Abstract
Interactions between proteins and nucleic acids are frequently analyzed using electrophoretic mobility shift assays (EMSAs). This technique separates bound protein:nucleic acid complexes from free nucleic acids by electrophoresis, most commonly using polyacrylamide gels. The current study utilizes recent advances in agarose gel electrophoresis technology to develop a new EMSA protocol that is simpler and faster than traditional polyacrylamide methods. Agarose gels are normally run at low voltages (~10 V/cm) to minimize heating and gel artifacts. In this study we demonstrate that EMSAs performed using agarose gels can be run at high voltages (≥ 20 V/cm) with 0.5× TB (Tris-borate) buffer, allowing for short run times while simultaneously yielding high band resolution. Several parameters affecting band and image quality were optimized for the procedure, including gel thickness, agarose percentage, and applied voltage. Association of the siRNA-binding protein p19 with its target RNA was investigated using the new system. The agarose gel and conventional polyacrylamide gel methods generated similar apparent binding constants in side-by-side experiments. A particular advantage of the new approach described here is that the short run times (5 – 10 minutes) reduce opportunities for dissociation of bound complexes, an important concern in non-equilibrium nucleic acid binding experiments.
Keywords: RNA binding, DNA binding, binding affinity, nucleic acid binding protein, electrophoretic mobility shift assay, gel shift assay
1. Introduction
The efficient separation of bound and unbound molecules is critical for accurate measurement of the binding affinity between a protein and its cognate ligand. Historically, dissociation constants (KD) for protein:nucleic acid interactions have been measured using both equilibrium-based and non-equilibrium-based methods. Examples of equilibrium-based approaches to measure dissociation constants include techniques such as isothermal titration calorimetry and sedimentation equilibrium [1–4]. Non-equilibrium methods include techniques such as native gel electrophoretic mobility shift assays (EMSAs) and filter-binding assays, which are affordable, fast, and measure apparent dissociation constants [5–7]. To most accurately capture the amounts of free and bound ligand at equilibrium, these latter methods require rapid separation of the free and bound species and quantitation of their relative amounts.
Because of their simplicity, low cost, and speed, EMSAs are among the most commonly used approaches for studying interactions between proteins and DNA or RNA. EMSAs also afford the ability to monitor the formation of multiple bound species, including supershifts and cooperative complexes [8]. Typical EMSA experiments involve separation of free nucleic acids from bound complexes using native polyacrylamide gel electrophoresis (PAGE). Polyacrylamide gels have historically been preferred over agarose gels because they exhibit greater resolution of the relatively small nucleic acids frequently used in the assays [9,10]. However, the use of polyacrylamide gels for conventional EMSAs involves several drawbacks. For example, the polyacrylamide gels used to separate free and bound ligands require the handling of unpolymerized acrylamide, a potent neurotoxin. Additionally, nondenaturing polyacrylamide gels must be prepared by performing free radical-catalyzed polymerization reactions that are more tedious than the simpler methods required to prepare agarose gels. Methods that avoid these approaches increase safety and reduce cost to the laboratory. Because of their ease of preparation and use, agarose gels have been employed in a number of past studies involving EMSAs. The first agarose-based EMSAs were generally restricted to larger nucleic acids, ranging in length from hundreds of nucleotides [11–17] to thousands of nucleotides [18–21] because of the poor resolution of small oligonucleotides in conventional agarose gels. To improve oligonucleotide resolution, researchers have successfully employed a variety of approaches, including higher percentage agarose gels, the incorporation of a commercial additive, a proprietary agarose matrix, and by altering the running buffer to contain lithium borate in place of conventional Tris or MOPS buffers [22–27]. These studies spanned a variety of protein:nucleic acid systems, including transcription factors, recombinases, transposases, internal ribosomal entry sites, and ribosomal proteins bound to pre-rRNAs. While nearly all of these studies observed distinct shifted bands, they all describe EMSAs that were run for over 30 min, and many experiments required up to 4 hours of electrophoresis.
Recently, improvements in agarose gel electrophoresis conditions have been described that allow for rapid (< 15 min) separation of small DNA and RNA species with high resolution using high percentage agarose (> 2.5%) and 0.5× Tris-borate (TB) buffer without EDTA [28]. By reducing the buffer concentration and eliminating EDTA, gels could be electrophoresed at higher voltages with less heating and with shorter run times. It is likely that faster runs in EMSA experiments, reducing opportunities for irreversible dissociation while in the presence of an electric field, will permit more accurate measurements of the initial equilibrium binding conditions. Faster run times also reduce diffusion of nucleic acids in gels, resulting in sharper band resolution [28]. Therefore, this method should be especially beneficial for separation of the small, highly-diffusible nucleic acids (< 50 nt) that are often used in traditional polyacrylamide EMSAs. The new approach has recently been used to measure the adsorption of 25 nt and 54 nt RNA molecules to clay nanoparticles [29].
