Abstract
Chronic cardiac hypertrophy is maladaptive and contributes to the pathogenesis of heart failure. The objective of this study was to identify small molecule inhibitors of pathological cardiomyocyte hypertrophy. High content screening was performed with primary neonatal rat ventricular myocytes (NRVMs) cultured on 96-well plates and treated with a library of 3241 distinct small molecules. Non-toxic hit compounds that blocked hypertrophy in response to phenylephrine (PE) and phorbol myristate acetate (PMA) were identified based on their ability to reduce cell size and inhibit expression of atrial natriuretic factor (ANF), which is a biomarker of pathological cardiac hypertrophy. Many of the hit compounds are existing drugs that have not previously been evaluated for benefit in the setting of cardiovascular disease. One such compound, the anti-malarial drug artesunate, blocked left ventricular hypertrophy (LVH) and improved cardiac function in adult mice subjected to transverse aortic constriction (TAC). These findings demonstrate that phenotypic screening with primary cardiomyocytes can be used to discover anti-hypertrophic lead compounds for heart failure drug discovery. Using annotated libraries of compounds with known selectivity profiles, this screening methodology also facilitates chemical biological dissection of signaling networks that control pathological growth of the heart.
Keywords: Cardiomyocyte, Hypertrophy, Small molecule, High throughput, High content, Screening
1. Introduction
The power of small molecule high throughput screening (HTS) in finding leads for drug discovery is well established [1]. HTS has historically been performed in industry and has largely focused on identifying modulators of distinct biochemical targets using simple in vitro assays or engineered reporter cell lines [2]. The standard approach has been to screen for modulators of a single target. Recently, phenotype-based screening has emerged as a more information-rich alternative to traditional target-based screening [3]. Phenotypic screening attempts to incorporate as much relevant biological information as possible and eliminate toxic hits, or hits with undesirable mechanisms-of-action, at an early stage of the discovery process. As such, phenotype-based screens have the potential to significantly lower the otherwise high attrition rates of lead compounds in the path of optimization and development into new drugs.
Heart failure affects 6 million people in the US alone, with 500,000 new diagnoses annually and a 5-year mortality rate of 42%, exceeding that of many cancers [4]. Cardiac hypertrophy is a hallmark of heart failure. Long-term suppression of cardiac hypertrophy is associated with reduced morbidity and mortality, and thus there is intense interest in developing novel therapeutics to target this growth response [5–7]. Current treatment of heart failure involves the use of drugs that inhibit signaling pathways triggered by cell surface receptors, such as the angiotensin receptor and the β-adrenergic receptor [8]. However, given the multitude of redundant signaling pathways capable of promoting pathological cardiac hypertrophy, it is hypothesized that increased efficacy will be obtained with therapies that target distal nodal points in the hypertrophic response. Phenotypic screening methods provide a unique opportunity to identify and target such nodal points [9].
Here, we describe results of an HTS campaign designed to discover small molecule inhibitors of pathological cardiomyocyte hypertrophy. This phenotypic screen employed primary neonatal rat cardiomyocytes and a high content screening platform, which enabled simultaneous image-based quantification of effects of compounds on cardiomyocyte cell area, biomarker expression, and viability. The results illustrate the power of phenotypic screening as a means to identify leads for heart failure drug discovery, and to yield compounds that can be employed as chemical biological probes to uncover nodal effectors of pathological cardiac hypertrophy.
2. Materials and methods
2.1. Chemical libraries
The NIH Clinical Collections I and II (446 and 281 compounds, respectively) were obtained from Evotec US (South San Francisco, CA). The Spectrum Collection (2320 compounds) was purchased from Microsource (Gaylordsville, CT). The Kinase Inhibitor Library (194 compounds) was obtained from SelleckChem (Houston, TX). All compounds were supplied as 10 mM stock solutions in DMSO.
2.2. Cardiomyocyte cell culture and compound treatment
Neonatal rat ventricular myocytes (NRVMs) were isolated from the hearts of 1–3 day-old Sprague Dawley rats (Charles River), as previously described [10]. Cell counting and viability was assayed using a Vi-Cell Cell Viability Analyzer (Beckman Coulter). Cells were dispensed using a 5 μL cassette in a MultiFlo dispenser (BioTek) onto 96-well clear-bottom plates (Greiner) coated with 0.2% gelatin (Sigma; G9391) in DMEM with 10% calf serum, 2 mM L-glutamine, and penicillin-streptomycin; each well received 10,000 cells in 100 mu;L of medium. The following morning, cells were washed with serum-free medium and maintained in DMEM supplemented with L-glutamine, penicillin-streptomycin and Neutridoma-SP (0.1%; Roche Applied Science). Cells were treated with PE (10 mu;M) or PMA (50 nM) for 48 h prior to fixation. Immediately after addition of hypertrophic agonist, compounds and controls were added to plates. A single column of negative control (DMSO, 8 wells) and a single column of positive control (Trichostatin-A (TSA), 200 nM, 8 wells) were included on all plates. Using a 96-tip MDT head on a Janus automated liquid handler (Perkin Elmer), 0.5 mu;L of each compound was delivered to each well at a final concentration of 10 mu;M (1 mu;M for the Kinase Inhibitor Library), yielding a residual DMSO concentration of 0.5%.
