test. The genes that changed over time from 12 to 72 hours for cell-free or cell-associated HIV infection were identified and compared.
Quantitative real time PCR for cytokine/chemokine mRNA level
Quantitative real time PCR (qRT-PCR) was performed to confirm the RNA sequencing data. Primer sets for human cytokines/chemokines as well as the control housekeeping gene GAPDH are list in Supplemental Table 3. To exclude the contamination of carryover donor cells, we purified target RTECs cells using flow sorting by gating the CMTMR-labeled RTECs population with exclusion of doublets. Flow sorted RTECs were cultured and cell pellets were obtained at 1, 2, 3 and 4 days post infection. Total cellular RNA was extracted using RNeasy Mini Kit, according to the manufacturer’s instructions (Qiagen, Cat #74104). DNA was removed by digestion with DNAse for 30 minutes at room temperature. Quantitative RT-PCR was performed using ABI7900 Real-Time PCR machine. Level of human cytokine/chemokine RNA was normalized to the expression of the housekeeping gene GAPDH.
Protein level of cytokine/chemokine by Flowcytomix
To confirm the expression of cytokines and chemokines at the protein level, we collected the supernatants from cultured RTECs at different time points post infection. A panel of selected cytokines/chemokines was measured using the FlowCytomix Multiple Analyte Detection System developed by eBioscience. 25ul of culture supernatant were incubated with a mixture of coated beads for each analyte to be measured according to manufacturer’s instructions (eBioscience, Cat#BMS8420FF). A biotin-conjugated 2nd Ab mixture is added and followed by streptavidin-PE allowing the fluorescent signal to be detected by flowcytometry. The protein level of each analyte was determined with multiplexing analysis software provided by eBioscience.
CD4+ T cells chemotaxis
CD4+ T cell chemotaxis was performed in transwell membrane inserts with 5um pore size (Corning Inc.). Supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) were added to the bottom chamber and to serve as chemoattractant stimuli. Purified human CD4+ T cells (PHA-activated or non-activated) were added to the upper chamber and transwell migration was conducted at 37°C for the indicated times. Migrated cells were counted by flow cytometry using absolute counting beads. Cell inputs and migrated cell (cell outputs) were also immunophenotyped as indicated using antibodies against cell surface markers and analyzed with flow cytometry.
RESULTS
Though the pathology HIV-1 infection is primarily associated with the depletion of CD4+ T cells that are the dominant target of infection, the influence of these infected cells and cell-free virus on non-immune tissues can also directly causes influence the state of these tissues. We therefore have established a model system to explore the intrinsic response of primary renal tubular epithelial cells (RTEC) to cell-free or cell-associated forms of HIV-1.
Differential gene expression in RTECs after HIV infection
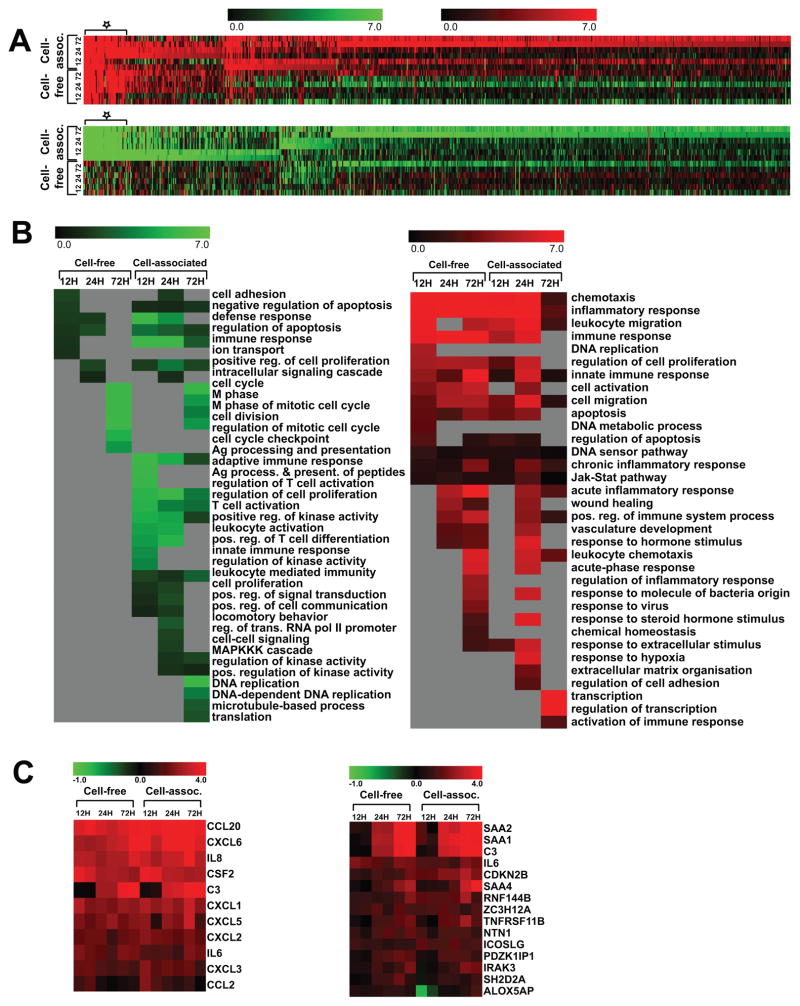
To investigate the cellular response of RTECs to HIV infection, we set up a cell co-culture system using HIV-infected human CD4+ T cells as donor and primary human renal cortical epithelial cells (HRCEpC) (Supplemental Figure 1A) [6]. We exposed HRCEpC to primary CD4+ T cells purified from healthy HIV-negative blood donors that were either infected with HIV or mock infected. We compared this cell co-culture system to direct exposure to cell-free HIV, or mock supernatants. To isolate the renal cell response to HIV and not the T cell response, the HIV-exposed HRCEpC were purified to exclude T cell contamination by flow cytometric sorting. The purified cells were cultured for 12, 24 and 72 hours and cellular RNAs were extracted for RNA deep sequencing. A time course of gene expression profiles of cell-free and cell-associated HIV-challenged RTECs along with the control cell lines (cell mock infection) were performed by Illumina RNA sequencing by multiplexing 6 samples into one lane, resulting in at least 20 million clean reads per sample of 50bp single ended reads. The data were processed and analyzed in the procedure depicted in Supplemental Figure 1B. After mapping the clean reads to human genome and RefSeq transcripts (UCSC hg19 build) using Burrows-Wheeler Alignment (BWA) algorithm, the differentially expressed