We show here that these rapid agarose gel methods can also be adapted for quantitatively measuring binding affinities between proteins and nucleic acids. More specifically, we have optimized the previously described agarose gel electrophoresis approach for use with EMSAs involving binding of proteins to small RNA oligonucleotides. As a model RNA:protein binding system, we employed the siRNA-binding protein p19 and its cognate dsRNA ligand [30]. This stable intermolecular interaction has a reported KD,app of 170 ± 20 pM, determined using conventional polyacrylamide gel EMSAs in conjunction with radiolabeled RNA ligands [31]. The binding affinity of a commercial form of p19 for its cognate RNA was measured using the new high voltage agarose method and the results were compared to those obtained using conventional polyacrylamide gels. The aggregate data demonstrate that EMSAs performed using agarose gels can produce results that are qualitatively and quantitatively comparable to those obtained with PAGE gels.
2. Materials & Methods
2.1 Binding reactions
RNA oligonucleotides (p19RNA-1, 5′-AUCUCAACCAGCCACUGCUAA, and p19RNA-2, 5′-AGCAGUGGCUGGUUGAGAUUU) were obtained from Integrated DNA Technologies. The 21-nt complementary strands were annealed by mixing equivalent molar amounts of each oligonucleotide, heating to 90 °C for 4 minutes, then allowing the solution to slow cool to room temperature over 30 minutes. Successful annealing was confirmed by agarose gel electrophoresis (Supp. Fig. 1A). The p19 siRNA binding protein (10 U/μL) used here was a commercially produced double-fusion protein, with a maltose binding protein fused to the N-terminus and a chitin-binding domain fused to the C-terminus (New England BioLabs). The binding properties of this double-fusion version of p19 have been previously described [32]. Twelve-microliter binding reactions were prepared with 20 ng of double-stranded RNA in 1× Binding Buffer (NEB) with varying protein concentrations. The reactions were briefly vortexed and then incubated at 25 °C for 1 hour. After incubation, 2 μL of 30% glycerol was added to each sample and, after brief vortexing, a total of 11 μL was loaded into each well of either an agarose or a polyacrylamide gel. Gels were prepared during incubation so that electrophoresis could immediately follow the binding reaction.
2.2 Agarose gel electrophoresis
Various percentages of agarose gel solutions were prepared with agarose LE powder (Gold Biotechnology) using 0.5× TB buffer (45 mM Tris, 45 mM boric acid) [28]. To ensure uniform depth of the viscous high-percentage gels, a taped 11×14 cm gel tray was pre-incubated at 60°C while the gel solution was prepared (~ 3 minutes). The gel was then poured into the pre-warmed tray in the incubator; a 10- or 12-well comb was inserted and the gel solution was allowed to settle for two minutes. The tray was then removed from the incubator and allowed to solidify at room temperature for at least 20 minutes. All gels contained 45, 50, or 55 mL total volumes and the solidified gels were approximately 6 – 8 mm thick. Gels were run in a submarine-style electrophoresis rig (Horizon 11–14 from Labrepco) in 0.5× TB buffer at the indicated voltages and times. Most electrophoreseis experiments were performed using a GE Healthcare EPS601 power supply. After electrophoresis, gels were stained for 40 minutes with mild shaking in 50 mL 2× SYBR Gold (Invitrogen), which was created by diluting 10 μL SYBR Gold stock into 50 mL 0.5× TB buffer (1:5000 dilution). To reduce background signal, the agarose gel was destained in ~150 mL 0.5× TB for 15 minutes with shaking.
2.3 Polyacrylamide gel electrophoresis
1.5 mm thick 6.7% native polyacrylamide gels were prepared using 29:1 acrylamide:bis-acrylamide (National Diagnostics Accugel) in 1× TBE (89 mM Tris, 89 mM boric acid 2 mM EDTA). Gels were run in a vertical electrophoresis rig (Bio-Rad Mini PROTEAN Tetracell) at 200 V for 20 minutes at room temperature using either a BioRad PowerPac HC or Life Technologies PowerEase 300W power supply. Each gel was stained for 10 minutes in 50 mL 2× SYBR Gold as described above. No destaining was performed for the polyacrylamide gels.