2.3. Cardiomyocyte staining and imaging
All plate washing steps were performed using the ELx 405 plate washer (BioTek). All dispense steps were done using a 5 mu;L cassette on the MultiFlo (BioTek). Cells were washed with Hank's Balanced Salt Solution (HBSS, Gibco) and fixed for 20 min with paraformaldehyde (3.2%, Electron Microscopy Sciences) in HBSS. Fixed cells were washed three times with HBSS and permeabilized with 0.1% IGEPAL with 3% BSA in HBSS for 20 min. Primary antibodies were added as a cocktail (1:1000 anti-ANF, Phoenix Pharmaceuticals, H-005-24; 1:750 anti-α-actinin, Sigma A-7811) in permeabilization buffer and incubated for 1–2 h at room temperature. Cells were washed with HBSS and incubated with a cocktail of secondary antibodies (donkey anti-mouse FITC, Jackson ImmunoResearch; goat anti-rabbit Cy3, Jackson ImmunoResearch) in permeabilization buffer and incubated for 1 h in the dark. Cells were washed and incubated with Hoechst 33342, 10 mu;M, in HBSS, for 10 min. Finally, plates were washed with HBSS and stored on sealed plates in HBSS until imaged. All imaging was performed on an Operetta (Perkin Elmer). Three fluorescence channels (Hoechst, FITC, Cy3) were measured for each field using a 20× objective. Twenty fields were typically imaged in each well. Image collection was focused on the perimeter of each round well. Imaged cell counts ranged from 0 cells for toxic compounds to 1200 cells, typically, for nontoxic compounds. The NIH Clinical Collection #1 was run in duplicate as a step in the assay validation; all subsequent compound libraries were run as n = 1.
2.4. High content analysis
A multistep algorithm was developed to quantitatively analyze myocyte morphology and biomarker expression using Harmony software (Perkin Elmer). Images were first segmented based on the Hoechst fluorescence. Cytoplasm was mapped around each nucleus based on the FITC channel (anti-α-actinin). A perinuclear mask was created for each cell that went from the outer edge of each nucleus to a nuclear radius out from the edge of the nucleus, forming a roughly donut-shaped mask. All partial objects (nuclei and/or cytoplasm) were filtered out of the final analysis. Quantitative parameters that were used for hit selection were the total cell area in mu;m2 (mean average value per well, α-actinin), and the sum of Cy3 fluorescence under the perinuclear masks (mean sum value per well, ANF).
2.5. Determination of hit compounds
All well-based data from the high content analysis were exported as text files and imported into an HTS database for secondary analysis, quality assurance, and hit selection (BioAssay HTS, CambridgeSoft). Statistical analysis was performed on a plate-by-plate basis. Z-primes were determined based upon positive and negative controls in plate columns one and twelve, respectively. Negative controls were vehicle control (0.5% DMSO) and positive controls were TSA (200 nM with 0.5% DMSO).
2.6. Experimental animals
Animal experiments were done in accordance with the Institutional Animal Care and Use Committee at the University of Colorado Denver. Ten week-old male C57BL/6 mice (Jackson Laboratories) were used for transverse aortic constriction (TAC). TAC and sham surgeries were performed as previously described, using a 27-gauge needle to guide suture constriction [11]. Artesunate (Sigma) was delivered daily via intra-peritoneal injection at a concentration of 50 mg/kg in a 1:4 DMSO:10% (2-hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich, 389145) vehicle, beginning one day post-TAC surgery. Control animals received vehicle alone. Treatment groups were weight-matched prior to the start of the study.
2.7. Hemodynamic analysis
Echocardiographic analyses were performed the day the animals were euthanized using a Vevo770 System equipped with a 30 mHz frequency mechanical transducer (VisualSonics). Hearts were imaged in the two-dimensional parasternal short axis. M-mode images were recorded to measure LV wall dimensions and internal diameter at the level of the papillary muscles. For analyses, animals were anesthetized using 2% isoflurane and their body temperature was maintained at 37 °C. For data from all in vivo studies, GraphPad Prism software was used to generate graphs and analyze data. One-way ANOVA with Newman-Keuls post hoc test (P < 0.05) was used to determine statistical differences between groups.
2.8. Determination of ventricular myocyte cross-sectional area
Myocyte cross-sectional area was quantified using latitudinal midsections of the LV treated with neuraminidase type V (Sigma), and stained with fluorescein-labeled peanut agglutinin (10 mg/mL; Vector Laboratories). Images were captured with an AxioVert 200 inverted microscope using an AxioCam MRc digital camera, and analyzed with AxioVision Release 4.8.1 imaging software (Zeiss, Germany). Approximately 100–125 myocytes were analyzed and averaged for each animal. Analysis focused on the epicardium and endocardium, where the best cross sections of myocytes were present. Data was analyzed using GraphPad Prism software, with a one-way ANOVA with Newman-Keuls post hoc test (P < 0.05) to determine statistical differences between groups.