genes in HIV-challenged cells compared to cell mock infection at 12, 24 or 72 hours post infection (hpi) were identified with a cutoff of p value of 0.01 and a minimum 1.5-fold change in both duplicate experiments (Figure 1A, Supplemental Table 1). We observed more genes changed at 72 hours than 12 or 24 hpi in both cell-free and cell-associated infection at higher magnitude. Interestingly, many more genes changed in cell-associated infection than cell-free HIV infection at all time points, which is even more significant at 72 hpi, with 1170 regulated genes in cell-associated infection versus only 214 genes for cell-free infection. (Supplemental Table 1). We have previously observed that cell-cell contact resulted in a much greater quantity of HIV transfer to RTECs than that achieved by exposure to large amounts of cell-free virus [6]. These data indicate that infection through exposure to HIV-infected T cells provoked stronger cellular responses than exposure to cell-free virus. Gene Ontology enrichment analysis of biological process terms was performed to further investigate the regulated genes and significant functions and data were presented by p value heatmaps (Figure 1B right: up-regulated GO terms, 34 in total; Left: down-regulated GO terms, 39 in total). As expected, a greater number of significant GO function terms were identified in cell-associated infection than cell-free infection for both up- and down-regulated GO. Strikingly, cell cycle-related GO terms such as cell cycle, M phase of mitotic cell cycle, cell division and cell cycle checkpoint are the most down-regulated in both groups at 72 hpi (Figure 1B, left). These gene expression patterns may relate to previous findings that HIV gene expression such as nef or vpr can cause cell cycle arrest and cell hypertrophy[9]. Interestingly, the genes involved in adaptive immune response, regulation of T cell activation, and cell proliferation were down regulated only in cell-associated infection (Figure 1B, left). Chemotaxis, inflammatory response, leukocyte migration and immune response are on top of the lists of up-regulated GO in both cell-free and cell-associated infection (Figure 1B, right). The same is true for DNA sensor, innate immune response, chronic inflammatory response and Jak-stat pathways, although with less prominence. Transcription, activation of immune response, response to extracellular stimulus, and regulation of cell adhesion are only revealed in cell-associated HIV infection at late time points (24 and 72 hpi, Figure 1B)
Figure 1. Heat map presentation of differentially regulated genes and GO terms in renal tubular epithelial cells in response to HIV infection.
(A) Heatmap representation of the log2 fold changes of genes modulated in response to cell-free HIV or cell-associated HIV compared to the control cells without HIV infection at 12, 24 or 72 hours after infection. The upregulated genes are in the upper panel and down-regulated genes in the lower panel. We further examined the top 100 differentially regulated genes in both cell-free and cell-associated infection (Supplemental Table 2 and demarcated by asterisk).
(B) The heatmap representation of −log10 p values of significant Gene Ontology biological process terms (p<0.05) for differentially expressed genes in cell-free HIV or cell-associated HIV compared to the control cells without HIV infection at 12, 24 or 72 hours. Significant GO terms for down-regulated genes are on the left and upregulated genes on the right.
(C) Heatmap of the log2 fold changes of top 11 up-regulated cytokines/chemokines (left) and 15 selected genes involved in inflammatory response (right). These genes were upregulated at all time points of HIV infection (free virus infection and cell-associated virus infection).
We further examined the top 100 differentially regulated genes in both cell-free and cell-associated infection (Supplemental Table 2 and demarcated by asterisk in Figure 1A). With full-length, non-pseudotyped HIV and using primary renal and immune cells, we found that among the top 100 up-regulated genes, the most up-regulated are cytokines/chemokines (Figure 1C, left). CCL20, CXCL6, IL8 (CXCL8) and CSF2 are the most prominent genes affected in our cell co-culture system. Several CXCL group chemokines CXCL1, CXCL2, CXCL3, CXCL5 and CXCL6 (IL-6) were also greatly up-regulated and persisted through 72 hours.
