Abstract
In Drosophila, as well as in many other plants and animals, pigmentation is highly variable both within and between species. This variability, combined with powerful genetic and transgenic tools as well as knowledge of how pigment patterns are formed biochemically and developmentally, have made Drosophila pigmentation a premier system for investigating the genetic and molecular mechanisms responsible for phenotypic evolution. In this chapter, we review and synthesize findings from a rapidly growing body of case studies examining the genetic basis of pigmentation differences in the abdomen, thorax, wings, and pupal cases within and between Drosophila species. A core set of genes, including genes required for pigment synthesis (e.g., yellow, ebony, tan, Dat) as well as developmental regulators of these genes (e.g., bab1, bab2, omb, Dll, and wg) emerge as the primary sources of this variation, with most genes having been shown to contribute to pigmentation differences both within and between species. In cases where specific genetic changes contributing to pigmentation divergence were identified in these genes, the changes were always located in noncoding sequences and affected cis-regulatory activity. We conclude this chapter by discussing these and other lessons learned from evolutionary genetic studies of Drosophila pigmentation and identify topics we think should be the focus of future work with this model system.
Keywords: melanin, evolution, gene expression, cis-regulation, yellow, ebony, tan, bric-a-brac, wingless
Introduction
Heritable changes in DNA sequence within and among species explain much of life’s diversity. Identifying these changes and understanding how they impact development to generate phenotypic differences remains a major challenge for evolutionary biology. A growing number of case studies have localized the specific genes involved in trait variation both within and among species, and some have described how individual mutations affect the developmental pathways underlying phenotypic differences. With a catalog of studies describing more than 1000 alleles contributing to morphological, physiological, or behavioral evolution of diverse traits in diverse species now available, researchers have begun to synthesize the genetic and developmental mechanisms underlying phenotypic evolution in search of genetic and molecular patterns that underlie the evolutionary process (Stern and Orgogozo 2008; Carroll 2008; Kopp 2009; Streisfeld and Rausher 2011; Martin and Orgogozo 2013).
One finding from this synthesis is that different types of traits tend evolve through different molecular mechanisms. For example, changes in cis-regulatory DNA sequences that regulate gene expression contribute to morphological differences within and among species more often than they contribute to differences in physiological traits, while the converse is true for changes in the amino acid sequence of proteins (Stern and Orgogozo 2008; 2009). Another finding to emerge from this synthesis is that some traits have evolved multiple times independently using the same genetic changes (e.g., xenobiotic resistance) whereas other traits have evolved similar changes using different mutations in the same gene (e.g., coat color) or using different genes (Martin and Orgogozo 2013). Differences in the genetic basis of phenotypic diversity also seem to exist within and between species, with changes in cis-regulatory sequences playing a larger role in interspecific than intraspecific differences (Stern and Orgogozo 2008; Wittkopp et al. 2008; Coolon et al. 2014).
In this chapter, we examine patterns in the genetic and molecular mechanisms responsible for phenotypic evolution that emerge from focusing on a collection of studies investigating changes in a single trait within and among species in the same genus. Specifically, we review and synthesize the collection of case studies dissecting the genetic basis of body color (pigmentation) in Drosophila, emphasizing a comparison of genetic and molecular mechanisms that vary within and among Drosophila species. Drosophila pigmentation is an ideal trait for such an analysis because (i) pigmentation is one of the most variable traits within and among species, (ii) much is known about the genes involved in pigment synthesis as well as those that control expression of these genes during Drosophila development (True 2003; Wittkopp et al. 2003a; Kopp 2009; Takahashi 2013), and (iii) specific genes and genetic changes contributing to differences in Drosophila pigmentation have been identified for changes in pigmentation that have evolved over multiple timescales and in multiple lineages (Table 1). These differences in pigmentation that have been dissected genetically include examples of trait divergence, convergent evolution, and evolutionary novelty.
Table 1.
The Loci of Pigmentation Evolution in Drosophila
| Level of variation | Pigmentation trait | Species | Gene(s) | Gene Function | Type of Mutation(s) | Reference |
|---|---|---|---|---|---|---|
| within species | abdominal | D. melanogaster | bab | TF | cis-regulatory | Rogers et al., 2013 |
| within species | abdominal | D. melanogaster | bab1 | TF | cis-regulatory | Bastide et al., 2013 |
| within species | abdominal | D. melanogaster | bab1 | TF | cis-regulatory | Endler et al., 2015 |
| within species | abdominal | D. melanogaster | bab | TF | unknown | Kopp et al., 2003 |
| within species | abdominal | D. melanogaster | bab1 | TF | cis-regulatory | Bickel et al., 2011 |
| within species | abdominal | D. melanogaster | bab2 | TF | cis-regulatory | Bickel et al., 2011 |
| within species | abdominal | D. melanogaster | ebony | enzyme | cis-regulatory | Johnson et al., 2015 |
| within species | abdominal | D. melanogaster | ebony | enzyme | cis-regulatory | Pool and Aquadro, 2007 |
| within species | abdominal | D. melanogaster | ebony | enzyme | cis-regulatory | Rebeiz et al. 2009 |
| within species | abdominal | D. polymorpha | omb | TF | unknown | Brisson et al. 2004 |
| within species | abdominal | D. melanogaster | tan | enzyme | cis-regulatory | Bastide et al., 2013 |
| within species | abdominal | D. melanogaster | tan | enzyme | cis-regulatory | Endler et al., 2015 |
| within species | abdominal | D. erecta | tan | enzyme | cis-regulatory | Yassin et al. 2016 |
| within species | abdominal | D. polymorpha | two genes | unknown | unknown | Martinez and Cordeiro, 1970 |
| within species | abdominal | D. melanogaster | 84 genes | multiple | cis-regulatory | Dembeck et al., 2015 |
| within species | abdominal | D. baimaii | unknown | NA | unknown | Ohnishi and Watanabe, 1985 |
| D. bicornuta | ||||||
| D. burlai | ||||||
| D. truncata | ||||||
| within species | abdominal | D. malerkotliana | at least three QTL | NA | unknown | Ng et al., 2008 |
|
| ||||||
| between species | abdominal | D. melanogaster | bab | TF | cis-regulatory | Rogers et al., 2013 |
| D. yakuba | ||||||
| D. fuyamai | ||||||
| D. auraria | ||||||
| between species | abdominal | D. willistoni | bab1 | TF | cis-regulatory | Williams et al. 2008 |
| D. melanogaster | ||||||
| between species | abdominal | D. auraria | ebony | enzyme | cis-regulatory | Johnson et al., 2015 |
| D. serrata | ||||||
| between species | abdominal | D. americana | ebony | enzyme | unknown | Wittkopp et al., 2009 |
| D. novamexicana | ||||||
| between species | abdominal | D. yakuba | ||||
| D. santomea | tan | enzyme | cis-regulatory | Jeong et al., 2008 | ||
| between species | abdominal | D. americana | tan | enzyme | cis-regulatory | Wittkopp et al., 2009 |
| D. novamexicana | ||||||
| between species | abdominal | D. melanogaster | yellow | unknown | cis-regulatory | Jeong et al., 2006 |
| D. kikkawai | ||||||
| between species | abdominal | D. prostipennis | yellow | unknown | cis-regulatory | Ordway et al. 2014 |
| D. melanogaster | ||||||
| between species | abdominal | D. yakuba | at least four QTL | NA | unknown | Carbone et al., 2005 |
| D. santomea | ||||||
| between species | abdominal | D. yakuba | at least five QTL | NA | unknown | Llolapart et al., 2002 |
| D. santomea | ||||||
| between species | abdominal | D. arawakan | unknown | NA | unknown | Hollocher et al., 2000 |
| D. nigrodunni | ||||||
| between species | abdominal | D. tenebrosa | at least two QTL | NA | unknown | Bray et al., 2014 |
| D. suboccidentalis | ||||||
|
| ||||||
| within species | thorax | D. melanogaster | ebony | enzyme | cis-regulatory | Miyagi et al., 2015 |
| within species | thorax | D. melanogaster | ebony | enzyme | cis-regulatory | Takahashi et al., 2007 |
| within species | thorax | D. melanogaster | ebony | enzyme | cis-regulatory | Takahashi and Takano-Shimizu, 2011 |
| within species | thorax | D. melanogaster | ebony | enzyme | cis-regulatory | Telonis-Scott et al., 2011 |
|
| ||||||
| between species | thorax | D. guttifera*** | wg | cell-cell signalling | cis-regulatory | Koshikawa et al., 2015 |
| D. melanogaster | ||||||
|
| ||||||
| between species | wing | D. biarmipes | yellow | uknown | cis-regulatory | Gompel et al., 2005 |
| D. melanogaster | ||||||
| between species | wing | D. elegans | yellow | unknown | cis-regulatory | Prud-homme et al., 2006 |
| D. gunungcola | ||||||
| D. mimetica | ||||||
| D. tristes | ||||||
| between species | wing | D. guttifera*** | wg | cell-cell signalling | cis-regulatory | Koshikawa et al., 2015 |
| D. melanogaster | ||||||
| between species | wing | D. elegans | at least three QTL | NA | unknown | Yeh and True, 2014 |
| D. gunungcola | ||||||
|
| ||||||
| between species | pupariam | D. virilis | Dat | enzyme | cis-regulatory | Ahmed-Braimah and Sweigart, 2015 |
| D. americana | ||||||
Development of Drosophila pigmentation
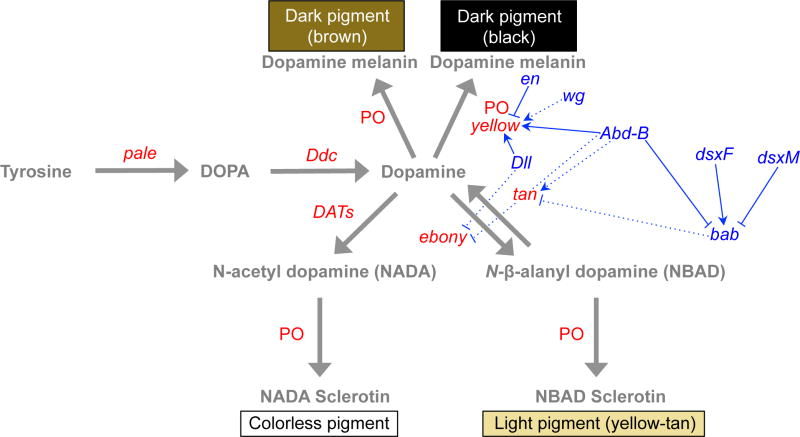