3. Results
3.1. Hypertrophy high throughput assay design and validation
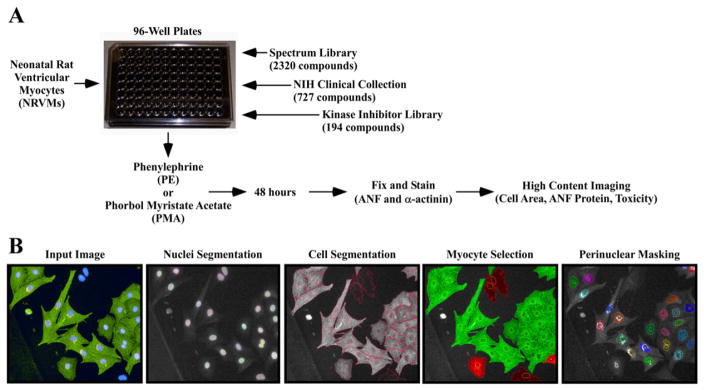
In order to develop an assay to screen chemical libraries for anti-hypertrophic compounds, a cell-based high content imaging approach was developed (Fig. 1A). Freshly isolated neonatal rat ventricular myocytes (NRVMs) were plated on gelatin-coated 96-well plates with optically clear bottoms. Subsequently, NRVMs were treated with phenylephrine (PE, 10 mu;M), or the diacylglycerol mimetic phorbol 12-myristate 13-acetate (PMA, 50 nM). PE elicits cardiomyocyte hypertrophy by stimulating the α1-adrenergic receptor (α1-AR), while PMA promotes cardiomyocyte growth by functioning intracellularly to trigger protein kinase C (PKC) signaling. By using two distinct agonists, we were able to identify compounds with generalizable anti-hypertrophic activity. Furthermore, the use of PMA enabled filtration of false positive compounds that non-selectively antagonized hypertrophy via promiscuous binding to the α1-AR.
Fig. 1.
A novel high throughput assay of cardiomyocyte hypertrophy. (A) Neonatal rat ventricular cardiomyocytes (NRVMs) were seeded on 96-well plates and treated with candidate compounds in the presence of PE or PMA for 48 h. Cells were fixed, subjected to indirect immunofluorescence with antibodies against ANF and α-actinin, and stained with Hoechst dye prior to high content imaging. (B) Images were analyzed using Harmony software. Cells were identified based on nuclei segmentation (Hoechst), and subsequently individual cells were segmented and cell area was calculated based on area of α-actinin fluorescence (FITC). Non-myocytes were removed from analysis based on the absence of α-actinin signal. Finally, ANF expression was measured using a perinuclear mask (Cy3).
Following 48 h of stimulation, cells were fixed and stained with antibodies against atrial natriuretic factor (ANF) and sarcomeric α-actinin, and nuclei were stained with Hoechst dye. High content imaging was employed to simultaneously quantify these signals (Fig. 1B). Using Harmony Image Analysis software, NRVMs were identified based on Hoechst signal, and cytoplasm was defined using the segmentation tool. Cardiomyocytes were selected for analysis on the basis of positive staining for α-actinin, which is not expressed in contaminating cells, such as cardiac fibroblasts. Following the creation of a perinuclear mask, the intensity of Cy3 (ANF) fluorescence was quantified within the mask. Cell area was calculated using the area of objects positive for FITC (α-actinin). Immunofluorescence artifacts were removed based on fluorescence intensity windows and object morphological properties. Finally, nuclei number was calculated to indicate cell number, thus enabling evaluation of cytotoxicity.
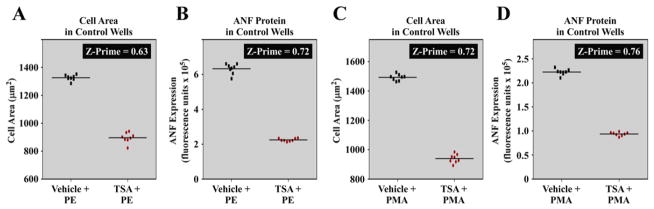
The Z-prime statistical method was employed to assess the quality of the hypertrophy assay prior to high content screening [12]. Z-prime is used to determine if an effect window is sufficient, and consistent enough, to enable efficient detection of a pharmacological effect in a given assay; Z-prime values between 0.5–1.0 are indicative of a high quality assay. As a positive control for validation, we employed the histone deacetylase (HDAC) inhibitor, trichostatin A (TSA), which has potent anti-hypertrophic activity [13]. When PE was used as the hypertrophic agonist, TSA robustly reduced NRVM cell area (hypertrophy) and ANF expression, and calculated Z-prime values for these effects were 0.63 and 0.72, respectively (Fig. 2A and B). Z-prime values are derived from the difference in the means and standard deviations of the positive and negative controls; a Z-prime of 0.63 indicates that the positive and negative controls were separated by >12 standard deviations. Statistically sound responses were also observed when PMA was employed as the hypertrophic agonist (Fig. 2C and D).
Fig. 2.