2.4 Gel imaging and quantitation
Both agarose and PAGE gels were imaged using a Bio-Rad Molecular Imager ChemiDoc XRS+ Imaging System set to the SYBR Gold Nucleic Acid Application with an exposure time of 0.6 seconds. Gels were quantitated with ImageLab (Bio-Rad, Inc.) using the externally-corrected, tight-binding method as previously described [33]. Band intensities were measured using the manual volume application within the program. All data used to calculate the KD,app employed the “adjusted volume” measures calculated by ImageLab, which are derived from the “local background” algorithm native to the program. The background signal from the agarose or the acrylamide gel was measured in an equal area of the gel that did not contain the binding samples. This background was subtracted from the sample volumes, and these background-corrected samples were plotted as the fraction of ligand bound versus protein concentration (Kaleidagraph, Synergy Software). KD,app values were calculated by fitting these data to the quadratic form of the binding isotherm
| (Eq. 1) |
in which [P]T is the total protein concentration, [L]T is the total RNA ligand concentration, [PL] is the concentration of protein-bound RNA, and KD,app is the apparent dissociation constant.
3. Results
Previous work established that high percentage agarose gels (3 – 4%) run in 0.5× TB at high voltages improved the resolution of dsDNAs between 20 and 150 bp as well as dsRNAs between 17 and 150 bp [28,29]. These studies suggested that high-percentage, high-voltage agarose electrophoresis is amenable to measuring the binding affinities of RNA/protein interactions. However, a concern is that high voltages may force molecules to migrate rapidly through the gel matrix, which may increase the risk of destroying the nucleic acid-protein complexes [34]. Several parameters were evaluated in the current study to identify the optimum conditions for separation and resolution of free RNA and protein-RNA complexes, using the p19/siRNA system as a model.
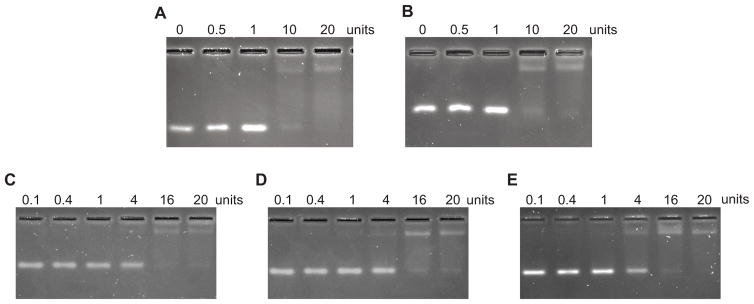
RNA was detected using SYBR Gold staining, a safe and sensitive detection method that is a widely-used alternative to the carcinogen ethidium bromide. Although 10 – 20 ng RNA could be readily detected (Supp. Fig. 1B), we observed that high concentrations of agarose caused a high level of background signal after SYBR Gold staining and destaining. To minimize background fluorescence caused by the retention of stain in the thick agarose matrix, we reduced both the concentration of agarose and the total volume of the gel (Fig. 1). Initial experiments that reduced the agarose concentration from 3.5% to 3.0% considerably reduced the background fluorescence (Fig. 1, panels A and B), even in a relatively large gel volume of 55 mL. Additional improvements in signal-to-noise were achieved by further reducing the agarose concentration to 2.5% while simultaneously reducing the total volume of the gel (and thus its thickness) from 55 mL to 45 mL (Fig. 1, panels C – E). The optimized conditions of 2.5% agarose and 45 mL gel volume maintained the excellent band resolution that was previously observed when running the gels at 20 – 25 V/cm. By minimizing the agarose concentration, we were also able to reduce the times needed to stain and destain the gel to 30 min and 15 min, respectively. In fact, longer incubations during staining and destaining were found to be counterproductive, because they allowed the RNA to diffuse in the gel, which significantly reduced both the resolution and signal-to-background ratio of the bands (Supp. Fig. 1C). From loading the sample to imaging the destained agarose gel, the total processing time using the optimized conditions was less than 1 hour.
Figure 1. Reduction of the fluorescent background improved the signal-to-noise ratio.
Agarose concentration and gel thickness were optimized to reduce the amount of fluorescence retained by the agarose gels. (A and B) 55 mL agarose gels were run at 25 V/cm for 10 min. Protein concentrations ranged from 0 to 1 U/μL. (C, D, and E) Agarose gels were run at 20 V/cm for 10 minutes at room temperature. Protein concentrations ranged from 0 to 0.8 U/μL. Gels were stained in SYBR Gold for 30 minutes and destained in 0.5× TB for 15 minutes before imaging.