In Drosophila (as well as in many other insects, (True 2003; Wittkopp and Beldade 2009; Zhan et al. 2010), body color results from a combination of dark black and brown melanins as well as light yellow-tan and colorless scelarotins (Wright 1987; True 2003; Wittkopp et al. 2003a). These four types of pigments are produced by a branched biochemical pathway that processes tyrosine obtained from the diet into these pigments (Figure 1). Tyrosine is first converted into DOPA (L-3,4-dihydroxyphenylalanine) by a tyrosine hydroxylase (TH) encoded by the pale gene. This DOPA is then converted into dopamine through a reaction catalyzed by the dopa decarboxylase enzyme encoded by the Ddc gene. Prior reviews have suggested that DOPA can also be polymerized into a black melanin through a process involving the Yellow protein (Kopp, 2009; Wittkopp et al., 2003), but recent data show that the formation of black pigment requires the function of Ddc (J.-M. Gibert, personal communication) and is thus likely produced from dopamine rather than DOPA, as has also been previously suggested (Riedel et al., 2011; Walter et al., 1996). Dopamine can then have one of four fates: it can be converted into a black melanin through a process involving the Yellow protein and phenol oxidases (POs); converted into a brown melanin through a process involving POs, but not yellow; into a yellow-tan sclerotin through the activity of Ebony converting dopamine into beta-alanyl dopamine (NBAD) and POs polymerizing it into NBAD sclerotin, or into a colorless pigment through the activity of dopamine-acetyl-transferases (DATs) converting dopamine into N-acetyl dopamine (NADA) and POs polymerizing it into NADA sclerotin. One of these reactions, the conversion of dopamine into NBAD, is reversible, with the reverse reaction catalyzed by the Tan protein. Disruption of the tan gene reduces the production of dark melanins, indicating that the conversion of NBAD back into dopamine is a necessary step in the development of pigmentation. Changing relative expression levels of yellow, ebony and/or tan can shift the balance between dark (black, brown) and yellow-tan pigments as this branched biochemical pathway produces more of one type at the expense of the other (Wittkopp et al. 2002a; 2009).
Figure 1. Developmental and biochemical control of pigmentation in Drosophila.
A simplified version of the biochemical pathway controlling pigment biosynthesis in insects is shown with regulators controlling expression of individual pigment synthesis genes in at least one Drosophila species overlaid. Genes colored red are part of the pigment biosynthesis pathway; metabolites are colored gray; and gray arrows indicate chemical reactions during pigmentation synthesis. Genes colored in blue are part of the regulatory network that directly (solid arrows) or indirectly (broken arrows) modulate enzyme expression during pigmentation development in Drosophila. Pointed and blunt arrows indicate positive and negative regulatory interactions, respectively. The pigment biosynthesis pathway is conserved among all Drosophila, but the regulatory relationships shown often function in only a subset of Drosophila species (Gompel et al. 2005; Arnoult et al. 2013).
Pigments produced by this biochemical pathway are deposited into the developing cuticle during late pupal and early adult stages (Kraminsky et al. 1980; Sugumaran et al. 1992; Walter et al. 1996; Wittkopp et al. 2003a). The spatial distribution of these pigments is determined in a nearly cell autonomous manner by spatially regulated transcription of genes such as yellow, tan, and ebony. As discussed in detail below, changes in the expression patterns of these genes often underlie evolutionary changes in pigmentation. Genes regulating expression of these pigment synthesis genes are thus also potential targets for genetic divergence contributing to pigmentation diversity. Six transcription factors (optomotor-blind (omb), bric a brac (bab), abdominal-B (Abd-B), doublesex (dsx), Distal-less (Dll), and Engrailed (en)) have been shown to regulate expression of pigment synthesis genes (yellow, ebony, tan, Ddc) in Drosophila either directly (by binding to transcription factor binding sites located in enhancers controlling the gene’s expression) or indirectly (by influencing abundance, activity, or binding of direct regulators) (Figure 1) (Kopp et al. 2000; Gompel et al. 2005; Jeong et al. 2006; Williams et al. 2008; Arnoult et al. 2013). For example, in at least one Drosophila species each, En (Gompel et al. 2005), Dll (Arnoult et al. 2013) and Abd-B (Jeong et al. 2006) have all been shown to directly bind to yellow enhancers, whereas Abd-B and Dsx (including both the male (dsxM) and female (dsxF) forms of dsx) have been shown to directly bind to enhancers of the bab gene (Williams et al. 2008). It is not yet known whether Bab proteins directly bind to enhancers of any pigment synthesis genes, but it is clear that Bab proteins affect expression of pigment synthesis genes in some manner (Kopp 2009). Similarly, Omb (Brisson et al. 2004) and Wingless (Wg, a ligand for a signal transduction pathway) (Werner et al. 2010; Koshikawa et al. 2015) have also been shown to influence expression of at least one pigment synthesis gene (Figure 1), although questions remain about the precise molecular mechanisms by which they do so. Additional transcription factors with effects on abdominal pigmentation in Drosophila melanogaster have been identified in recent RNAi screens (Kalay 2012; Rogers et al. 2013a), but the ways in which they alter expression of pigment synthesis genes remains unknown. Elucidating the structure and complexity of the gene network regulating expression of pigment synthesis genes (and hence pigmentation) remains one of the biggest challenges for understanding the development and evolution of Drosophila pigmentation within and between species.
Tissue-specific regulation of pigmentation
Null mutations disrupting the function of proteins required for pigment synthesis such as TH, DDC, Yellow, Tan, Ebony, DATs, and POs alter pigmentation throughout the fly, whereas mutations in specific enhancers of these genes and mutations affecting transcriptional regulators of these genes typically alter pigmentation in only some parts of the fly. Evolutionary changes in pigmentation are often restricted to specific body parts, suggesting that such changes are likely to result from these latter types of mutations. One reason for this may be that null mutations in pigment synthesis genes often also alter behavior and/or other phenotypes in addition to pigmentation (True 2003; Wittkopp and Beldade 2009; Takahashi 2013), making null mutations unlikely to survive in natural populations. The presence of tissue-specific enhancers for pigment synthesis genes coupled with differences in the sets of regulators that interact with each enhancer provide genetic mechanisms for overcoming these pleiotropic constraints and altering pigmentation independently in different body parts. Because the developmental control of pigmentation in different body regions often involves different regulatory genes, we have chosen to structure our review of the genetic mechanisms underlying pigmentation differences within and between Drosophila species by body part, examining the evolution of abdominal pigmentation, thorax pigmentation, wing pigmentation, and pupal pigmentation in Drosophila separately below.
Abdominal pigmentation
Abdominal pigment patterns (especially those on the dorsal side of the abdomen) are conspicuous and highly variable within and among species. It is not surprising then that most studies of genetic mechanisms underlying pigmentation differences in Drosophila have attempted to explain differences in intra- and interspecific abdominal pigmentation. These abdominal pigment patterns are displayed in a series of overlapping tergites that can vary in pigment color, pattern, and intensity among individuals and sexes in the same population, different populations, and different species (Wittkopp et al. 2003a; Kronforst et al. 2012). For example, in D. melanogaster females, the most prominent abdominal tergites (A2-A6) show a “stripe” of dark melanins at the posterior edge of the segment as well as a peak of this dark color along the dorsal midline (Figure 2, left). In male D. melanogaster, this pattern is seen in the A2, A3, and A4 tergites, but A5 and A6 are much more completely covered by dark melanins (Figure 2, right). Sexually dimorphic pigmentation is absent in many species, however, with both sexes showing the same pigmentation pattern in all segments (Kopp et al. 2000). The pattern of pigmentation within each segment can also vary, with modifications to the shape of the stripe, unique patterns such as spots, and melanins distributed evenly throughout the abdomen seen in different species (Wittkopp et al. 2003a). Differences in abdominal pigmentation are generally assumed to result from adaptation, but the selection pressures responsible for the evolution of a particular pattern in a particular species remain unclear. Potential selection pressures proposed for divergent abdominal pigmentation include sexual selection resulting from mate choice as well as environmental factors that differ across gradients of altitude, latitude, temperature, humidity, and UV radiation (Capy et al. 1988; Kopp et al. 2000; True 2003; Wittkopp et al. 2011; Clusella-Trullas and Terblanche 2011; Matute and Harris 2013; Bastide et al. 2014).
Figure 2. Abdominal pigmentation in D. melanogaster.
The dorsal abdomen of D. melanogaster is shown for wild-type adult females (left) and males (right). Note the dark pigment stripe visible at the posterior edge of abdominal segments A2-A6 in females and A2-A4 in males as well as the more complete melanization in tergites A5 and A6 of males relative to females.