Statistical assessment of the cardiomyocyte hypertrophy high throughput assay. The HDAC inhibitor TSA was used as a positive control to block NRVM hypertrophy and ANF expression in the presence of PE or PMA. Data shown are from two experimental plates: one with each hypertrophic agonist. Individual data points representing mean values per well (total value divided by cell count) are shown. (A and B: Plate one) Z-prime values for cell size, and ANF expression in the presence of PE were 0.63 and 0.72, respectively. (C and D: Plate two) Z-prime values for cell size and ANF expression in the presence of PMA were 0.72 and 0.76, respectively. These values indicate that the differences between the negative controls (PE + DMSO or PMA + DMSO) and the positive controls (PE + TSA or PMA + TSA) are sufficient, and consistent enough, to enable efficient high throughput screening for inhibitors of hypertrophy.
3.2. Determination of hit compounds that block cardiomyocyte hypertrophy
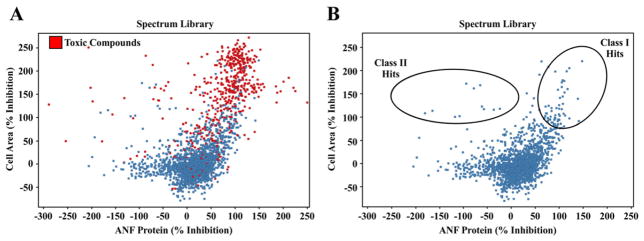
High throughput screening was performed following validation of the hypertrophy assay. Using a robotic liquid handling system, each NRVM-containing well was treated with a distinct compound from the Microsource Spectrum Library, the NIH Clinical Collection, or a Kinase Inhibitor Library, on duplicate plates; 3241 compounds were screened in total (Fig. 1A). Percent inhibition of cell size and ANF expression was calculated for each compound relative to the positive control, TSA, which was set to 100% inhibition for each plate. Compounds that decreased cell area >70% in PE- and PMA-treated NRVMs were classified as hits. These hits were further segregated based on ANF expression, with “Class I” hits defined as those that also reduced ANF expression >70%, and “Class II” hits defined as those that reduced cell area (hypertrophy) but increased ANF expression. Cytotoxic compounds were removed from subsequent analyses (Fig. 3A and B).
Fig. 3.
Graphical representation of hit compounds that block cardiomyocyte hypertrophy. (A) Compounds from the Spectrum Library are depicted based on percent inhibition of ANF expression (X-axis) and cell area (Y-axis) relative to the positive control HDAC inhibitor, TSA; % inhibition by TSA was set to 100% for each plate. Reduction in the number of identified cells (nuclei) compared to the positive control TSA was used as an indicator of toxicity. Toxic compounds are indicated in red, and were removed from subsequent analysis. (B) Class I hits are those compounds that significantly reduce cell area and ANF expression. Class II hits are compounds that significantly reduce cell area, but increase ANF expression.
3.3. Artesunate blocks cardiomyocyte hypertrophy in vitro and in vivo
Class I hits from the Spectrum and NIH Clinical Collections are listed in Table 1, while Class II hits from these libraries are listed in Table 2. Hits from the Kinase Inhibitor Library are listed in Table 3. Some of the anti-hypertrophic compounds target proteins that have previously been shown to regulate cardiomyocyte growth, such as mTOR (Table 3). For other compounds, there was no a priori reason to suspect that they would have the ability to block cardiomyocyte hypertrophy. For example, artesunate, a Class I hit from the screen, is a member of the artemisinin class of antimalarial drugs, which are considered to be the most essential antimalarial agents worldwide due to their potency and rapid onset of action [14].
Table 1.
Inhibitors of cardiomyocyte hypertrophy: class I hits.
| Compound | % Inhibition PE
|
% Inhibition PMA
|
Pharmacological class
|
Target
|
||
|---|---|---|---|---|---|---|
| Cell area | ANF | Cell area | ANF | |||
| Bromocriptine | >100 | >100 | >100 | >100 | Central nervous system (CNS) | Dopamine D2 agonist |
| Phenelzine | >100 | >100 | >100 | 94 | CNS | MAOi |
| SKF-83566 | >100 | >100 | 72 | 59 | CNS | Dopamine D1 antagonist |
| Securinine | >100 | 86 | >100 | >100 | CNS, NP | GABA (A) receptor |
| Clozapinea | >100 | 80 | 60 | 61 | CNS/antipsychotic | Multiple GPCRs |
| Perospirone | >100 | >100 | 97 | 73 | CNS/antipsychotic | Multiple GPCRs |
| Loxapine | 90 | 84 | 70 | 52 | CNS/antipsychotic | Multiple GPCRs |