Renal epithelial cells are thought to play an active role in the inflammatory process especially after engagement with interstitial immune cells [19]. Inflammatory genes with the greatest up-regulation in response to cell-free or cell-associated HIV, at all time points (12, 24 and 72 hpi) (Figure 1C, right) included SAA1, SAA2, C3, and SAA4. SAA1 and SAA2 were up-regulated at 24h and 72 hpi while SAA4 was only up-regulated at late time (72 hpi). Serum amyloid protein A (SAA1, 2, 3 and 4) induction is observed in response to inflammation and tissue injury. The cellular response of renal epithelial cells upon HIV infection is cytokines/chemokines, chemotaxis, leukocyte taxis and inflammatory response.
Confirmation of altered cytokines/chemokines gene expression
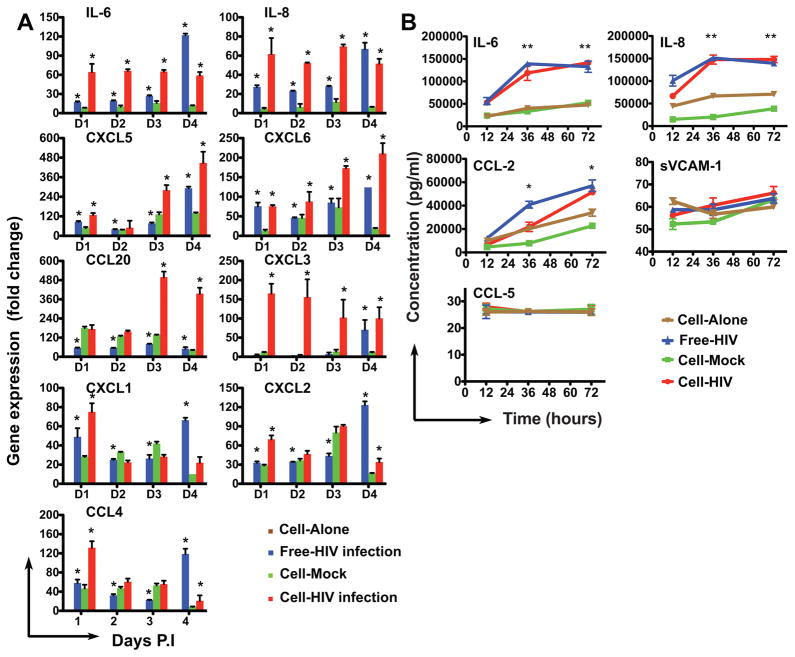
Since we found that HIV infection induced marked upregulation of cytokines/chemokines and inflammatory immune response genes in primary renal epithelial cells, we performed quantitative real-time PCR (qRT-PCR) to confirm the changes in expression of candidate cytokines/chemokines (Figure 2A). Expression of IL-6 and IL-8 increased at each time point after cell-associated HIV infection, but only increased significantly at the latest time point (4 dpi) following infection with cell-free virus. We observed a steady increase of CXCL5 and CXCL6 after both cell-free and cell-associated infection. In agreement with our RNA sequencing data, we found that following cell-associated infection, CCL20 and CXCL3 were the most strongly up regulated cytokines, particularly at the late time point of infection 3 dpi. Statistical analysis of free-hiv infection (free-hiv vs cell-alone) reveals significant changes of all cytokine (except CXCL3) RNA level at all times post infection (paired t-test, * indicates P<0.05). For cell-HIV infection, similar results were found except for cytokine CXCL1 (at D2, D3, D4 PI), CCL20, CXCL2 and CCL4 (at D2, D3 P.I) Increases in CXCL1, CXCL2 and CCL2 gene expression were also confirmed. In nearly all cases, cell-associated viral infection induced stronger cytokine gene expression than cell-free virus infection. It is important to note that we also observed cytokine gene expression in the mock infected T cell exposed control group in which RTEC were exposed to T cells that were not infected with HIV. This may relate to an intrinsic response of renal epithelial cells to infiltrating T cells [19].
Figure 2. Measurement of RNA and protein levels of genes induced in RTEC following exposure to HIV or HIV infected T cells.
A. Quantitative real time PCR measurement of RNA level in HIV infected RTECs. RNA extraction was from RTECs infected with HIV (free-HIV or cell-HIV) at 1, 2, 3 and 4 days post infection, indicated as D1–D4 respectively. Gene expression levels of individual human cytokines/chemokines were normalized to the expression of GAPDH and are represented as fold change relative to GAPDH. HIV infection (cell-free infection, cell-associated infection and cell-mock infection) were normalized to cell alone which is set to default (fold change = 1).
B. Flowcytomix measurement of protein level from HIV-infected RTECs supernatants. Supernatants were collected from HIV infected RTECS at 12, 36 and 72 hours post infection. Protein level was determined by multiplexed analysis using standard references included in the detection kit.
CD4+ T cell chemotaxis driven by HIV infection induced soluble factors
Our differential gene expression analysis and qRT-PCR confirmed that most of the genes up-regulated by HIV infection are cytokines/chemokines. We next examined whether key inflammatory/chemokine genes that were up-regulated at the mRNA level corresponded with enhanced secretion of soluble protein. Protein levels of candidate cytokines/chemokines released into the supernatant from cultured RTECs exposed to cell-free or cell-associated HIV were measured by a flow cytometry-based ELISA Flowcytomix assay as described in the methods. We found that between 36 and 72 hours a significant amount of IL-6, IL-8 and CCL-2 in the supernatants from HIV-infected cells (cell free or cell-associated infection), higher than that from mock infected control group (IL-6 and IL-8: paired t-test, p<0.001; CCL-2: paired t-test, p<0.05) (Figure 2B).