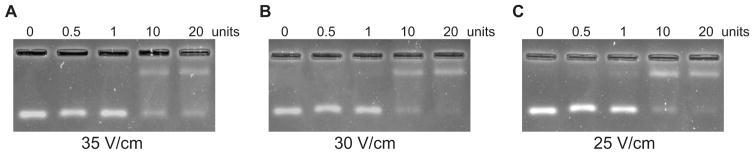
Next, we identified the optimal voltage for separating the free and bound RNAs while retaining high resolution of both the free and bound species. Using a 3.0% agarose gel, we tested several voltages (35, 30, 25, and 20 V/cm, where cm refers to the total length of the gel) (Fig. 2). With decreasing voltage, we increased the run time from 6 min to 10 min to ensure that the high molecular weight protein/RNA complex entered the gel matrix. Higher voltages of 35 (data not shown) and 30 V/cm rapidly and cleanly resolved the free RNA. However, the protein-RNA complex was not stable at these voltages, as demonstrated by the presence of free RNA bands at higher protein concentrations as well as diffuse shifted RNA bands and increased smearing throughout the lane (Fig. 2, panels A–B). Reducing the voltage to 25 or 20 V/cm and running for 10 min allowed for clear separation of free and bound RNAs with excellent resolution of both species (Fig. 2, panel C).
Figure 2. Optimization of voltage improves the resolution of both free and protein-bound RNA species.
30 ng of dsRNA was incubated with increasing concentrations of p19 protein (0 – 1 U/μL) and then the bound and free complexes were separated on 2.5% agarose gels at varying voltages at room temperature. (A) 30 V/cm; (B) 25 V/cm; and (C) 20 V/cm. Gels were stained in SYBR Gold for 30 minutes and destained in 0.5× TB for 15 minutes before imaging.
The robust resolution that was achieved with the new high-percentage, high-voltage agarose gel conditions enabled quantification of the free and bound fractions of the RNA ligand, which can be used to determine the binding constants for the p19/RNA interaction. To directly compare the agarose system to a conventional polyacrylamide EMSA, we first optimized polyacrylamide EMSA conditions for the p19/RNA system. Briefly, 6.7% polyacrylamide minigels were run in 1× TBE at 200 V for 20 min to separate free RNA from protein-bound RNA. Traditionally, polyacrylamide gels are often run in a cold environment to minimize heating from the electrophoretic field and promote protein stability [34]. However, in the p19 system, this approach was counterproductive, as the 2-nucleotide 3′ ssRNA overhang of the p19 dsRNA ligand has a TM of 4 °C [35]. Therefore, the dsRNAs were able to anneal and form multimeric complexes when the gels were run in the cold (Supp. Fig. 2). As observed in the agarose gel system, the RNA binding activity of p19 was stable at room temperature (Fig. 1 & 2). No significant heating was observed in the polyacrylamide gel system despite the high voltage (data not shown), which allowed the conventional native PAGE system to be run at room temperature as well.
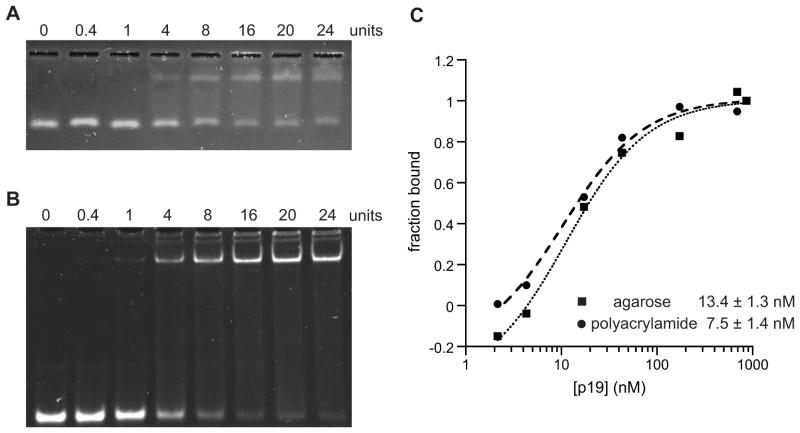
Apparent binding constants were measured using the same reaction conditions followed by electrophoresis using both the agarose and the polyacrylamide gel systems in a head-to-head comparison. A range of p19 protein concentrations was incubated with 20 ng of dsRNA ligand, spanning well below and well above the published KD,app [31]. Half of each reaction was run on an agarose gel, while the other half was run on a polyacrylamide gel (Fig. 3, panels A and B). Three independent sets of agarose and polyacrylamide gel pairs were executed in this manner. After staining with SYBR Gold, the free and bound RNA bands were quantitated as described [33]. Because the dsRNA concentration in the binding reactions (~100 nM) was above the reported KD,app values for p19 in the mid-picomolar to low-nanomolar range [30,32], the data were fit with the quadratic form of the binding isotherm, which does not make any assumptions about the concentration of free protein relative to the concentration of total protein (Eq. 1).