| Imipraminea | >100 | 89 | >100 | 82 | CNS/antidepressant | Multiple GPCRs |
| Doxepina | >100 | 96 | 79 | 51 | CNS/antidepressant | Multiple GPCRs |
| Fluvoxaminea | >100 | 68 | >100 | 86 | CNS/antidepressant | Serotonin transporter, σ1R |
| Escitalopram | 94 | 71 | 71 | 52 | CNS/antidepressant | Serotonin transporter |
| Nortriptyline | >100 | 93 | 93 | 63 | CNS/antidepressant | Multiple GPCRs |
| Atomoxetine | >100 | 56 | >100 | 87 | CNS/ADHD | Norepinephrine transporter |
| Eticlopride | >100 | 66 | >100 | 68 | CNS | Dopamine-D2 receptor |
| Nefazodone | >100 | >100 | >100 | 86 | CNS/antidepressant | Multiple GPCRs |
| Bicalutamide | 80 | 73 | 63 | 62 | Nuclear hormone receptors (NHR) | Androgen receptor |
| RU-486 | >100 | 66 | >100 | >100 | NHR | Progesterone receptor |
| Melengestrol | 62 | 65 | 64 | 71 | NHR | Progesterone receptor? |
| Oxymetholone | 57 | 53 | 62 | 80 | NHR/Anabolic steroid | Androgen receptor? |
| Tretinoin | >100 | 82 | >100 | 83 | NHR? | Retinoic acid receptors? |
| Isotretinoin | 73 | 79 | >100 | 75 | NHR? | Retinoic acid receptors? |
| Tiratricol | >100 | >100 | >100 | >100 | NHR? | Thyroid hormone receptors? |
| Ethopropazine | >100 | 92 | >100 | 68 | Anticholinergic | Multiple GPCRs |
| Mebeverine | >100 | 97 | >100 | 67 | Antimuscarinic | Muscarinic receptors |
| Clemizole | >100 | >100 | >100 | 83 | Antihistamine | Histamine H1 receptor |
| Naftopidilb | >100 | >100 | 82 | 87 | Cardiovascular | α-1 adrenergic receptorb |
| Labetalol | 69 | 69 | 55 | 64 | Cardiovascular | Mixed α/β blocker |
| Cloperastine | >100 | >100 | >100 | 93 | Antitussive | Multiple GPCRs? |
| Candesartan | >100 | 100 | >100 | >100 | Cardiovascular | Angiotensin receptor |
| Penbutolol | 85 | 81 | 60 | 64 | Cardiovascular | β-adrenergic receptors |
| Nicardipine | >100 | 99 | 91 | 83 | Cardiovascular | Calcium channel blocker |
| Verapamil | 74 | 71 | 65 | 67 | Cardiovascular | Calcium channel blocker |
| Propafenonea | >100 | 81 | >100 | 89 | Cardiovascular/anti-arrhythmic | Sodium channel blocker |
| Benoxinate | >100 | 84 | 96 | 56 | CNS | Sodium channel blocker |
| Vorinostat | >100 | 95 | 99 | 95 | Cancer | HDAC inhibitor |
| Atorvastatin | >100 | 80 | 82 | 92 | Cardiovascular/dyslipidemia | HMG-CoA reductase |
| Cerivastatin | >100 | >100 | >100 | >100 | Cardiovascular/dyslipidemia | HMG-CoA reductase |
| Mevastatin | >100 | 60 | >100 | >100 | Cardiovascular/dyslipidemia | HMG-CoA reductase |
| Lovastatina | >100 | 60 | >100 | >100 | Cardiovascular/dyslipidemia | HMG-CoA reductase |
| Ezetimibe | >100 | 62 | 89 | 78 | Dyslipidemia | NPC1L1 |
| Diphenoxylate | >100 | >100 | >100 | >100 | Antidiarrheal | Opioid receptor(s) |
| GR 89696 | 98 | 67 | 69 | 58 | Analgesic? | β-opioid receptor agonist |
| Phenazopyridine | >100 | 70 | 88 | 79 | Analgesic | Unknown |
| Mycophenolate mofetil | 73 | 76 | 79 | 89 | Immune suppressant | IMDH inhibitor |
| Ketoconazole | >100 | 64 | 63 | 64 | Antifungal | 14-α demethylase, PXR |
| Itraconazole | >100 | 78 | >100 | >100 | Antifungal | 14-α demethylase |
| Artesunate | >100 | 86 | >100 | >100 | Antimalarial | Unknown |
| Rifabutin | >100 | 44 | 110 | 107 | Antibiotic | RNA polymerase |
| Irinotecan | 81 | 56 | 71 | 57 | Antineoplastic | Topoisomerase |
| Carmofur | 76 | 51 | 79 | 91 | Antineoplastic | Pyrimidine analog |
| 5-Azacytidine | 85 | 79 | 80 | 67 | Antineoplastic | DNA methyltransferase |
| Dihydrogedunin | 93 | 84 | 98 | 82 | Natural products (NP) | Unknown |
| α-Dihydrogedunol | >100 | 88 | 69 | 71 | NP | Unknown |
| Khayanthone | 93 | 75 | 78 | 68 | NP | Unknown |
| Gangaleoidin | >100 | 82 | >100 | >100 | NP | Unknown |
| 4′-Hydroxychalcone | >100 | 95 | 88 | 99 | NP | Unknown |
| Avocadyne | >100 | 97 | 100 | 74 | NP | Unknown |
| Abietic acid | 94 | 81 | 58 | 73 | NP | Unknown |
Some compounds with structures and/or mechanisms of action that are unlikely to be of therapeutic interest were intentionally omitted from this summary table.
Scored as hits in both NIHcc and Spectrum collections.
Other α-adrenergic antagonists were not effective against PMA-mediated hypertrophy. Naftopidil has also been reported to inhibit TGF-β signaling.