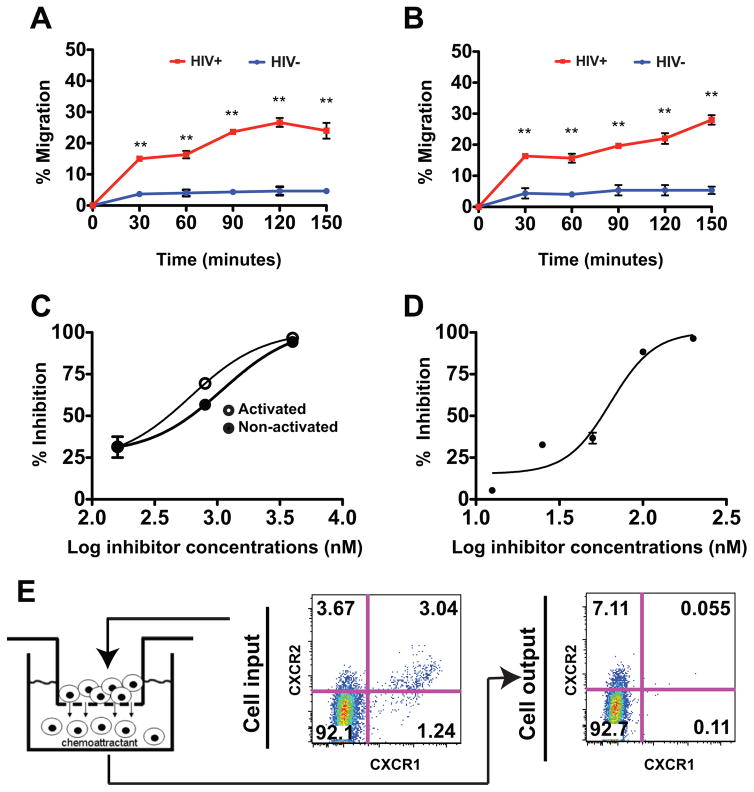
We next examined the ability of epithelial cell supernatants to induce chemotaxis of human peripheral blood CD4+ T cells. Cell supernatants released into the culture media by RTECs exposed to HIV-infected CD4+ T cells were compared to supernatants from normal RTECs for their ability to induce T cell migration in a transwell migration assay. Transwell migration of CD4+ T cells (either quiescent or PHA-activated) in response to chemoattractants released by RTEC was dependent on exposure to HIV and increased over time (paired t-test, p<0.005) (Figure 3A, 3B). We also observed chemotaxis activity of supernatants collected from cell-free HIV-infected RTECs at 1, 2 and 3 dpi (data not shown).
Figure 3. Transwell chemotaxis of human CD4+ T cells in response to supernatants from HIV infected RTEC.
(A & B) Time course chemotaxis of PHA activated (A) or non-activated (B) CD4+ T cells. Supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) were served as chemoattractants. Human CD4+ T cells were placed in the upper chamber and transwell migration proceeded at 37°C for the indicated time. Migrated cells were collected and counted by flow cytometry using absolute count beads. The percentage of migrated cells was calculated by dividing the number of migrated cells by the total the number of input cells.
(C) Pertussis toxin (PT) inhibits CD4+ T cell chemotaxis. Supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) served as chemoattractants. PHA-activated or non-activated CD4+ T cells were pretreated with inhibitor, PT, at indicated concentrations at 37°C for 30min before placed at the top chamber for migration. Migrated cells were collected at 3 hours and percentage inhibition was calculated relative to control without PT treatment.
(D) CXCR2 antagonist SB225002 blocks CD4+ T cell chemotaxis to RTEC supernatants. Supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) were used as chemoattractants. PHA activated CD4+ T cells were pretreated with CXCR2 antagonist at 37°C for 30min before addition to the top chamber. Percentage inhibition of migratory response was calculated as described in C.
(E) Enrichment of CXCR1−/CXCR2+ cells induced by migration toward HIV infected RTEC supernatants. The supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) were served as chemo attractants. Migrated cells were collected and stained with CXCR1 and CXCR2 antibodies at 4°C for 45 min. Cell surface expression of CXCR1 and CXCR2 was analyzed by flow cytometry using FlowJo software.
Chemokine-directed cell migration or chemotaxis requires the interaction with its receptor on target cells and can be blocked by receptor antagonists or inhibitors. To test if the migration of the T cells was dependent upon G-protein coupled receptors (GPCR), we pretreated target CD4+ T cells with pertussis toxin, a global inhibitor of chemotaxis mediated by GPCR. We found PT effectively blocks quiescent or PHA-activated CD4+ T cells migration with an IC50 at 200nM (Figure 3C). Many of chemokines that we observed were induced by HIV, including CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8 (IL-8) are IL-8 related chemokines that signal through CXCR2. We therefore examined the ability of SB225002 [24], a selective non-peptide CXCR2 antagonist that inhibits cytokines binding to CXCR2 to block migration of CD4+ cells in response to HIV induced RTEC superanants. We observed potent inhibitory effect of SB225002 on migration of activated CD4+ T cells, with an IC50 at 60nM (Figure 3D). To examine whether the chemotaxis response preferentially recruited particular T cell subsets, we examined the surface expression of CXCR1 and CXCR2 on the T cells before and after migration. We found that migrating CXCR1−/CXCR2+ CD4+ T cells were enriched in cells that transmigrated in response to HIV infected RTECs (increased from 3.67% to 7.11%). Surface expression of CXCR1 was diminished in the transmigrated CD4+ T cells in response to the HIV-exposed RTEC supernatants, which reflect the lack of recruitment of these cells, or the effect of receptor internalization after it binds to IL-8 related chemokines (Figure 3E). These findings may indicate that chemotaxis induced by HIV infected RTECs may preferentially recruit subsets of T cells with distinct chemokine receptor surface expression.