Figure 3. High-voltage, high-percentage agarose gels yield KD,app values comparable to conventional PAGE.
Representative EMSA data using either (A) an agarose or (B) a polyacrylamide gel to separate bound and free RNA ligand. (C) The gels shown in (A) and (B) were quantitated and subjected to multivariate fitting using the Langmuir binding isotherm to determine the KD,app. (D) The average KD,app values (± SEM) for three independent replicate experiments analyzed by the two gel systems.
To directly compare the ability of the native agarose gel shift to provide accurate measures of binding affinity, we carried out direct comparisons with a conventional native polyacrylamide gel. A set of binding reactions were incubated, and then divided between the high-voltage agarose gel and a conventional polyacrylamide gel for analysis (Fig. 3). Both systems yielded similar data quality and the fitted curves had similar shapes (Fig. 3, panels A, B, and C). The average Kd,app of three independent replicates was determined. Using the standard polyacrylamide system, the Kd,app was found to be 7.5 ± 1.4 nM, consistent with previous reports for the MBP-p19-CBD fusion protein [32]. Similarly, the new agarose system produced an average Kd,app of 13.4 ± 1.3 nM. While the standard errors of the means do not overlap, the less than twofold difference in average Kd,app values is within the generally accepted range of measurements in the field when using different methods.
4. Discussion
We have developed a native agarose gel system that enables rapid quantitation of RNA binding activity by proteins. We found that a 45 mL 2.5% agarose gel (approximately 7 mm thick) was ideal for optimal staining and destaining using SYBR Gold. Agarose gels are normally run in 1× TAE or 1× TBE buffer at ~10 V/cm to minimize artifacts caused by buildup of current and asymmetric heating effects [26,28]. Using a lower-conductivity buffer (0.5× TB), gels could be electrophoresed at 20 – 25 V/cm, which effectively and efficiently separated the unbound 21-nt dsRNA ligand (~12 kDa) from the RNA bound by the 40 kDa dimeric p19 protein in less than 10 minutes, with minimal smearing and good band resolution. When quantitated, this system produced measurements that were comparable to those obtained by conventional native polyacrylamide gel electrophoresis with minimal exposure to potentially toxic reagents. This represents a significant advance in electrophoresis methodology that is highly adaptable to a variety of protein-nucleic acid complexes, and permits the use of very short oligonucleotides.
One of the reasons that polyacrylamide gels have been used historically for EMSAs is the higher resolving power of polyacrylamide matrices, which allow for single-nucleotide resolution of oligonucleotides [10]. In contrast, agarose gels have a limited ability to resolve small oligonucleotides, especially those that differ in size by only a few nucleotides [10]. Several different approaches have been taken in an effort to improve agarose gels for use in EMSAs with shorter DNAs. Increasing the percentage of agarose to 1 – 1.5% in one study improved band resolution, but run times were still fairly long which allowed for diffusion of the DNA [24]. The innate resolving power of an agarose matrix has also been improved by manipulating the gel composition through the addition of other chemicals, such as metals and amines [26] or polysaccharides [25,36]. Still other approaches employed chemically-modified agarose, in the form of hydroxyethylated agarose molecules, in an effort to reduce pore size and thereby improve resolution [22,27].
However, the most promising and widely-applicable alteration to the agarose system involves simply altering the running buffer [23]. In particular, it was recently demonstrated that reducing the conductivity of the running buffer could improve the resolving potential of an agarose gel by allowing for the use of higher voltages and significantly shorter gel run times [28]. The work described here has continued to develop the applications of these high-voltage agarose gel systems. Gel shift assays like EMSAs substantially benefit from the potential for shorter gel run times, which minimizes the time during which irreversible dissociation of the protein/ligand complex can occur. This allows for a more accurate quantitation of the initial equilibrium binding states, thereby yielding robust measurements of the apparent equilibrium dissociation constant (KD,app).
The adaptation of the agarose gel system for protein/RNA EMSAs benefitted from optimizing several parameters, including agarose content, gel thickness, and voltage. We reduced the fluorescence background produced after staining with SYBR Gold by minimizing agarose concentration and gel thickness, as well as optimizing the destaining protocol by incubating in TB instead of water. These gel parameters can be adjusted for the protein:RNA system of interest, as well as each individual lab’s gel and detection systems of choice. Possible detection methods that are more sensitive than SYBR Gold staining, such as fluorescent labels, radiolabels, and biotinylation, may be considered [3,10,17]. An additional advantage to optimizing the agarose concentration of the gels along with screening multiple voltages was to improve band resolution and band signal intensity, while simultaneously reducing the amount of smearing between the shifted and unshifted ligand bands. Therefore, particular attention should be paid to these parameters when establishing this method for an individual protein:RNA system, as the unique binding affinities and binding rates of each system will likely require individual optimization.