Table 2.
Inhibitors of cardiomyocyte hypertrophy: class II hits.
| Compound | % Inhibition PE
|
% Inhibition PMA
|
Pharmacological class | Target | ||
|---|---|---|---|---|---|---|
| Cell area | ANF | Cell area | ANF | |||
| Conessine | >100 | −23 | >100 | 51 | Natural product | Histamine H3 |
| Dipyridimole | >100 | −10 | 66 | 47 | Cardiovascular | Phosphodiesterases |
| Imatinib | >100 | −9 | 98 | 41 | Anti-neoplastic | BCR-Abl, other kinases |
| Biperiden | 94 | −6 | 80 | 31 | CNS/anti-Parkinsonian | Cholinergic receptors, others |
| Halometasone | 77 | −2 | 62 | 46 | Corticosteroid (topical) | Corticosteroid |
| Deoxysappanone B 7,3′-dimethyl ether acetate | >100 | −70 | 51 | 16 | Natural product | Unknown; ANF mislocalization |
| Colchicine | >100 | −64 | >100 | −35 | Natural product | Microtubule poison; ANF mislocalization |
| Oxibendazole | >100 | −82 | 76 | −21 | Anti-helminthic | Unknown; ANF mislocalization |
| Picropodophyllin | >100 | −124 | >100 | −16 | Anti-neoplastic | IGF-1R; possible ANF mislocalization |
| 4′-Demethylepi-podophyllotoxin | >100 | −112 | >100 | −4 | Anti-neoplastic | Topoisomerase; ANF mislocalization |
| Podofilox | >100 | −74 | >100 | −54 | Anti-neoplastic | Topoisomeras; ANF mislocalization |
| Vindesine | >100 | −7.5 | >100 | 16 | Anti-neoplastic | Microtubule poison; Some ANF mislocalization |
| Vincristine | >100 | −35 | >100 | 3.9 | Anti-neoplastic | Microtubule poison; Some ANF mislocalization |
Table 3.
Inhibitors of cardiomyocyte hypertrophy: kinase inhibitor hits.
| Compound | % Inhibition PE
|
% Inhibition PMA
|
Target | ||
|---|---|---|---|---|---|
| Cell area | ANF | Cell area | ANF | ||
| AZD8055 | >100 | >100 | >100 | >100 | mTOR |
| WYE-125132 | >100 | >100 | >100 | >100 | mTOR |
| GDC-0980 | >100 | 84 | >100 | 84 | mTOR/PI3K |
| PF-04691502 | >100 | 75 | >100 | 97 | mTOR/PI3K |
| BEZ235 | >100 | 88 | >100 | >100 | mTOR/PI3K |
| Pelitinib | >100 | >100 | >100 | >100 | EGFR |
| WZ3146 | >100 | >100 | >100 | >100 | EGFR |
| WZ4002 | >100 | >100 | >100 | >100 | EGFR |
| AEE788 | 97 | 82 | 82 | 79 | EGFR/Her1-2/VEGFR |
| OSI-930 | >100 | >100 | >100 | 85 | c-Kit/VEGFR |
| Hesperadin | >100 | >100 | 82 | >100 | Aurora |
| Aurora A Inhibitor I | >100 | >100 | >100 | >100 | Aurora |
| ENMD-2076 | 81 | 79 | 92 | 76 | Aurora/Flt-3/Src |
| TG101209 | >100 | 100 | >100 | 98 | FLT-3/JAK |
| NVP-BSK805 | >100 | 79 | >100 | 93 | JAK |
| PHA-793887 | >100 | >100 | >100 | >100 | CDK |
| AZD7762 | >100 | >100 | 71 | 96 | CHK |
| Crizotinib | >100 | >100 | >100 | >100 | c-Met/ALK |
| BI 2536 | >100 | >100 | >100 | >100 | PLK |
| Bosutinib | >100 | >100 | >100 | >100 | Src |
| Vinorelbinea | >100 | −11 | >100 | 24 | P38 MAPK |
| ON-01910a | >100 | −1 | 88 | −28 | PLK |
| KX2-391a | >100 | −60 | 74 | −39 | Src |
Kinase inhibitors were screened at 1 mu;M.
Class II hits (no inhibition of ANF expression).
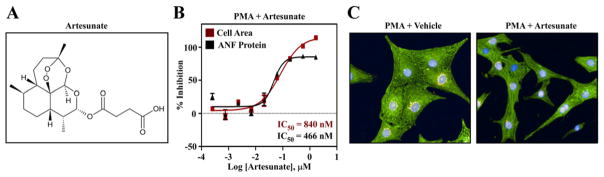
Given the extensive clinical experience with artesunate, this compound was chosen for follow-up analyses to address its potential as an agent to target pathological cardiac hypertrophy, and to assess whether the cell-based hypertrophy assay yields compounds that have the ability to block cardiac remodeling in vivo. Artesunate is a hemisuccinate derivative of artemisinin, a compound obtained from the sweet wormwood plant, which has been used extensively in traditional herbal medicine (Fig. 4A). Artesunate dose-dependently suppressed NRVM hypertrophy, with sub-micromolar half maximal inhibitory concentrations (IC50s) for blockade of myocyte growth and ANF expression (Fig. 4B). The compound was well tolerated by NRVMs, as illustrated by the representative images of cells stained with antibodies for α-actinin and ANF (Fig. 4C).