Genetic basis of abdominal pigmentation differences within a species
In D. melanogaster, the most studied of all Drosophila species, abdominal pigmentation often varies within and among populations. For example, in sub-Saharan Africa, D. melanogaster collected from low elevations showed lighter abdominal pigmentation than D. melanogaster collected from high elevations (Pool and Aquadro 2007); these differences persisted when rearing these flies in the lab, demonstrating that the differences in pigmentation were caused by genetic differences rather than phenotypic plasticity. Genetic analysis implicated one or more loci on the X and 3rd chromosomes in this pigmentation difference, and analysis of the pigment synthesis gene ebony, which is required for the synthesis of yellow-tan pigments and is located on the 3rd chromosome, revealed distinct haplotypes in populations from different altitudes that correlated with these differences in abdominal pigmentation. Nucleotide diversity levels within this region suggested that natural selection has elevated the frequency of dark ebony alleles in one of the populations sampled from Uganda, possibly facilitating adaptation to different altitudes (Pool and Aquadro 2007). Further analysis identified a cis-regulatory element in this region that controls ebony expression in the abdomen, and showed that the allele of this sequence from a lightly pigmented fly drives higher levels of ebony expression than the allele of this sequence found in a more darkly pigmented fly (Rebeiz et al. 2009a), consistent with Ebony’s function in the synthesis of light colored sclerotin (Walter et al. 1996). This region was also found to have recently accumulated multiple mutations in the Uganda population that appear to have given rise to an allele of large effect that contributes to divergence of abdominal pigmentation (Rebeiz et al. 2009a).
Genetic differences in ebony cis-regulatory sequences also appear to contribute to variable abdominal pigmentation in other populations of D. melanogaster and other species (Bastide et al. 2013; Johnson et al. 2015; Dembeck et al. 2015b; Endler et al. 2016). For example, an association study using the Drosophila Genetic Reference Panel (DGRP) of D. melanogaster strains isolated from a population in Raleigh, North Carolina (Mackay et al. 2012) found a significant correlation between a noncoding variant located within a known cis- regulatory element of ebony and pigmentation variation within this population (Dembeck et al. 2015b). Weak associations with noncoding SNPs in ebony cis-regulatory elements were also observed for European populations of D. melanogaster (Bastide et al. 2013; Dembeck et al. 2015a; Endler et al. 2016), with the most highly ranked SNP associated with ebony in Bastide et al. (2013) located in a sequence that inhibits ebony expression in male abdominal segments during development. Outside of D. melanogaster, genetic variation linked to ebony has been shown to be associated with polymorphic abdominal pigmentation within Drosophila americana (Wittkopp et al. 2009) and Drosophila auraria (Johnson et al. 2015). In this latter species, specific alleles of ebony cis-regulatory sequences were identified in light and dark individuals, and transgenic analyses of reporter genes were used to demonstrate the effects of these variable sites on ebony expression (Johnson et al. 2015).
The tan gene, which plays the opposite role of ebony in pigment synthesis, promoting production of dark brown melanin at the expense of yellow-tan sclerotin, also contributes to variation within Drosophila species. In fact, the study that found evidence of an association between ebony genotype and abdominal pigmentation within the DGRP collection also identified multiple SNPs within non-coding regions near tan that were associated with differences in abdominal pigmentation in this population of D. melanogaster (Dembeck et al. 2015b). Three of these non-coding SNPs were also found to be associated with abdominal pigmentation in European populations and an African population of D. melanogaster (Bastide et al. 2013; Endler et al. 2016). These SNPs were located within a cis-regulatory element known as the male-specific enhancer (MSE, (Jeong et al. 2008)) that drives expression in D. melanogaster in the abdominal stripes as well as throughout the A5 and A6 abdominal segments with male-specific pigmentation. tan cis-regulatory evolution at the MSE was also recently implicated in a sex-specific color dimorphism involving abdominal pigmentation differences within Drosophila erecta (Yassin et al. 2016). This final case study is particularly interesting because ancient balancing selection was shown to likely be responsible for maintaining alternative alleles at the tan MSE and thus both light and dark morphs of female D. erecta (Yassin et al. 2016).
Genetic changes contributing to polymorphic pigmentation are not always caused by pigmentation synthesis genes such as ebony and tan, however; changes in regulatory genes upstream of the pigmentation synthesis pathway contribute to pigmentation differences segregating within a species as well. These sources of variation include genetic changes at the bab locus, a locus originally discovered to be an important regulator of abdominal pigmentation differences between sexes in D. melanogaster (Robertson et al. 1977). Null mutations in bab cause the development of a male-like pigmentation pattern in the A5 and A6 abdominal segments of female D. melanogaster, suggesting that bab acts to repress male-specific abdominal pigmentation in females (Kopp et al. 2000). Using quantitative trait locus (QTL) mapping coupled with quantitative complementation tests to examine the genetic basis of abdominal pigmentation differences in a population of D. melanogaster from Winters, California, Kopp et al. (2003) found genetic variation at bab had a major effect on abdominal pigmentation differences in females. The bab locus includes two genes, bab1 and bab2, each of which acts as a transcriptional regulator, and it was unclear in Kopp et al. (2003) if variation affecting bab1 and/or bab2 was responsible for variation in abdominal pigmentation. To address this uncertainty, Bickel et al. (2011) sequenced the bab region in multiple inbred lines from the California population and found that non-coding SNPs at both bab1 and bab2 were associated with abdominal pigmentation differences. Specifically, SNPs associated with pigmentation were found in the first intron of bab1 and near the promoter region of bab2. In the DGRP collection, European populations, and an African population of D. melanogaster, only SNPs in the first intron of bab1 were associated with abdominal pigmentation variation (Bastide et al. 2013; Dembeck et al. 2015a,b; Endler et al. 2016). A cis-regulatory element controlling sex-specific expression of bab1 in the A5-A7 segments in D. melanogaster males (repression) and females (induction) was also identified in the first intron of bab1 (Williams et al. 2008) and overexpression of bab1 during late pupal development was shown to be sufficient to suppress dark pigmentation (Salomone et al. 2013), suggesting that the associated sites might alter pigmentation by altering expression of bab1. Indeed, Rogers et al. (2013a) found that different alleles of this element were present in lightly and darkly pigmented D. melanogaster that drove different patterns of gene expression that correlate with pigmentation in the manner expected given bab’s role as a repressor of dark pigmentation. A small number of derived sequence changes were found to be responsible for these differences in cis-regulatory activity (Rogers et al. 2013a). Genetic variation linked to another regulator of pigmentation, omb, has also been found to be associated with polymorphic body color in Drosophila polymorpha, but much less is known about this association, including whether coding or noncoding changes are more likely to be responsible for the association (Brisson et al. 2004).
Together, the studies described above demonstrate that genetic variation contributing to variable abdominal pigmentation within a species has repeatedly accumulated at non-coding regions near the ebony, tan, and bab1 genes. In fact, in the European D. melanogaster population studied in Bastide et al. (2013), 79% of the most strongly associated SNPs mapped to non-coding regions linked to ebony, tan, and bab1. Other loci also clearly contribute to polymorphic abdominal pigmentation, however (Ng et al. 2008; Dembeck et al. 2015a,b), and some of these loci have recently begun to be identified in D. melanogaster (Dembeck et al. 2015a,b). The developmental role that these newly identified genes (e.g. pinstripe, triforce, plush, and farmer) play in pigment patterning remains unknown.
Genetic basis of abdominal pigmentation differences between species
Differences in pigmentation between species have evolved over longer time-scales than differences in pigmentation within a species, suggesting that even phenotypically similar changes in pigmentation might have a distinct genetic basis within and between species (Orr 2001). For example, different genes and/or different types of changes in the same genes might tend to contribute to phenotypic differences that have evolved over longer evolutionary timescales (Orr 2001; Stern and Orgogozo 2009). By directly comparing the genetic basis of intra- and interspecific pigmentation differences, we can better understand how the variants underlying polymorphism within a species give rise to divergence between species. In this section, we review what is known about the genetic basis of abdominal pigmentation differences between species.
The genetic basis of pigmentation differences between species can be dissected genetically using the same methods used to identify genes contributing to intraspecific polymorphism if two species with differences in pigmentation are closely related enough that they can still be crossed and produce viable offspring in the laboratory. One such species pair is D. yakuba and D. santomea, which are estimated to have begun diverging ~400,000 years ago (Cariou et al. 2001) (Figure 3). D. yakuba exhibits stripes of dark melanins in A2-A6 in both sexes as well as more complete dark pigmentation in segments A5 and A6 of males similar to D. melanogaster (Figure 2B), whereas D. santomea lacks dark melanin in these regions in both sexes (Jeong et al. 2008). QTL mapping was used to identify regions of the genome contributing to abdominal pigmentation divergence between these two species. In Llopart et al. (2002), five QTLs were identified, one of which was on the X chromosome and explained nearly 90% of the species differences. Using a slightly different phenotyping procedure, Carbone et al. (Carbone et al. 2005) identified four QTL, two located on the X chromosome (one with a much larger effect on pigmentation than the other) and two located on autosomes.