The commercially-produced recombinant p19 protein employed in this study was previously reported to have low nanomolar affinity for miRNAs [32] (NEB Technical Support, personal communication). A direct comparison between the new agarose system and a conventional polyacrylamide system was carried out by analyzing the same binding reactions in both systems. We determined that the two methods produced similar measurements of binding affinity. There was less than a twofold difference in the measured KD,app values using the two methods (13.4 ± 1.3 nM using agarose versus 7.5 ± 1.4 nM with polyacrylamide). As noted above, these values are consistent with published data for the MBP-p19-CBD fusion protein, and demonstrate that the new agarose-based system is able to quantitatively measure high-affinity and physiologically relevant interactions between proteins and RNAs.
4.1 Conclusions
In summary, we have developed an electrophoretic mobility shift assay method using an agarose gel and low-conductivity buffer system that requires only standard molecular biology reagents and equipment. The assay allows for rapid, quantitative analysis of protein/RNA interactions, requiring less than 90 minutes from gel pouring to data acquisition.
Supplementary Material
(A) Successful annealing of p19 ssRNAs to create dsRNA ligand. 500 ng of each indicated RNA was run on a 3.5% agarose gel at 350 V for 17 min in 0.5× TB to distinguish the ssRNA 21-mers from the annealed dsRNA complex (p19RNA-1/2). (B) The lower detection limit of p19 dsRNA ligand in high-percentage agarose gels is ~20 ng. The indicated amounts of annealed p19 dsRNA ligand were analyzed on a 3.5% agarose gel at 350 V for 7 min to determine the optimal amount of dsRNA for the binding experiments. (C) High-percentage agarose gels should be briefly destained in 0.5× TB. Incubating the gel in destaining solution for 2.5 hr is too long, and allows the RNA to diffuse within the gel. This significantly reduces the resolution of the RNA bands and precludes accurate quantitation. Destaining for 15 min was found to reduce background fluorescence and improve image quality with minimal band diffusion.
A set of RNA molecules were separated on a 6.7% native polyacrylamide gel run in 1× TBE at 200 V for 20 min at either room temperature (22 °C) or with ice packs (2 °C). Samples were as follows: 1, dsRNA ladder (New England BioLabs); 2, p19RNA-1; 3, p19RNA-2; 4, dsRNA stored at −20 °C; 5, freshly prepared dsRNA; 6, 48-nt RNA stem-loop; 7, 25mer ssRNA. Oligonucleotides p19 RNA-1 and p19 RNA-2 are described in Methods. The 48-nt stem-loop and 25-nt RNAs were not predicted to form intermolecular complexes. At 2 °C, the quality of the bands was significantly reduced in two ways. First, RNA ladders are observed in all p19 RNA lanes but not the control RNAs. Second, those RNAs show smearing and tailing, as seen in the dsRNA ladder and the p19 RNA lanes. This indicated that the two-nucleotide 3′ single-stranded overhang forms multimeric complexes at 2 °C, which was two degrees below the TM of the isolated dinucleotide.
Acknowledgments
This work was supported by a grant from the National Science Foundation (NSF 1407736) that supports the Houston Louis-Stokes Alliance for Minority Participation Scholars Program (J.A.R., participating scholar), and by the National Institutes of Health – National Institute for General Medical Sciences (GM099049 to L.K.L. and GM119096 to K.A.L.). All authors declare no conflict of interest.