Fig. 4.
The anti-malarial compound artesunate blocks hypertrophy of cultured NRVMs. (A) Chemical structure of artesunate. (B) Quantitative high content imaging demonstrates dose-dependent inhibition of PMA-mediated NRVM hypertrophy and ANF expression by artesunate, with sub-micromolar IC50 values for each endpoint. (C) Representative images of NRVMs treated with PMA in the absence or presence of artesunate.
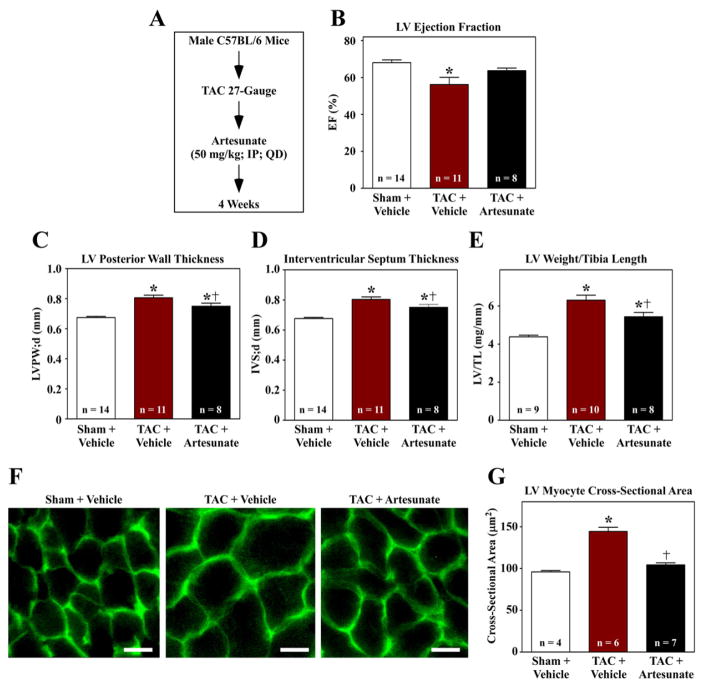
To address the potential of artesunate to block cardiac remodeling in vivo, mice were subjected to transverse aortic constriction (TAC) and administered artesunate or vehicle control every day for four weeks (Fig. 5A). Echocardiographic assessment revealed mild LV systolic dysfunction in animals subjected to TAC, and this functional impairment was prevented by artesunate (Fig. 5B); the impact of artesunate on diastolic function was not determined. Ultrasound imaging also demonstrated that artesunate normalized LV free wall and septal wall thickness in mice with TAC, suggesting that the compound reduced cardiac hypertrophy (Fig. 5C and D). Consistent with this, artesunate reduced LV weight-to-tibia length ratio (Fig. 5E), and also normalized myocyte cross-sectional area in LVs exposed to pressure overload (Fig. 5F and G). These findings demonstrate that artesunate has the ability to block cardiac hypertrophy in vivo, and demonstrate the utility of the NRVM-based screening assay for identifying novel anti-hypertrophic compounds.
Fig. 5.
Artesunate blocks cardiac hypertrophy in vivo. (A) Ten week-old mice were subjected to transverse aortic constriction (TAC) and treated with vehicle or artesunate for four weeks. (B–D) Echocardiography revealed that artesunate blunted TAC-induced systolic dysfunction, and reduced TAC-mediated thickening of the LV posterior wall and intraventricular septum. (E) LV weight-to-tibia length ratios were determined at necropsy. (F) Images of LV sections stained with fluorescein-conjugated peanut agglutinin were used to assess myocyte cross-sectional area. Scale bar = 10 mu;m. (G) Quantification of myocyte cross-sectional area (mu;m2); >100 myocytes were quantified per the indicated number of LVs. *P < 0.05 vs. sham; †P < 0.05 vs. TAC + vehicle.
4. Discussion
We discovered multiple small molecule inhibitors of cardiomyocyte hypertrophy by employing primary cardiomyocytes and high throughput, high content imaging. The findings establish the feasibility of using phenotypic screens to initiate heart failure drug discovery programs. Furthermore, since the protein targets of many of the small molecule hits are known, this work has uncovered the potential involvement of novel effectors that control pathological cardiac growth.
Based on current guidelines, most patients with heart failure are treated with three classes of drugs: β-blockers, drugs targeting the angiotensin II type I receptor (ACE inhibitors or ARBs), and a mineralocorticoid antagonist. Collectively referred to as neurohormonal blockade, this treatment regimen clearly improves outcomes in heart failure patients [8]. Furthermore, LCZ696, which is a dual angiotensin receptor blocker/neutral endopeptidase inhibitor, was recently found to be efficacious in a clinical trial with systolic heart failure patients [15–17]. Indeed, the primary outcome of cardiovascular death or a first hospitalization for heart failure occurred in 21.8% patients treated with LCZ696, compared with 26.5% of patients in the control treatment arm. While these findings are encouraging, they also serve to illustrate that disease progression persists in many patients treated with the full armamentarium of heart failure drugs, including LCZ696. Thus, heart failure remains a major unmet medical need, and novel therapeutic approaches are needed for patients with this prevalent and deadly syndrome.