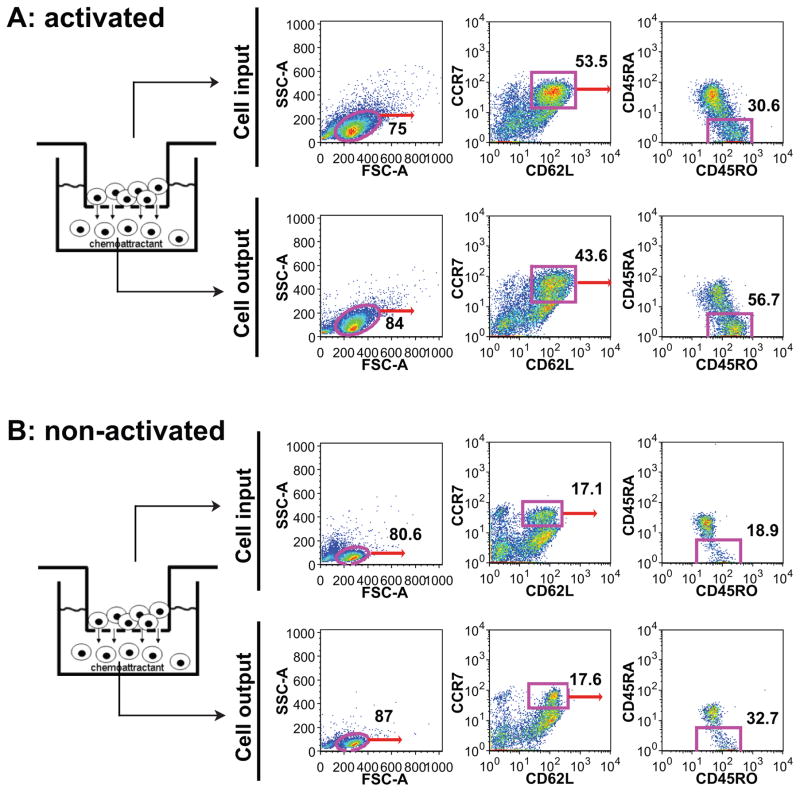
Preferential migration of central memory T cells induced by HIV infection
Recent studies indicate that memory T cells contain distinct populations of central memory (TCM) and effector memory (TEM) cells characterized by distinct homing capacity and effector function. Memory T cells efficiently traffic into inflamed tissues where they can exert effector functions if they receive the appropriate antigenic signals[25]. To investigate the possibility of preferential migration of distinct subset of CD4+ T cells, we collected supernatants from cultured RTECs at Day 3 after infection with cell-associated HIV, added it to the bottom of the transwell chamber as a chemoattractant. Using a panel of Abs (Supplemental Table 4) by flowcytometry, we examined the cell surface phenotype of central memory and effector memory T cells that migrated toward the chemoattractant. Migrated cells (cell output) as well as cells before migration (cell input) were examined for CD62L, CCR7, CD45RO and CD45RA staining. The CD62L+CCR7+CD45RO+CD45RA− cells were designated as TCM and CD62L−CCR−CD45RO+CD45RA− cells were designated as TEM Cells [26]. We found that the percentage of CD62L+ and CCR7+ double positive population is similar in cell input (53.5% vs. 43.6% for activated CD4+ T cell and 17.1% vs. 17.6% for resting CD4+ T cell) (Figure 4). However the percentage of TCM from cell output was almost doubled as compared to that from cell input (56.7% vs. 30.6% for activated CD4+ T cell and 32.7% vs. 18.9% for resting CD4+ T cell). In contrast when we examined the TEM population, we observed equal distributions of TEM in cell input and the cell output, for both the activated or non-activated CD4+ T cells (Supplemental Figure 2). These data may indicate that there is some preferential recruitment of TCM in response to chemoattractants released from HIV-infected RTECs.
Figure 4. HIV infection induced migration of central memory CD4+ T cell chemotaxis.
Supernatants collected from cultured RTECs at day 3 post HIV infection (cell-associated infection) served as chemoattractants. PHA activated (A) or non-activated (B) CD4+ T cells were placed at the top chamber. Cell migration was terminated after incubation at 37°C for three hours. Migrated cells were washed once with PBS and stained with a panel of cell marker antibodies TCM (CCR7+CD62L+CD45RO+CD45RA−) and analyzed by flow cytometry using FlowJo software.