Abbreviations used
- EMSA
electrophoretic mobility shift assay
- PAGE
polyacrylamide gel electrophoresis
- KD,app
apparent dissociation constant
- dsRNA
double-stranded RNA
- ssRNA
single-stranded RNA
- nt
nucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: A tutorial review. Prot Sci. 2009;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ragazzon PA, Garbett NC, Chaires JB. Competition dialysis: A method for the study of structural selective nucleic acid binding. Methods. 2007;42:173–182. doi: 10.1016/j.ymeth.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Pagano JM, Clingman CC, Ryder SP. Quantitative approaches to monitor protein–nucleic acid interactions using fluorescent probes. Rna. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freyer MW, Lewis EA. Isothermal Titration Calorimetry: Experimental Design, Data Analysis, and Probing Macromolecule/Ligand Binding and Kinetic Interactions. Meth Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 5.Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried MG, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Nat Acad Sci USA. 1993;90:5428. doi: 10.1073/pnas.90.12.5428. http://www.ncbi.nlm.nih.gov/pubmed/?term=8516284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senear D, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J Biol Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 9.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4. Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 10.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vossen KM, Fried MG. Sequestration Stabilizes lac Repressor–DNA Complexes during Gel Electrophoresis. Analytical Biochemistry. 1997;245:85–92. doi: 10.1006/abio.1996.9944. [DOI] [PubMed] [Google Scholar]
- 12.Frank EG, Hauser J, Levine AS, Woodgate R. Targeting of the UmuD, UmuD″, and MucA″ mutagenesis proteins to DNA by RecA protein. Proc Nat Acad Sci USA. 1993;90:8169. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemon BD, Fondell JD, Freedman LP. Retinoid X Receptor:Vitamin D3 Receptor Heterodimers Promote Stable Preinitiation Complex Formation and Direct 1,25-Dihydroxyvitamin D3-Dependent Cell-Free Transcription. Mol Cell Biol. 1997;17:1923–1937. doi: 10.1128/mcb.17.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fojta M, Pivonkova H, Brazdova M, Kovarova L, Palecek E, Pospisilova S, et al. Recognition of DNA modified by antitumor cisplatin by “latent” and ‘active’ protein p53. Biochem Pharmacol. 2003;65:1305–1316. doi: 10.1016/s0006-2952(03)00078-9. [DOI] [PubMed] [Google Scholar]
- 15.Frontini M, Imbriano C, diSilvio A, Bell B, Bogni A, Romier C, et al. NF-Y Recruitment of TFIID, Multiple Interactions with Histone Fold TAFIIs. J Biol Chem. 2002;277:5841–5848. doi: 10.1074/jbc.M103651200. [DOI] [PubMed] [Google Scholar]
- 16.Parsons RL, Harshey R. Autoregulation of phage mu transposase at the level of translation. Nucleic Acids Res. 1988;16:11285. doi: 10.1093/nar/16.23.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodgers JT, Patel P, Hennes JL, Bolognia SL, Mascotti DP. Use of Biotin-Labeled Nucleic Acids for Protein Purification and Agarose-Based Chemiluminescent Electromobility Shift Assays. Analytical Biochemistry. 2000;277:254–259. doi: 10.1006/abio.1999.4394. [DOI] [PubMed] [Google Scholar]
- 18.Chen TY, Hsu CT, Chang KH, Ting CY, Whang-Peng J, Hui CF, et al. Development of DNA delivery system using Pseudomonas exotoxin A and a DNA binding region of human DNA topoisomerase I. Appl Microbiol Biotechnol. 2000;53:558–567. doi: 10.1007/s002530051657. [DOI] [PubMed] [Google Scholar]
- 19.Yoo S. Characterization of Drosophila Rad51/SpnA protein in DNA binding and embryonic development. Biochem Biophys Res Comm. 2006;348:1310–1318. doi: 10.1016/j.bbrc.2006.07.211. [DOI] [PubMed] [Google Scholar]
- 20.Brázda V, Čechová J, Coufal J, Rumpel S, Jagelská EB. Superhelical DNA as a preferential binding target of 14-3-3γ protein. J Biomol Struct Dyn. 2012;30:371–378. doi: 10.1080/07391102.2012.682205. [DOI] [PubMed] [Google Scholar]
- 21.Yeh L-CC, Lee JC. Yeast ribosomal proteins L4, L17, L20, and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA. Biochim Biophys Acta. 1998;1443:139–148. doi: 10.1016/s0167-4781(98)00202-4. [DOI] [PubMed] [Google Scholar]