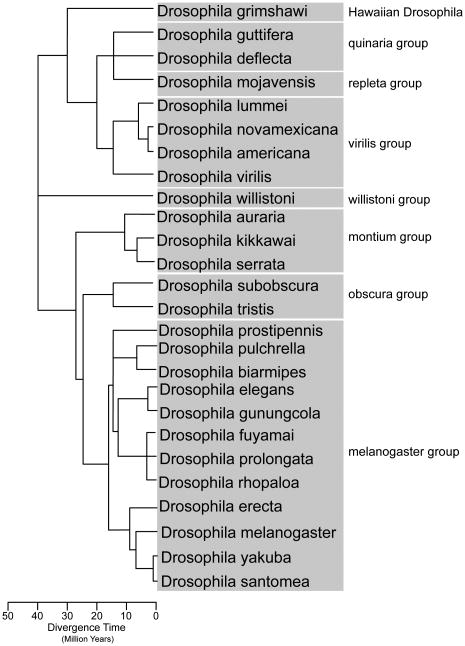
Figure 3. Phylogeny of Drosophila species used to study the genetic basis of pigmentation evolution.
Phylogenetic relationships shown were inferred using the online Interactive Tree of Life (iTOL) (Letunic and Bork 2007; 2011), with branch lengths estimated using data from the online Time Tree website (Hedges et al. 2006).
To identify the specific gene(s) that might be responsible for the X-linked QTLs contributing to pigmentation differences between D. yakuba and D. santomea, Jeong et al. (2008) took a candidate gene approach. Specifically, they examined the pigment synthesis genes tan and yellow, which were located within the large and small effect X-linked QTLs respectively, and found differences in expression of both genes that correlated with differences in abdominal pigmentation between D. yakuba and D. santomea (Jeong et al. 2008). Analysis of yellow and tan expression in F1 hybrids from reciprocal crosses showed that only the expression difference in tan was caused by cis-acting genetic changes on the X chromosome; the difference in yellow expression appeared to be caused by one or more trans-acting autosomal loci (Jeong et al. 2008). To further localize the genetic changes responsible for divergent tan expression and presumably pigmentation, transgenic reporter genes were used to compare enhancer activity of sequences from D. yakuba and D. santomea in D. melanogaster. Mutations within a male-specific enhancer located 5’ of tan in the genome were found to have likely caused, in part, loss of abdominal pigmentation in D. santomea (Jeong et al. 2008). This role of tan in pigmentation divergence between D. santomea and D. yakuba was further supported by introgressing the D. yakuba allele of tan into D. santomea and directly demonstrating this gene’s contribution to the evolution of abdominal pigmentation differences between these two species (Rebeiz et al. 2009b).
A similar story has emerged for pigmentation differences between the interfertile sister species D. americana and D. novamexicana, which are also thought to have diverged approximately 400,000 years ago (Morales-Hojas et al. 2008) (Figure 3). D. americana has an overall dark body color typical for a member of the virilis species group, whereas D. novamexicana displays a derived light body color with greatly reduced abundance of dark melanins (Wittkopp et al. 2003b). Analysis of F1 hybrids from reciprocal crosses again showed a large contribution of the X-chromosome to pigmentation divergence (Wittkopp et al. 2003b), at least some of which was attributable to loci linked to the tan gene (Wittkopp et al. 2009). Fine-scale genetic mapping confirmed that divergence at tan was indeed a contributor to pigmentation divergence and localized the functionally divergent sites within tan to the first intron (Wittkopp et al. 2009). Subsequent work has shown small, but significant differences in cis-regulatory activity of the D. americana and D. novamexicana tan alleles that presumably contribute to pigmentation differences (Cooley et al. 2012). The contribution of tan to pigmentation divergence between these two species was further confirmed when the D. americana tan allele caused darker pigmentation than the D. novamexicana tan allele when each was put into a common D. melanogaster genetic background using transgenes (Wittkopp et al. 2009). Variation linked to the ebony gene is also an important source of pigmentation divergence between these two species, with introgression of chromosomal regions containing tan and ebony from D. americana into D. novamexicana together explaining 87% of the difference in abdominal pigmentation seen between D. americana and D. novamexicana (Wittkopp et al. 2009). Effects of ebony have yet to be separated from linked loci, however, because ebony is located within a region of the genome inverted between these two species, preventing recombination-based mapping. In all, genetic mapping between D. americana and D. novamexicana has identified five regions of the genome that contribute to the difference in abdominal pigmentation (Wittkopp et al. 2003b; 2009).
Variation at ebony also appears to be important for abdominal pigmentation differences between the montium subgroup species D. auraria and D. serrata in the melanogaster group (Johnson et al. 2015), which last shared a common ancestor approximately as long ago as D. melanogaster and D. simulans (Nikolaidis and Scouras 1996), or ~1.5 million years ago (Cutter 2008) (Figure 3). In D. auraria, males have a stripe of pigment in each abdominal segment similar to D. melanogaster, but the more complete pigmentation of male abdominal segments is seen only on A6 rather than in A5 and A6 (Johnson et al. 2015). By contrast, males of D. serrata have an abdomen that is more yellow in color overall and lacks dark melanins almost completely in both A5 and A6 (Johnson et al. 2015). Using in situ hybridization, expression of ebony was found to be higher in the A5 and A6 segments of D. serrata than D. auraria, consistent with the role of ebony in the formation of yellow-tan sclerotins at the expense of dark melanins (Wittkopp et al. 2002a). This evolutionary change in expression appears to have resulted from changes in a cis-regulatory element located upstream of ebony that controls its expression in the A5 and A6 abdominal segments of males (Johnson et al. 2015).
Another montium subgroup species, D. kikkawai, which is estimated to have diverged from D. melanogaster ~20 million years ago (Prud'homme et al. 2006) (Figure 3), has also lost the dark male-specific pigmentation in A5 and A6, but in this case, changes in a male-specific enhancer of yellow that reduce its expression in these segments seems to have played a role (Jeong et al. 2006). Changes in yellow expression caused by cis-regulatory divergence have also been implicated in an expansion of male-specific abdominal pigmentation to include segments A3 and A4 in D. prostipennis relative to D. takahashi, two members of the oriental lineage in the melanogaster subgroup (Ordway et al. 2014) (Figure 3). Interestingly, D. prostipennis also showed changes in ebony and tan expression that correlated with the expanded male-specific pigmentation (decrease in ebony expression and increase in tan expression), but these changes in gene expression were found to be caused by divergence of trans-acting loci rather than cis-regulatory changes at ebony and tan (Ordway et al. 2014). cis-regulatory changes altering yellow expression have also been found to correlate with differences in abdominal pigmentation among the much more distantly related group of species, D. melanogaster, D. subobscura, D. willistoni, D. mojavensis, D. virilis and D. grimshawi (Wittkopp et al. 2002b; Kalay and Wittkopp 2010) (Figure 3). Observing these changes in cis-regulation and gene expression that correlate with divergent abdominal pigmentation for pigment synthesis genes yellow, tan, and ebony strongly suggests that these changes have contributed to pigmentation divergence, although their relative contributions in any individual case remain unknown.
Pigment synthesis genes are not the only source of abdominal pigmentation divergence between species; divergence in a transcription factor regulating expression of pigmentation genes, bab1, also plays a role in interspecific differences. In D. melanogaster, Bab1 expression represses development of dark pigmentation in segments A5 and A6 of males (Kopp et al. 2000). By contrast, in D. willistoni, a species without sexually dimorphic pigmentation in which males and females both have only a stripe of dark melanin near the posterior edge of each tergite, bab1 is expressed in segments A2-A6 in both sexes (Kopp et al. 2000; Williams et al. 2008). Sex-specific differences in abdominal bab1 expression seen in D. melanogaster were found to be controlled by a dimorphic cis-regulatory element containing binding sites for the transcription factors Abd-B and Dsx (Williams et al. 2008). Changes in the binding sites for these transcription factors as well as other changes in the cis-regulatory sequence were found to be responsible for the differences in bab1 cis-regulatory activity between D. melanogaster and D. willistoni (Williams et al. 2008). Divergence in this sexually dimorphic cis-regulatory element was also found to contribute to interspecific differences in bab expression that correlate with differences in female abdominal pigmentation among D. melanogaster, D. yakuba, D. fuyamai, and D. auraria (Rogers et al. 2013b).
Thorax pigmentation
Like abdominal pigmentation, thorax pigmentation varies widely in intensity and patterning within and among Drosophila species. Species like D. guttifera, for example, possess distinctive stripes of black melanin along their thorax that D. melanogaster and most other Drosophila species lack (Koshikawa et al. 2015). In D. melanogaster populations, individuals often vary in the intensity of black and brown melanins that fill a “trident” pattern on the thorax, and variation in this pattern tends to follow altitudinal or latitudinal clines around the world (David and Capy 1988; Parkash and Munjal 1999; Telonis-Scott et al. 2011). Intensity of UV radiation was also recently shown to be a good predictor of thorax pigmentation in D. melanogaster for clinal variation in Africa, with more darkly pigmented flies found to inhabit regions with higher levels of UV radiation (Bastide et al. 2014). This finding suggests that increased levels of melanin in the thorax may play a protective role for D. melanogaster in the wild; however, D. yakuba shows the opposite relationship between the intensity of UV radiation and abdominal pigmentation (which is often correlated with thorax pigmentation, (Rajpurohit and Gibbs 2012)) (Matute and Harris 2013), indicating that this is not a general relationship for all Drosophila. Regardless of the selective forces driving diversity of thorax pigmentation in Drosophila, the variety of pigment patterns seen within and among species provides the raw material needed to further investigate the genetic basis of phenotypic evolution.