DISCUSSIONS
We report here that exposure of primary RTEC to HIV-infected primary CD4+ T cells promotes a functional T cell chemoattractant response. This potent response occurred without the requirement for viral pseudotyping to enhance the transduction of cells, or transfection of viral DNAs as has been studied in related model systems in the past. RNA-seq analysis revealed a prominent representation of inflammatory response and immune response genes at late time after infection (24 and 72 hpi). Detailed analysis revealed that a majority of the most highly upregulated genes belong to chemokine/cytokine families, such as CCL20 and IL6, as well as IL8-related chemokines CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8 (IL8). These are involved in inflammation and immune response, and reinforce previous findings by Ross, et al[16], in which VSV-G pseudotyped, replication-defective HIV was used to enhance transduction of renal epithelial cells. We report for the first time using non-pseudotyped, replication-competent virus delivered by infected primary CD4+ T cells to initiate HIV infection in human primary RTECs through cell coculture. Productive HIV infection and HIV gene expression in primary renal epithelial cells can be achieved either by inoculation of large amount of cell-free HIV, or by coculture with HIV-infected T cells[6]. As compared to cell-free HIV infections, the cell-associated viral inocula induced stronger up-regulation of chemokine/cytokine and inflammatory immune response genes.
We performed transwell cell migration assays using the supernatant from primary RTECs infected with HIV and observed strong chemotaxis activity on CD4+ T cells. The migration appeared to be GPCR-dependent based on pertussis sensitivity, and also potently blocked by the CXCR2-selective small molecule antagonist SB225002. It is notable that a large fraction of the HIV induced chemokines in the RTE cells are IL-8 related chemokines that signal through CXCR2, including CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8 (IL-8). Though these chemokines are better known for their role in attracting neutrophils, these are also expressed on T cells [27] and thus could be involved in promoting the mononuclear infiltrates observed in HIVAN. It may be interesting to explore chemokine antagonists directed against CXCR2 as a strategy to inhibit influx of inflammatory cells into the kidney in response to HIV infection.
When examining the surface phenotype, we found evidence of enrichment for migration of central memory CD4+ T cells. The results support a model whereby cocultured primary RTECs with HIV infected T cells result in efficient virus transfer and virus gene expression[6], leading to the differential expression of genes related to immune and inflammatory response and release of cytokines/chemokines from RTECs that can act as strong T cell chemoattractants. The recruitment of cells with unique T cell migratory and effector phenotypes may contribute to the further amplification of HIV infection in the kidney.
Interstitial infiltration and tubular injury is a hallmark of renal allograft rejection and many renal inflammatory diseases[17, 18]. T cell recruitment into kidney could potentially result in increased risk of acute rejection in HIV positive transplant recipients[21]. RTECs are thought to play a central role in the local inflammatory response via cytokine and chemokine production[28]. But a major question is how this is regulated. Previous investigations have suggested a potential role of T cells in this process[29, 30]. T cells may stimulate RTECs either via soluble factors or by direct cell-to-cell contact. Cell-to-cell contact mediates important biologic effects of T cells that, in some cases, are distinct from those mediated by soluble factors[31, 32]. Activation of renal tubular cells by infiltrating T cells can amplify and perpetuate local inflammatory responses through chemokine production mediated by both soluble and cell contact-dependent factors[33]. Recent study of HIV-transduced renal epithelial cells and analyses of kidneys from HIV transgenic mice revealed that the most prominent response was production of proinflammatory mediators including cytokines/chemokines[16]. A goal of this study was to examine how renal epithelial cells will differentially respond to productive HIV infection initiated by cell-free virus or cell-associated virus, a process more relevant to physiological conditions where infected infiltrating T cells can interact with epithelial cells. Our data revealed renal epithelial cells respond to both free virus infection and cell-associated infection by augmented expression of genes close related to inflammatory immune responses (Fig. 1). As compared to cell-free infection, cell-associated HIV infection provoked stronger and prolonged responses from epithelial cells, presumably due to the high efficiency of cell-cell contact mediated viral transfer and the internalization of larger amounts of virus particles through this route [6]
Memory T lymphocytes contain distinct populations of central memory (TCM) and effector memory (TEM) cells that are characterized by distinct homing capacity and effector function[34–37]. TCM recirculate preferentially to the T cell areas of lymphoid organs conducting surveillance for specific Ag whereas TEM preferentially migrate to inflamed peripheral tissues[35, 38]. These studies suggest that TCM and TEM have access to different environments through their unique migratory potential. For example, CXCL-10 (IL-10) stimulates rapid trans endothelial migration of human TEM but not TCM CD4+ T cells[39]. Infiltration of leukocytes into the interstitial compartment of kidney is an important factor for tubular injury leading to progressive renal failure in HIVAN. Here we begin to examine what T cell populations may be recruited to the HIV exposed kidney. We show here that in response to chemoattractants released from HIV infected RTECs, TCM CD4+ T cells migrated preferentially in response to the chemokine gradient (Fig. 5 and Supplemental Fig. 5). TCM are of particular interest because they have a greater capacity than TEM to persist in vivo and they are also more efficient at mediating protective immunity[40].
The expression profiling data provide evidence for HIV upregulation of a DNA sensor pathway in renal tubular cells. HIV infection can trigger the innate host defense mechanisms through DNA sensor pathways that can induce type I interferon (IFN) or other inflammatory cytokines in immune cells[41, 42]. The antiviral DNA sensor IFI16, can trigger CD4+ T cell death during abortive HIV-1 infection[43, 44]. DDX58, a DEAD box protein, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD) associated with pathogen sensor pathways was also upregulated. This protein has a caspase recruitment domain (CARD) and is involved in viral double-stranded dsRNA recognition and the regulation of immune response[45]. Future studies may examine the innate immune signaling pathways that are required to trigger this virus-induced inflammatory state.