- 22.Vossen KM, Fried MG. Stability of lac repressor-operator complexes in a new agarose-based gel matrix. Nucleic Acids Res. 1995;23:2346. doi: 10.1093/nar/23.12.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellorini M, Dantonel JC, Yoon JB, Roeder RG, Tora L, Mantovani R. The major histocompatibility complex class II Ea promoter requires TFIID binding to an initiator sequence. Mol Cell Biol. 1996;16:503–512. doi: 10.1128/MCB.16.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz-Norton JR, Ziegler YS, Likhite VS, Nardulli AM. Methods in Molecular Biology (Clifton, NJ) Humana Press; Totowa, NJ: 2009. Isolation of Proteins Associated with the DNA-Bound Estrogen Receptor α; pp. 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrasekhar S, Souba WW, Abcouwer SF. Use of Modified Agarose Gel Electrophoresis to Resolve Protein-DNA Complexes for Electrophoretic Mobility Shift Assay. BioTechniques. 1998;24:216–218. doi: 10.2144/98242bm09. [DOI] [PubMed] [Google Scholar]
- 26.Brody JR, Calhoun ES, Gallmeier E, Creavalle TD, Kern SE. BioTechniques - Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. BioTechniques. 2004;37:598–602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- 27.Svingen R, Takahashi M, Åkerman B. Gel-Shift Assays:3 Migrative Dissociation of a RecA–Oligonucleotide Complex during Electrophoresis in Hydroxyethylated Agarose Gels. J Phys Chem B. 2001;105:12879–12893. [Google Scholar]
- 28.Sanderson BA, Araki N, Lilley JL, Guerrero G, Lewis LK. Modification of gel architecture and TBE/TAE buffer composition to minimize heating during agarose gel electrophoresis. Analytical Biochemistry. 2014;454:44–52. doi: 10.1016/j.ab.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez BV, Pescador J, Pollok N, Beall GW, Maeder C, Lewis LK. Impact of size, secondary structure, and counterions on the binding of small ribonucleic acids to layered double hydroxide nanoparticles. Biointerphases. 2015;10:041007. doi: 10.1116/1.4936393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakatos L, Szittya G, Silhavy D, Burgyán J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. Embo J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargason JM, Szittya G, Burgyán J, Hall TMT. Size Selective Recognition of siRNA by an RNA Silencing Suppressor. Cell. 2003;115:799–811. doi: 10.1016/S0092-8674(03)00984-X. [DOI] [PubMed] [Google Scholar]
- 32.Jin J, Cid M, Poole CB, McReynolds LA. BioTechniques - Protein-mediated miRNA detection and siRNA enrichment using p19. BioTechniques. 2010;48 doi: 10.2144/000113364. [DOI] [PubMed] [Google Scholar]
- 33.Altschuler SE, Lewis KA, Wuttke DS. Practical strategies for the evaluation of high-affinity protein/nucleic acid interactions. J Nucleic Acids Investig. 2013;4:19–28. doi: 10.4081/jnai.2013.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laniel MA, Béliveau A, Guérin SL. Electrophoretic mobility shift assays for the analysis of DNA-protein interactions. Methods Mol Biol. 2001;148:13–30. doi: 10.1385/1-59259-208-2:013. [DOI] [PubMed] [Google Scholar]
- 35.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–6. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanek PG, Fabian SJ, Fisher CL, Chirikjian JG, Collier GB. Alternative to polyacrylamide gels improves the electrophoretic mobility shift assay. BioTechniques. 1995;18:704–706. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Successful annealing of p19 ssRNAs to create dsRNA ligand. 500 ng of each indicated RNA was run on a 3.5% agarose gel at 350 V for 17 min in 0.5× TB to distinguish the ssRNA 21-mers from the annealed dsRNA complex (p19RNA-1/2). (B) The lower detection limit of p19 dsRNA ligand in high-percentage agarose gels is ~20 ng. The indicated amounts of annealed p19 dsRNA ligand were analyzed on a 3.5% agarose gel at 350 V for 7 min to determine the optimal amount of dsRNA for the binding experiments. (C) High-percentage agarose gels should be briefly destained in 0.5× TB. Incubating the gel in destaining solution for 2.5 hr is too long, and allows the RNA to diffuse within the gel. This significantly reduces the resolution of the RNA bands and precludes accurate quantitation. Destaining for 15 min was found to reduce background fluorescence and improve image quality with minimal band diffusion.
A set of RNA molecules were separated on a 6.7% native polyacrylamide gel run in 1× TBE at 200 V for 20 min at either room temperature (22 °C) or with ice packs (2 °C). Samples were as follows: 1, dsRNA ladder (New England BioLabs); 2, p19RNA-1; 3, p19RNA-2; 4, dsRNA stored at −20 °C; 5, freshly prepared dsRNA; 6, 48-nt RNA stem-loop; 7, 25mer ssRNA. Oligonucleotides p19 RNA-1 and p19 RNA-2 are described in Methods. The 48-nt stem-loop and 25-nt RNAs were not predicted to form intermolecular complexes. At 2 °C, the quality of the bands was significantly reduced in two ways. First, RNA ladders are observed in all p19 RNA lanes but not the control RNAs. Second, those RNAs show smearing and tailing, as seen in the dsRNA ladder and the p19 RNA lanes. This indicated that the two-nucleotide 3′ single-stranded overhang forms multimeric complexes at 2 °C, which was two degrees below the TM of the isolated dinucleotide.