Genetic basis of thorax pigmentation differences within a species
In natural populations of D. melanogaster and D. simulans, variation in a pigmented thorax trident pattern is often seen in which individuals differ in the intensity of darkness in trident shape and size (Capy et al. 1988; David and Capy 1988). A similar darkening of this trident pattern is also readily observed in D. melanogaster ebony loss-of-function mutants (Lindsley and Zimm 1992), suggesting that variation in ebony expression and/or activity might underlie this intraspecific diversity. Consistent with this hypothesis, Takahashi et al. (2007) found that a chromosomal region containing the ebony locus was most strongly associated with differences in trident pigmentation intensity between inbred lines of D. melanogaster isolated from West Africa and Taiwan. Complementation tests combined with differences in ebony expression levels between strains further suggested that regulatory changes at ebony contributed to these differences in trident pigmentation (Takahashi et al. 2007). Natural variation in trident intensity within a D. melanogaster population collected from Japan was also found to be associated with genetic variants in ebony enhancer regions located on the cosmopolitan inversion, In(3R)Payne (Takahashi and Takano-Shimizu 2011). Interestingly, none of the 19 nucleotide sites found to be in complete association with trident pigment intensity in this study overlapped with sites associated with differences in abdominal pigmentation in African populations described above (Pool and Aquadro 2007; Rebeiz et al. 2009a). Genetic variants associated with thoracic pigmentation in this Japanese population do still appear to affect cis-regulation of ebony, however, because differences in relative allelic expression were observed for ebony in F1 hybrids produced by crossing lightly and darkly pigmented lines of D. melanogaster from this population (Takahashi and Takano-Shimizu 2011). Variable sites located within an enhancer that drives expression in both the thorax and abdomen (Rebeiz et al. 2009b) failed to cause differences in cis-regulatory activity when tested in a common genetic background using reporter genes, however (Takahashi and Takano-Shimizu 2011). cis-regulatory variation affecting ebony expression also seems to contribute to variable thoracic pigmentation observed among the DGRP lines of D. melanogaster used in the Dembeck et al. (2015b) study of abdominal pigmentation, with the most strongly associated SNPs again unique to this population (Miyagi et al. 2015). Significant associations were also observed between genetic variants in known enhancers of tan and allele-specific tan expression levels, but not with variation in thoracic pigmentation (Miyagi et al. 2015). Taken together, these studies indicate that ebony cis-regulatory sequences are often variable in natural populations of D. melanogaster, with different genetic variants contributing to differences in thoracic pigmentation in different populations.
Genetic basis of thorax pigmentation differences between species
The best studied difference in thoracic pigmentation between species is that seen between D. guttifera and D. melanogaster. In D. guttifera, a member of the quinaria species group (Figure 3), males and females possess a distinct pattern of darkly pigmented stripes along their thorax in addition to the “polka-dot” deposits of black melanin seen on their abdomen and wings. To identify genes involved in the evolution of D. guttifera thoracic pigmentation, Koshikawa et al. (2015) examined the regulation of wingless expression, which was previously shown to be spatially correlated with the black polka-dots in the wings during development (Werner et al. 2010). After testing many non-coding sequences in and around wingless for activity in the thorax, an enhancer driving expression in this part of the body was finally located in an intron of the Wnt10 gene, two genes away from wingless (Koshikawa et al. 2015). This enhancer, called “gutTS” for D. guttifera thorax stripes, was sufficient to activate wingless expression during pupal stages of D. guttifera that mirrors the thoracic pigment stripes seen in adult D. guttifera (Koshikawa et al. 2015). In D. melanogaster, this cis-regulatory element drove weaker thoracic stripes, indicating that some trans-acting regulators of this wingless enhancer had diverged between species (Koshikawa et al. 2015). The orthologous enhancer from D. melanogaster was also tested for activity in both D. melanogaster and D. guttifera and failed to drive expression in thoracic strips in either species, indicating that cis-regulatory divergence had occurred between D. melanogaster and D. guttifera within the gutTS wingless enhancer (Koshikawa et al. 2015). These results suggest that the evolution of a novel cis-regulatory element affecting wingless expression contributes to the derived thoracic stripe pigment pattern seen in D. guttifera.
Wing pigmentation
D. melanogaster wings are evenly pigmented throughout the wing blade, but many other species of Drosophila (especially Hawaiian and Oriental species) have wing spots of dark melanins that vary in size, shape, and position on the wing (O'Grady and DeSalle 2000; Wittkopp et al. 2003a; Prud'homme et al. 2006; Edwards et al. 2007). These darkly pigmented wing patterns are often sexually dimorphic and thought to be the result of sexual selection. Males that possess wing spots in the Oriental melanogaster species group, for example, perform an elaborate wing display behavior in front of females during courtship, whereas males without wing spots tend to perform courtship from behind the female (Yeh and True 2006). Developmentally, these complex wing pigment patterns result from a two step process in which i) spatial pre-patterns of enzymes involved in the pigmentation synthesis pathway are laid down in the developing wing during the Drosophila pupal stage and ii) precursors for melanin such as dopa and dopamine are transported to the wing through the hemolymph and diffuse from the wing veins post-eclosion, polymerizing to form black and/or brown melanins in the shape of the enzymatic pre-patterns (True et al. 1999). The precise size and shape of wing spots often varies within species, but the genetic basis of this variation has yet to be determined. Several studies have, however, elucidated genetic mechanisms underlying interspecific differences in wing spot size and patterning, and these are reviewed below.
Genetic basis of wing pigmentation differences between species
In the melanogaster group of Drosophila, several species possess a darkly pigmented male-specific spot at the distal tip of their wing. Phylogenetic reconstructions suggest that the common ancestor of the melanogaster group lacked a wing spot and that the current distribution of this trait in this species resulted from at least one gain followed by multiple losses in independent lineages (Prud'homme et al. 2006). The best studied of these spotted species is D. biarmipes, a member of the Oriental lineage within the melanogaster species group (Figure 3), that has a single spot of dark pigmentation at the distal tip of the wing in males. This spot has been shown to be prefigured by expression of the Yellow protein and the absence of the Ebony protein during pupal stages (Wittkopp et al. 2002a). For yellow, the novel pattern of expression is caused by cis-regulatory changes in a pre-existing wing enhancer of yellow, suggesting that cis-regulatory evolution at yellow contributed to the evolution of the wing spot pattern (Gompel et al. 2005). Further investigation revealed that cis-regulatory changes affecting yellow expression had arisen independently in multiple lineages, with different pre-existing wing enhancers co-opted to create the novel patterns of wing spot expression (Prud'homme et al. 2006). In the case of spot divergence between the two sister species D. elegans (spotted) and D. gunungcola (spotless), which are also members of the Oriental lineage of the melanogaster group (Figure 3), the spot of yellow expression present in D. elegans is controlled by sequences orthologous to the spot enhancer in D. biarmipes and divergence of only a few nucleotides in this sequence is responsible for the loss of this yellow expression pattern (and presumably at least part of the wing spot) in D. gunungcola (Prud'homme et al. 2006). In another spotted species, however, D. tristis, which is a member of the obscura group, a wing spot prefigured by yellow expression has evolved using a novel cis-regulatory element that co-opted a different pre-existing wing enhancer of yellow (Prud'homme et al. 2006). Taken together, these studies suggest that the cis-regulatory sequences of yellow have evolved repeatedly to cause changes in gene expression that contribute to the gain and loss of wing spots in multiple Drosophila species.
To better understand how yellow expression is regulated and evolves, Arnoult et al. (2013) performed an RNAi screen in a strain of D. melanogaster that carried a reporter gene reflecting activity of the D. biarmipes spot enhancer. Among the ~350 screened transcription factors, five candidates emerged as potential activators of the D. biarmipes spot enhancer. One of these genes was Distalless (Dll), which has previously been shown to be important in Drosophila wing development (Cohen et al. 1991). Using RNAi knockdown, overexpression, and electrophoretic mobility shift assays, Dll was shown to be both necessary and sufficient for driving activity of the yellow spot enhancer in the wings of D. melanogaster (Arnoult et al. 2013). Moreover, manipulating Dll expression in D. biarmipes itself lead to a gain and loss of wing pigmentation when Dll was over- and under-expressed, respectively (Arnoult et al. 2013).
Changes in wing pigmentation were not observed when Dll expression was modified in D. ananassae, a species without a wing spot, indicating that the regulatory connection between Dll and yellow had evolved in the lineage leading to D. biarmipes since it last shared a common ancestor with D. ananassae (Arnoult et al. 2013). This regulatory link does not appear to be restricted to D. biarmipes, however, as correlations between Dll expression, yellow expression, and wing spots were also observed in D. pulchrella, D. elegans, D. rhopaloa, and D. prolongata (Arnoult et al. 2013). These data suggest an evolutionary trajectory in which Dll regulation of yellow was gained and then changes in Dll expression evolved to produce a variety of wing spot patterns. While the second step of this model remains to be tested, it is clear from these data that divergent expression patterns of Dll (as well as potentially other transcription factors) has contributed to the divergence of wing pigment patterns through the direct (and likely also indirect) modulation of genes in the pigmentation synthesis pathway.
In other Drosophila species, wing pigmentation is not limited to males and involves more than a single spot. For example, in D. guttifera, both males and females develop a polka-dot pattern of 16 dark melanin spots and 4 melanized areas across their wings (Werner et al. 2010; Koshikawa et al. 2015). Yellow expression during pupal stages again mirrors the final adult wing pigment pattern (Werner et al. 2010), as does expression of Ebony expression, which is reduced in regions with wing spots (Gompel et al. 2005). To identify cis-regulatory regions of yellow responsible for this spotted expression pattern, non-coding regions surrounding yellow were tested for cis-regulatory activity using a reporter gene introduced into D. melanogaster.