In other studies, we have found that primary kidney epithelial cells can support productive HIV infection evidenced by detection of integrated viral DNA that can initiate a new infection in cocultured T cells [7]. The inflammatory influx of T cells that support HIV infection into the kidney may amplify local viral replication and may contribute to a non-immune cell reservoir that can exchange with incoming T cells. Our in vitro data support a model whereby HIV infected renal epithelial cells serve as a chemoattractant source to further promote lymphocyte infiltration. To further test this hypothesis future studies may examine the ability of particular chemokines to recruit T cells to the kidney using humanized HIV mouse model. Although antiretroviral therapy can largely prevent HIVAN, improved survival of HIV patients results in an increase in end stage renal disease among HIV infected individuals. Whetehr this renal pathology is the result of residual replication or the sporadic activation of latent virus is unknown. An autopsy study of advanced HIV patients without chronic kidney disease, found evidence of subclinical renal inflammation and arteriosclerosis [46], which may indicate that residual inflammation may still cause pathology in the kidney during cART. It will therefore be important to understand the how interactions between infected lymphocytes and renal epithelial cells may persist during highly effective antiretroviral therapy and whether anti-inflammatory treatments may be beneficial to long term renal health.
Supplementary Material
Acknowledgments
This work was supported by NIH AI074420 and a Burroughs Wellcome Fund Investigator Award to B.K.C. and P01DK056492 to M.E.K. and B.K.C.
We thank Dr. Michael Ross for critical reading and comments. We thank Dr. Bala Balakumaran for coordinating RNA sequencing at Duke sequencing center. This work was supported by NIH grant DK056492 to M.E.K. and B.K.C.
Footnotes
STATEMENT of COMPETING FINANCIAL INTERESTS
Authors declare no competing financial interests.
References
- 1.Wyatt CM, Klotman PE, D’Agati VD. HIV-associated nephropathy: clinical presentation, pathology, and epidemiology in the era of antiretroviral therapy. Semin Nephrol. 2008;28:513–522. doi: 10.1016/j.semnephrol.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourgoignie JJ, Meneses R, Ortiz C, Jaffe D, Pardo V. The clinical spectrum of renal disease associated with human immunodeficiency virus. Am J Kidney Dis. 1988;12:131–137. doi: 10.1016/s0272-6386(88)80008-8. [DOI] [PubMed] [Google Scholar]
- 3.Rao TK, Filippone EJ, Nicastri AD, Landesman SH, Frank E, Chen CK, et al. Associated focal and segmental glomerulosclerosis in the acquired immunodeficiency syndrome. N Engl J Med. 1984;310:669–673. doi: 10.1056/NEJM198403153101101. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt CM, Meliambro K, Klotman PE. Recent progress in HIV-associated nephropathy. Annu Rev Med. 2012;63:147–159. doi: 10.1146/annurev-med-041610-134224. [DOI] [PubMed] [Google Scholar]
- 5.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 6.Chen P, Chen BK, Mosoian A, Hays T, Ross MJ, Klotman PE, et al. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J Am Soc Nephrol. 22:496–507. doi: 10.1681/ASN.2010040379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasi M, Balakumaran B, Chen P, Negri DR, Cara A, Chen BK, et al. Renal epithelial cells produce and spread HIV-1 via T-cell contact. AIDS. 2014;28:2345–2353. doi: 10.1097/QAD.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruggeman LA, Dikman S, Meng C, Quaggin SE, Coffman TM, Klotman PE. Nephropathy in human immunodeficiency virus-1 transgenic mice is due to renal transgene expression. J Clin Invest. 1997;100:84–92. doi: 10.1172/JCI119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenstiel PE, Chan J, Snyder A, Planelles V, D’Agati VD, Klotman PE, et al. HIV-1 Vpr activates the DNA damage response in renal tubule epithelial cells. AIDS. 2009;23:2054–2056. doi: 10.1097/QAD.0b013e32833088a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husain M, D’Agati VD, He JC, Klotman ME, Klotman PE. HIV-1 Nef induces dedifferentiation of podocytes in vivo: a characteristic feature of HIVAN. AIDS. 2005;19:1975–1980. doi: 10.1097/01.aids.0000191918.42110.27. [DOI] [PubMed] [Google Scholar]
- 11.Medapalli RK, He JC, Klotman PE. HIV-associated nephropathy: pathogenesis. Curr Opin Nephrol Hypertens. 2011;20:306–311. doi: 10.1097/MNH.0b013e328345359a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang ST, Sova P, Peng X, Weiss J, Law GL, Palermo RE, et al. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. MBio. 2011:2. doi: 10.1128/mBio.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, et al. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1) PLoS One. 2011;6:e22730. doi: 10.1371/journal.pone.0022730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Conaldi PG, Biancone L, Bottelli A, Wade-Evans A, Racusen LC, Boccellino M, et al. HIV-1 kills renal tubular epithelial cells in vitro by triggering an apoptotic pathway involving caspase activation and Fas upregulation. J Clin Invest. 1998;102:2041–2049. doi: 10.1172/JCI3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray PE, Liu XH, Henry D, Dye L, 3rd, Xu L, Orenstein JM, et al. Infection of human primary renal epithelial cells with HIV-1 from children with HIV-associated nephropathy. Kidney Int. 1998;53:1217–1229. doi: 10.1046/j.1523-1755.1998.00900.x. [DOI] [PubMed] [Google Scholar]
- 16.Ross MJ, Fan C, Ross MD, Chu TH, Shi Y, Kaufman L, et al. HIV-1 infection initiates an inflammatory cascade in human renal tubular epithelial cells. J Acquir Immune Defic Syndr. 2006;42:1–11. doi: 10.1097/01.qai.0000218353.60099.4f. [DOI] [PubMed] [Google Scholar]
- 17.Becker GJ, Hewitson TD. The role of tubulointerstitial injury in chronic renal failure. Curr Opin Nephrol Hypertens. 2000;9:133–138. doi: 10.1097/00041552-200003000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Remuzzi G, Ruggenenti P, Benigni A. Understanding the nature of renal disease progression. Kidney Int. 1997;51:2–15. doi: 10.1038/ki.1997.2. [DOI] [PubMed] [Google Scholar]
- 19.van Kooten C, Daha MR. Cytokine cross-talk between tubular epithelial cells and interstitial immunocompetent cells. Curr Opin Nephrol Hypertens. 2001;10:55–59. doi: 10.1097/00041552-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Parkhie SM, Fine DM, Lucas GM, Atta MG. Characteristics of patients with HIV and biopsy-proven acute interstitial nephritis. Clin J Am Soc Nephrol. 2010;5:798–804. doi: 10.2215/CJN.08211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canaud G, Dejucq-Rainsford N, Avettand-Fenoel V, Viard JP, Anglicheau D, Bienaime F, et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol. 2014;25:407–419. doi: 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Hubner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White JR, Lee JM, Young PR, Hertzberg RP, Jurewicz AJ, Chaikin MA, et al. Identification of a potent, selective non-peptide CXCR2 antagonist that inhibits interleukin-8-induced neutrophil migration. J Biol Chem. 1998;273:10095–10098. doi: 10.1074/jbc.273.17.10095. [DOI] [PubMed] [Google Scholar]
- 25.Mueller SN. Effector T-cell responses in non-lymphoid tissues: insights from in vivo imaging. Immunol Cell Biol. 2013;91:290–296. doi: 10.1038/icb.2012.75. [DOI] [PubMed] [Google Scholar]
- 26.Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook KW, Letley DP, Ingram RJ, Staples E, Skjoldmose H, Atherton JC, et al. CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut. 2014;63:1550–1559. doi: 10.1136/gutjnl-2013-306253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kooten C, Daha MR, van Es LA. Tubular epithelial cells: A critical cell type in the regulation of renal inflammatory processes. Exp Nephrol. 1999;7:429–437. doi: 10.1159/000020622. [DOI] [PubMed] [Google Scholar]
- 29.Deckers JG, Van Der Woude FJ, Van Der Kooij SW, Daha MR. Synergistic effect of IL-1alpha, IFN-gamma, and TNF-alpha on RANTES production by human renal tubular epithelial cells in vitro. J Am Soc Nephrol. 1998;9:194–202. doi: 10.1681/ASN.V92194. [DOI] [PubMed] [Google Scholar]
- 30.Prodjosudjadi W, Gerritsma JS, Klar-Mohamad N, Gerritsen AF, Bruijn JA, Daha MR, et al. Production and cytokine-mediated regulation of monocyte chemoattractant protein-1 by human proximal tubular epithelial cells. Kidney Int. 1995;48:1477–1486. doi: 10.1038/ki.1995.437. [DOI] [PubMed] [Google Scholar]
- 31.McInnes IB, Leung BP, Liew FY. Cell-cell interactions in synovitis. Interactions between T lymphocytes and synovial cells. Arthritis Res. 2000;2:374–378. doi: 10.1186/ar115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rezzonico R, Burger D, Dayer JM. Direct contact between T lymphocytes and human dermal fibroblasts or synoviocytes down-regulates types I and III collagen production via cell-associated cytokines. J Biol Chem. 1998;273:18720–18728. doi: 10.1074/jbc.273.30.18720. [DOI] [PubMed] [Google Scholar]
- 33.Kuroiwa T, Schlimgen R, Illei GG, McInnes IB, Boumpas DT. Distinct T cell/renal tubular epithelial cell interactions define differential chemokine production: implications for tubulointerstitial injury in chronic glomerulonephritides. J Immunol. 2000;164:3323–3329. doi: 10.4049/jimmunol.164.6.3323. [DOI] [PubMed] [Google Scholar]
- 34.Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 35.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 36.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 37.Weninger W, Crowley MA, Manjunath N, von Andrian UH. Migratory properties of naive, effector, and memory CD8(+) T cells. J Exp Med. 2001;194:953–966. doi: 10.1084/jem.194.7.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell JJ, Murphy KE, Kunkel EJ, Brightling CE, Soler D, Shen Z, et al. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 39.Manes TD, Pober JS, Kluger MS. Endothelial cell-T lymphocyte interactions: IP[corrected]-10 stimulates rapid transendothelial migration of human effector but not central memory CD4+ T cells. Requirements for shear stress and adhesion molecules. Transplantation. 2006;82:S9–14. doi: 10.1097/01.tp.0000231356.57576.82. [DOI] [PubMed] [Google Scholar]
- 40.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 41.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 46.Wyatt CM, Morgello S, Katz-Malamed R, Wei C, Klotman ME, Klotman PE, et al. The spectrum of kidney disease in patients with AIDS in the era of antiretroviral therapy. Kidney Int. 2009;75:428–434. doi: 10.1038/ki.2008.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.