Unlike in other studies of yellow cis-regulatory elements (Wittkopp et al. 2002b; Gompel et al. 2005; Prud'homme et al. 2006; Kalay and Wittkopp 2010; Arnoult et al. 2013), the unique expression pattern of D. guttifera yellow could not be recapitulated by reporter genes in D. melanogaster, indicating that changes in trans-regulatory factors controlling yellow expression in D. guttifera had diverged between these two species. Transforming these reporter genes into D. guttifera did, however, drive spotted patterns of expression similar those seen for endogenous yellow (Werner et al. 2010). Through careful examination of the reporter constructs assayed in D. melanogaster, phenotypes observed in a spontaneous D. guttifera mutant, and prior knowledge of wing development, Werner et al (2010) identified wingless as a potential regulator of D. guttifera yellow. Ectopic expression of wingless in D. guttifera resulted in ectopic wing pigmentation, providing evidence that wingless does indeed regulate wing spot pigmentation in D. guttifera (Werner et al. 2010). Additional reporter gene experiments using an orthologous spot enhancer from a closely related species lacking wing spots, D. deflecta, also showed that D. guttifera had evolved a novel pattern of wingless expression that contributed to the evolution of its polka-dotted wings (Werner et al. 2010).
The novel expression pattern of wingless in D. guttifera could have evolved through changes in its cis-regulatory sequences, changes in one or more trans-acting regulators of wingless, or both. To determine whether cis-regulatory changes were responsible for divergent wingless expression, Koshikawa et al. (2015) tested noncoding sequences in and around the wingless gene for cis-regulatory activity in pupal wings. A cis-regulatory element located 3’ of D. guttifera wingless was found to drive expression in D. guttifera-like spots near the distal tip of the wing, an activity that seems to have evolved by co-opting activity of pre-existing cis-regulatory elements driving expression in the cross-veins and/or wing margin (Koshikawa et al. 2015). Two more cis-regulatory elements that appear to drive novel patterns of wingless expression in D. guttifera were also identified more than 69 kb away from wingless in introns of the Wnt10 gene (Koshikawa et al. 2015). Testing the activity of these cis-regulatory regions using transgenes inserted into D. melanogaster showed that changes in the cis-regulatory elements of wingless were largely sufficient to explain divergent wingless expression and presumably thus contribute to the evolution of novel wing pigmentation in D. guttifera (Koshikawa et al. 2015).
Because of the candidate gene approaches used to study the evolution of wing spots in the species described above, the contribution of cis-regulatory changes observed in yellow and wingless relative to changes that likely exist at other loci in the genome remain unknown. Two studies investigating the genetic basis of a difference in wing spot between interfertile species in the Oriental lineage of the melanogaster subgroup, D. elegans and D. gunungcola (Figure 3), begin to address this issue (Yeh and True 2006; 2014). D. elegans has a male-specific wing spot of dark pigment similar to that seen in D. biarmipes, whereas its sister species D. gunungcola has no spots of dark pigment on its wing (Prud'homme et al. 2006). The similarity of wing spots seen in D. biarmipes and D. elegans is consistent with the proposed inheritance from a common ancestor that also had a wing spot (Prud'homme et al. 2006), suggesting that the roles of yellow (Prud'homme et al. 2006) and Dll (Arnoult et al. 2013) in the development of the D. biarmipes wing spot described above are likely conserved in D. elegans. Genetic mapping of loci contributing to the difference in wing spot between D. elegans and D. gunungcola identified three QTL affecting the wing spot (Yeh and True 2006; 2014). Although each of these QTL encompass many genes, the inclusion of yellow in one QTL and Dll in another is consistent with prior studies suggesting that divergence at these loci contributes to the loss of the wing spot in D. gunungcola (Prud'homme et al. 2006; Arnoult et al. 2013). The QTL overlapping yellow provides more circumstantial evidence that the cis-regulatory divergence of yellow identified between D. elegans and D. gunungcola using reporter genes (Prud'homme et al. 2006) impacts pigmentation. In addition, the QTL overlapping Dll suggests that differences in Dll expression might exist between D. elegans and D. gunungcola and be caused by cis-regulatory changes at Dll itself, similar to observations for divergent wingless expression in D. guttifera (Koshikawa et al. 2015). The third QTL does not include any obvious candidate genes.
Pupal pigmentation
In addition to the highly variable pigment patterns of the Drosophila abdomen, thorax, and wings, differences in pigmentation are also seen among some species in the pupal cases from which the adult flies emerge. For example, in the virilis group of Drosophila (Figure 3), D. virilis has a distinctly darker pupal case color than its closest relatives, D. americana, D. lummei, and D. novamexicana (Stalker 1942). The D. virilis pupal case appears almost completely black, whereas pupal cases in the other species are lighter shades of brown and tan (Ahmed-Braimah and Sweigart 2015). The virilis species group is amenable to genetic dissection of this trait because D. americana, D. novamexicana, and D. virilis all produce fertile hybrids when crossed with each other (Heikkinen 1992). Early studies investigating the genetic basis of this difference in pupal color between D. virilis and D. americana suggested that it was was due to a large effect locus on chromosome 5 as well as other loci, possibly linked to chromosomes 2 and 3 (Stalker 1942). To identify the molecular basis of pupal color divergence between D. virilis and D. americana more precisely, Ahmed-Braimah and Sweigart (2015) analyzed a backcross population between these two species and scored more than 30,000 recombinant offspring for pupal case color. This experimental design allowed them to identify an ~11-kb sequence on chromosome 5 that contributes to the difference in pupal case color. This region contains the first exon and non-coding regions of the Dat gene (Ahmed-Braimah and Sweigart 2015). Dat, as described above and in Figure 1, is required for the conversion of dopamine to NADA, which is then polymerized into a colorless pigment. Expression differences were observed for Dat at the onset of pupation between D. americana (high expression) and D. virilis (low expression) (Ahmed-Braimah and Sweigart 2015) that suggest reduced expression of Dat in D. virilis creates an excess of dopamine that allows production of more dark melanins and thus a much darker pupal case. Pupal expression of Dat in D. novamexicana, which has a lighter body color than D. americana but a similarly colored pupal case, was similar to that observed for D. americana (Ahmed-Braimah and Sweigart 2015). Genetic variation linked to Dat did not explain any of the difference in body color between D. americana and D. novamexicana, consistent with prior work identifying ebony and tan as the primary drivers of divergent body color between these two species (Wittkopp et al. 2009).
Lessons learned from Drosophila pigmentation
With the rapid growth of studies identifying genes and genetic changes contributing to pigmentation differences within and between Drosophila species during the last 10 years, the time is ripe to step back and take an integrative look at the findings from these case studies. What have we learned about the genetic basis of pigmentation evolution and hopefully phenotypic evolution more generally? What questions remain unanswered?
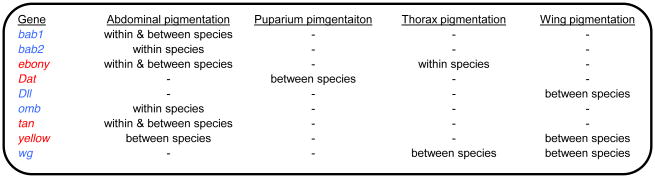
First and foremost, we’ve learned that the same handful of genes have been modified over and over again in different lineages to give rise to polymorphic pigmentation within a species as well as divergent pigmentation between species (Table 1). A similar pattern has also been seen for other types of evolutionary changes (Stern and Orgogozo 2009; Martin and Orgogozo 2013), suggesting that evolutionary trajectories are sometimes predictable. For pigmentation, genes harboring polymorphism and divergence that affects body color include genes that encode developmental regulators (blue in Figure 4) as well as enzymes required for pigment biosynthesis (red in Figure 4). The apparent reuse of these genes has likely been biased by the use of candidate gene approaches that limited analysis to these genes in some studies (Wittkopp et al. 2002b; Gompel et al. 2005; Prud'homme et al. 2006; Werner et al. 2010; Johnson et al. 2015); however, the same conclusion emerges if only studies using unbiased genetic mapping approaches are considered (Pool and Aquadro 2007; Wittkopp et al. 2009; Bastide et al. 2013; Dembeck et al. 2015a,b; Endler et al. 2016). Despite this repeatability, the set of nine genes implicated in pigmentation diversity thus far is clearly not exhaustive; Dembeck et al. (2015a) found SNPs in 84 loci that had significant associations with variable abdominal pigmentation in a single population of D. melanogaster.
Figure 4. The loci of pigmentation evolution.
A summary of genes implicated in pigmentation differences within and/or between species is shown. Genes labeled in blue are regulators of pigmentation development. Genes labeled in red are involved in the pigment biosynthesis pathway. All genetic changes identified as likely to be contributing to a pigmentation difference either within or between species thus far affect cis-regulatory sequences.
A second lesson learned results from the striking consistency seen in the types of functional genetic changes observed in genes contributing to pigmentation diversity: cis-regulatory changes in non-coding sequences appear responsible for a gene’s effects on pigmentation in all cases where the type of mutation is known (Table 1). This observation holds for both developmental regulators and genes in the pigment synthesis pathway (Table 1). cis-regulatory changes have been proposed to be the predominant source of evolutionary change in genes with pleiotropic effects on multiple traits because they allow one function of the gene to be modified without affecting others (Wray et al. 2003; Carroll 2008; Stern and Orgogozo 2008). All of the genes implicated in pigmentation diversity thus far are indeed pleiotropic and are regulated by multiple cis-regulatory elements that subdivide their functions. In addition to pigmentation, bab1 and bab2 also affect development of mechanosensory organs (Godt et al. 1993; Kopp et al. 2000); ebony, tan, yellow, and Dat also impact behavior (Shaw et al. 2000; Drapeau et al. 2003; True et al. 2005), and Dll, omb, and wg have widespread effects on development (DrysdaleFlyBase Consortium 2008). The genetic basis of pigmentation differences in vertebrates reveals a different pattern, however, with changes in pigmentation attributed more equally to cis-regulatory changes and changes in amino acid sequence affecting protein function (Hubbard et al. 2010).
A final message to emerge from these studies is that intra- and interspecific sources of pigmentation diversity share some properties but not others. For example, nearly all genes shown to contribute to differences in abdminal pigmentation within a species also contribute to pigmentation differences that exist between species (Figure 4). One notable exception is yellow. Changes in yellow expression often accompany changes in pigmentation between Drosophila species, but they have yet to be implicated in intraspecific variation. This might be because overexpression of yellow has more subtle effects on pigmentation than overexpression of ebony, tan, or bab1 (Wittkopp et al. 2002a; Jeong et al. 2008; Wittkopp et al. 2009; Salomone et al. 2013), such that changes in yellow expression arising alone within a species are insufficient for altering pigmentation in most populations (but see Wittkopp et al. 2002b). Genetic changes in the same cis-regulatory regions have been observed within and between species, but the scope of these changes differs. Within a species, genetic variants typically modulate activity of existing cis-regulatory elements, with different variants affecting cis-regulatory activity in different populations. By contrast, divergent sites that differ between species are much more likely to have given rise to a novel enhancer that co-opts pre-existing developmental regulators. Differences between alleles contributing to intra- and interspecifc pigmentation variation are not always apparent, however, as the alleles of tan and ebony contributing to divergent pigmentation in D. novamexiana were found to also contribute to clinal variation in pigmentation within D. americana (Wittkopp et al. 2009).
As illustrated in this chapter, detailed studies of pigmentation divergence within and among Drosophila species have provided an unprecedented look at the genetic mechanisms underlying phenotypic evolution over various timescales. There is still much more to be learned from studying this system, however. For example, many QTLs contributing to pigmentation differences within and between species have been identified for which the causative genes remain unknown. Identifying these genes might alter our view of the types of genes most likely to harbor genetic changes affecting pigmentation. Many direct and indirect regulators of genes in the pigment synthesis pathway are also yet to be identified. Knowing the identity of these factors and the sequences they bind to will help us understanding why some non-coding changes alter pigmentation while others do not. Important questions also remain about how the complementary changes in expression of pigmentation genes such as yellow and ebony that are often observed between species have evolved. Finally, improving our understanding of both the ecological functions of pigmentation in specific taxa as well as the pleiotropic effects of pigmentation genes will help us better understand the role natural selection might play in shaping the genetic basis of pigmentation evolution. Ultimately, understanding the genetic and molecular mechanisms underlying pigmentation diversity has the potential to answer questions not only about evolution, but also about ecology, biochemistry, and neuroscience.
References
- Ahmed-Braimah YH, Sweigart AL. A single gene causes an interspecific difference in pigmentation in Drosophila. Genetics. 2015;200:331–342. doi: 10.1534/genetics.115.174920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult L, Su KFY, Manoel D, Minervino C, Magriña J, Gompel N, Prud'homme B. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science. 2013;339:1423–1426. doi: 10.1126/science.1233749. [DOI] [PubMed] [Google Scholar]
- Bastide H, Betancourt A, Nolte V, Tobler R, Stöbe P, Futschik A, Schlötterer C. A genome-wide, fine-scale map of natural pigmentation variation in Drosophila melanogaster. PLoS Genetics. 2013;9:e1003534. doi: 10.1371/journal.pgen.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H, Yassin A, Johanning EJ, Pool JE. Pigmentation in Drosophila melanogaster reaches its maximum in Ethiopia and correlates most strongly with ultra-violet radiation in sub-Saharan Africa. BMC Evolutionary Biology. 2014;14:179. doi: 10.1186/s12862-014-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel RD, Kopp A, Nuzhdin SV. Composite effects of polymorphisms near multiple regulatory elements create a major-effect QTL. PLoS Genetics. 2011;7:e1001275. doi: 10.1371/journal.pgen.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MJ, Werner T, Dyer KA. Two genomic regions together cause dark abdominal pigmentation in Drosophila tenebrosa. Heredity. 2014;112:454–462. doi: 10.1038/hdy.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Templeton AR, Duncan I. Population genetics of the developmental gene optomotor-blind (omb) in Drosophila polymorpha: evidence for a role in abdominal pigmentation variation. Genetics. 2004;168:1999–2010. doi: 10.1534/genetics.104.032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, David JR, Robertson A. Thoracic trident pigmentation in natural populations of Drosophila simulans: a comparison with D. melanogaster. Heredity. 1988;61:263–268. [Google Scholar]
- Carbone MA, Llopart A, deAngelis M, Coyne JA, Mackay TFC. Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea. Genetics. 2005;171:211–225. doi: 10.1534/genetics.105.044412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou ML, Silvain JF, Daubin V, Da Lage JL, Lachaise D. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Molecular Ecology. 2001;10:649–660. doi: 10.1046/j.1365-294x.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Clusella-Trullas S, Terblanche JS, et al. Local adaptation for body color in Drosophila americana: commentary on Wittkopp. Heredity. 2011;106:904–905. doi: 10.1038/hdy.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, Wimmer EA, Cohen SM. Early development of leg and wing primordia in the Drosophila embryo. Mechanisms of development. 1991;33:229–240. doi: 10.1016/0925-4773(91)90030-a. [DOI] [PubMed] [Google Scholar]
- Cooley AM, Shefner L, McLaughlin WN, Stewart EE, Wittkopp PJ. The ontogeny of color: developmental origins of divergent pigmentation in Drosophila americana and D. novamexicana. Evolution & Development. 2012;14:317–325. doi: 10.1111/j.1525-142X.2012.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, Mcmanus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. Tempo and mode of regulatory evolution in Drosophila. Genome Research. 2014;24:797–808. doi: 10.1101/gr.163014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Molecular Biology and Evolution. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- da Cunha AB. Genetic analysis of the polymorphism of color pattern. Drosophila polymorpha Evolution. 1949;3:239–251. doi: 10.1111/j.1558-5646.1949.tb00024.x. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends in Genetics. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- Dembeck LM, Huang W, Carbone MA, Mackay TFC. Genetic basis of natural variation in body pigmentation in Drosophila melanogaster. Fly. 2015a;9:75–81. doi: 10.1080/19336934.2015.1102807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck LM, Huang W, Magwire MM, Lawrence F, Lyman RF, Mackay TFC. Genetic Architecture of Abdominal Pigmentation in Drosophila melanogaster. PLoS Genetics. 2015b;11:e1005163. doi: 10.1371/journal.pgen.1005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau MD, Radovic A, Wittkopp PJ, Long AD. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. Journal of Neurobiology. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- Drysdale R, FlyBase Consortium FlyBase : a database for the Drosophila research community. Methods in Molecular Biology. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Doescher LT, Kaneshiro KY, Yamamoto D. A database of wing diversity in the Hawaiian Drosophila. PloS ONE. 2007;2:e487. doi: 10.1371/journal.pone.0000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler L, Betancourt AJ, Nolte V, Schlötterer C. Reconciling Differences in Pool-GWAS Between Populations: A Case Study of Female Abdominal Pigmentation in Drosophila melanogaster. Genetics. 2016;202:843–855. doi: 10.1534/genetics.115.183376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D, Couderc JL, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Heikkinen E. Genetic basis of reduced eyes in the hybrids of Drosophila virilis phylad species. Hereditas. 1992;117:275–285. doi: 10.1111/j.1601-5223.1992.tb00025.x. [DOI] [PubMed] [Google Scholar]
- Hollocher H, Hatcher JL, Dyreson EG. Genetic and developmental analysis of abdominal pigmentation differences across species in the Drosophila dunni subgroup. Evolution. 2000;54:2057–2071. doi: 10.1111/j.0014-3820.2000.tb01249.x. [DOI] [PubMed] [Google Scholar]
- Hubbard JK, Uy JAC, Hauber ME, Hoekstra HE, Safran RJ. Vertebrate pigmentation: from underlying genes to adaptive function. Trends in Genetics. 2010;26:231–239. doi: 10.1016/j.tig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Johnson WC, Ordway AJ, Watada M, Pruitt JN, Williams TM, Rebeiz M. Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Drosophila Species. PLoS Genetics. 2015;11:e1005279. doi: 10.1371/journal.pgen.1005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay G. Rapid evolution of cis-regulatory architecture and activity in the Drosophila yellow gene. 2012 [Google Scholar]
- Kalay G, Wittkopp PJ. Nomadic enhancers: tissue-specific cis-regulatory elements of yellow have divergent genomic positions among Drosophila species. PLoS Genetics. 2010;6:e1001222. doi: 10.1371/journal.pgen.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kopp A. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution. 2009;63:2771–2789. doi: 10.1111/j.1558-5646.2009.00761.x. [DOI] [PubMed] [Google Scholar]
- Kopp A, Graze RM, Xu S, Carroll SB, Nuzhdin SV. Quantitative trait loci responsible for variation in sexually dimorphic traits in Drosophila melanogaster. Genetics. 2003;163:771–787. doi: 10.1093/genetics/163.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, Carroll SB. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7524–7529. doi: 10.1073/pnas.1509022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraminsky GP, Clark WC, Estelle MA, Gietz RD, Sage BA, O'Connor JD, Hodgetts RB. Induction of translatable mRNA for dopa decarboxylase in Drosophila: an early response to ecdysterone. Proc. Natl. Acad. Sci. U.S.A. 1980;77:4175–4179. doi: 10.1073/pnas.77.7.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst MR, Barsh GS, Kopp A, Mallet J, Monteiro A, Mullen SP, Protas M, Rosenblum EB, Schneider CJ, Hoekstra HE. Unraveling the thread of nature's tapestry: the genetics of diversity and convergence in animal pigmentation. Pigment Cell & Melanoma Research. 2012;25:411–433. doi: 10.1111/j.1755-148X.2012.01014.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research. 2011;39:W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. 1992 [Google Scholar]
- Llopart A, Elwyn S, Lachaise D, Coyne JA. Genetics of a difference in pigmentation between Drosophila yakuba and Drosophila santomea. Evolution. 2002;56:2262–2277. doi: 10.1111/j.0014-3820.2002.tb00150.x. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RRH, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu L-L, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu Y-Q, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. The Loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- Martinez MN, Cordeiro AR. Modifiers of Color Pattern Genes in DROSOPHILA POLYMORPHA. Genetics. 1970;64:573–587. doi: 10.1093/genetics/64.3-4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, Harris A. The influence of abdominal pigmentation on desiccation and ultraviolet resistance in two species of Drosophila. Evolution. 2013;67:2451–2460. doi: 10.1111/evo.12122. [DOI] [PubMed] [Google Scholar]
- Miyagi R, Akiyama N, Osada N, Takahashi A. Complex patterns of cis-regulatory polymorphisms in ebony underlie standing pigmentation variation in Drosophila melanogaster. Molecular Ecology. 2015;24:5829–5841. doi: 10.1111/mec.13432. [DOI] [PubMed] [Google Scholar]
- Morales-Hojas R, Vieira CP, Vieira J. Inferring the evolutionary history of Drosophila americana and Drosophila novamexicana using a multilocus approach and the influence of chromosomal rearrangements in single gene analyses. Molecular Ecology. 2008;17:2910–2926. doi: 10.1111/j.1365-294X.2008.03796.x. [DOI] [PubMed] [Google Scholar]
- Ng CS, Hamilton AM, Frank A, Barmina O, Kopp A. Genetic basis of sex-specific color pattern variation in Drosophila malerkotliana. Genetics. 2008;180:421–429. doi: 10.1534/genetics.108.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Scouras ZG. The Drosophila montium subgroup species. Phylogenetic relationships based on mitochondrial DNA analysis. Genome. 1996;39:874–883. doi: 10.1139/g96-110. [DOI] [PubMed] [Google Scholar]
- O'Grady PM, DeSalle R. How the fruit fly changed (some of) its spots. Current Biology. 2000;10:R75–7. doi: 10.1016/s0960-9822(00)00281-5. [DOI] [PubMed] [Google Scholar]
- Ohnishi S, Watanabe T. Genetic analysis of color dimorphism in the Drosophila montium subgroup. Jpn J Genet. 1985;60:355–358. [Google Scholar]
- Ordway AJ, Hancuch KN, Johnson W, Wiliams TM, Rebeiz M. The expansion of body coloration involves coordinated evolution in cis and trans within the pigmentation regulatory network of Drosophila prostipennis. Developmental Biology. 2014;392:431–440. doi: 10.1016/j.ydbio.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Orr HA. The genetics of species differences. Trends in Ecology & Evolution. 2001 doi: 10.1016/s0169-5347(01)02167-x. [DOI] [PubMed] [Google Scholar]
- Parkash R, Munjal AK. Phenotypic variability of thoracic pigmentation in Indian populations of Drosophila melanogaster. Journal of Zoological Systematics and …. 1999 [Google Scholar]
- Pool JE, Aquadro CF. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Molecular Ecology. 2007;16:2844–2851. doi: 10.1111/j.1365-294X.2007.03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh S-D, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Rajpurohit S, Gibbs AG. Selection for abdominal tergite pigmentation and correlated responses in the trident: a case study in Drosophila melanogaster. Biological Journal of the Linnean Society. 2012;106:287–294. [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009a;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Ramos-Womack M, Jeong S, Andolfatto P, Werner T, True J, Stern DL, Carroll SB, et al. Evolution of the tan locus contributed to pigment loss in Drosophila santomea: a response to Matute. Cell. 2009b;139:1189–1196. doi: 10.1016/j.cell.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Robertson A, Briscoe DA, Louw JH. Variation in abdomen pigmentation in Drosophila melanogaster females. Genetica. 1977;47:73–76. [Google Scholar]
- Rogers WA, Grover S, Stringer SJ, Parks J, Rebeiz M, Williams TM. A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Developmental Biology. 2013a doi: 10.1016/j.ydbio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Rogers WA, Salomone JR, Tacy DJ, Camino EM, Davis KA, Rebeiz M, Williams TM. Recurrent modification of a conserved cis-regulatory element underlies fruit fly pigmentation diversity. PLoS Genetics. 2013b;9:e1003740. doi: 10.1371/journal.pgen.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomone JR, Rogers WA, Rebeiz M, Williams TM. The evolution of Bab paralog expression and abdominal pigmentation among Sophophora fruit fly species. Evolution & Development. 2013;15:442–457. doi: 10.1111/ede.12053. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Stalker HD. The Inheritance of a Subspecific Character in the Virilis Complex of Drosophila. The American Naturalist. 1942;76:426–431. [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution. 2011;65:629–642. doi: 10.1111/j.1558-5646.2010.01165.x. [DOI] [PubMed] [Google Scholar]
- Sugumaran M, Giglio L, Kundzicz H, Saul S, Semensi V. Studies on the enzymes involved in puparial cuticle sclerotization in Drosophila melanogaster. Archives of insect biochemistry and physiology. 1992;19:271–283. doi: 10.1002/arch.940190406. [DOI] [PubMed] [Google Scholar]
- Takahashi A. Pigmentation and behavior: potential association through pleiotropic genes. Drosophila Genes & Genetic Systems. 2013;88:165–174. doi: 10.1266/ggs.88.165. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Takahashi K, Ueda R, Takano-Shimizu T. Natural variation of ebony gene controlling thoracic pigmentation in Drosophila melanogaster. Genetics. 2007;177:1233–1237. doi: 10.1534/genetics.107.075283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Takano-Shimizu T. Divergent enhancer haplotype of ebony on inversion In(3R)Payne associated with pigmentation variation in a tropical population of Drosophila melanogaster. Molecular Ecology. 2011;20:4277–4287. doi: 10.1111/j.1365-294X.2011.05260.x. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Hoffmann AA, Sgro CM. The molecular genetics of clinal variation: a case study of ebony and thoracic trident pigmentation in Drosophila melanogaster from eastern Australia. Molecular Ecology. 2011;20:2100–2110. doi: 10.1111/j.1365-294X.2011.05089.x. [DOI] [PubMed] [Google Scholar]
- True JR, Edwards KA, Yamamoto D, Carroll SB. Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Current Biology. 1999;9:1382–1391. doi: 10.1016/s0960-9822(00)80083-4. [DOI] [PubMed] [Google Scholar]
- True JR. Insect melanism: the molecules matter. Trends in Ecology & Evolution. 2003;18:640–647. [Google Scholar]
- True JR, Yeh S-D, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou S-R, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MF, Zeineh LL, Black BC, McIvor WE, Wright TR, Biessmann H. Catecholamine metabolism and in vitro induction of premature cuticle melanization in wild type and pigmentation mutants of Drosophila melanogaster. Archives of insect biochemistry and physiology. 1996;31:219–233. doi: 10.1002/(SICI)1520-6327(1996)31:2<219::AID-ARCH9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Smith-Winberry G, Arnold LL, Thompson EM, Cooley AM, Yuan DC, Song Q, McAllister BF. Local adaptation for body color in Drosophila americana. Heredity. 2011;106:592–602. doi: 10.1038/hdy.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Seminars in Cell & Developmental Biology. 2009;20:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends in Genetics. 2003a;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nature Genetics. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, Thompson EM, Akhras S, Smith-Winberry G, Shefner L. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science. 2009;326:540–544. doi: 10.1126/science.1176980. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002a;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Current Biology. 2002b;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2003b;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Molecular Biology and Evolution. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Wright TR. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Advances in genetics. 1987;24:127–222. [PubMed] [Google Scholar]
- Yassin A, Bastide H, Chung H, Veuille M, David JR, Pool JE. Ancient balancing selection at tan underlies female colour dimorphism in Drosophila erecta. Nature communications. 2016;7:10400. doi: 10.1038/ncomms10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S-D, True JR. The Genetic Architecture of Coordinately Evolving Male Wing Pigmentation and Courtship Behavior in Drosophila elegans and D. gunungcola. Heredity. 2006;96:383–395. doi: 10.1038/sj.hdy.6800814. [DOI] [PubMed] [Google Scholar]
- Yeh S-D, True JR. The genetic architecture of coordinately evolving male wing pigmentation and courtship behavior in Drosophila elegans and Drosophila gunungcola. G3. 2014;4:2079–2093. doi: 10.1534/g3.114.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Guo Q, Li M, Li M, Li J, Miao X, Huang Y. Disruption of an N-acetyltransferase gene in the silkworm reveals a novel role in pigmentation. Development. 2010;137:4083–4090. doi: 10.1242/dev.053678. [DOI] [PubMed] [Google Scholar]