Hypertrophy per se is not considered an approvable endpoint by the FDA. However, measures of reverse remodeling (e.g. LVEF, which is based on ventricular volumes) have been used in Phase 2 trials in patients with heart failure with reduced ejection fraction (HFrEF) as primary and secondary endpoints to assist in dose selection, and as surrogates of clinical efficacy. This is because the mechanism of action of multiple drugs or devices that improve HFrEF is reverse remodeling, or a reduction in hypertrophy [18]. Furthermore, although a reduction in hypertrophy has not been used in Phase 2 or 3 trials of heart failure with preserved ejection (HFpEF) patients, LV wall thickness or mass could also be employed in this setting to assess the effects of an anti-hypertrophic therapeutic modality.
Two additional imaging-based screens of cardiomyocytes have been reported. NRVMs were employed to screen 230 synthetic microRNAs (miRs) for effects on cell size [19]. Additionally, 15 agonists were screened for their ability to alter NRVM shape and gene expression patterns [20]. The current work represents the first reported screen for small molecule inhibitors of NRVM hypertrophy. Adaptation of this hypertrophy assay to 384-well plates will facilitate expanded screening efforts. Furthermore, this approach could be employed with iPS-derived cardiac myocytes [21], although optimized culture methods will likely be needed to maximize the responses of these cells to hypertrophic agonists [22].
Our screen yielded two types of hit compounds, which we refer to as Class I and Class II hits. Class I hits were shown to reduce cell size (hypertrophy) as well as agonist-induced expression of ANF, without causing cytotoxicity. Some of the targets of Class I hits were previously shown to regulate hypertrophy. For example, mTOR (5 hits), epidermal growth factor receptor (4 hits), HMG-CoA reductase (4 hits), and HDACs (1 hit) have all been demonstrated to promote hypertrophy [13,23–27]. Conversely, other targets of Class I hit compounds, such as Aurora Kinase, have not previously been linked to the regulation of cardiac hypertrophy. Additional pharmacologic and genetic gain- and loss-of-function studies will be required to validate roles for these other putative targets in the control of cardiac hypertrophy.
Many of the Class I hits target G protein-coupled receptors (GPCRs), and have previously been shown to have activity within the central nervous system (Table 1). In this regard, it is important to note that we employed PE to stimulate cardiomyocyte hypertrophy through the α1-AR, which is a GPCR. To filter out hits that blocked hypertrophy through promiscuous binding to the α1-AR, we also screened compounds for their ability to block PMA-induced hypertrophy, which is governed by direct activation of intracellular PKC. Our discovery that multiple pharmacological regulators of diverse cell surface receptors modulate cardiomyocyte growth suggests the possibility that stimulation of NRVMs triggers complex GPCR-governed autocrine pathways that are required to sustain the hypertrophic response.
The Class I hit, artesunate, which is a derivative of artemisinin, was chosen for follow-up evaluation because of its rich clinical history. Indeed, half of the 2015 Nobel Prize in Physiology and Medicine was awarded to the investigator who discovered artemisinin, a drug that is estimated to reduce malaria-associated mortality by 20% overall, and by >30% in children [28]. Artemisinin was previously shown to reduce LV hypertrophy in rat models of pressure overload and myocardial infarction, in association with blunted NF-κB signaling in the heart [29,30]. Unlike artemisinin, which is a natural product, artesunate is a semisynthetic derivative that has improved solubility, bioavailability and anti-malarial activity [31]. Our data demonstrate that, in addition to blocking NRVM hypertrophy, artesunate effectively reduced pressure overload-induced LVH in mice and improved LV systolic function (Fig. 5). Based on these findings, expanded evaluation of artesunate in pre-clinical models of heart failure is warranted, and should include assessment of the effects of the anti-malarial compound on both systolic and diastolic cardiac function. Given the favorable safety profile of this compound in humans, findings from these studies could lead to rapid translation of artesunate to the clinic to assess efficacy in humans with heart failure. Identification of the pro-hypertrophic molecular target(s) of artesunate in cardiomyocytes awaits further investigation. However, components of the NF-κB, NRF2 and MAP kinase signaling cascades are viable candidates, since all have been shown to be regulated by artesunate [32,33], and have also been implicated in the control of cardiac hypertrophy [34,35].
ANF and the related protein, brain natriuretic peptide (BNP), are markers of pathological cardiac remodeling. However, signaling initiated by these natriuretic peptides is not pathological, and instead represents a compensatory response within stressed cardiomyocytes. Indeed, numerous reports have shown that deletion of the gene encoding ANF (nppa) or the ANF receptor (npra) leads to exaggerated cardiac hypertrophy rather than reduced growth of the heart [36]. Furthermore, at least part of the protective mechanism-of-action of LCZ696 is via inhibition of the metallopeptidase, neutral endopeptidase, which normally degrades ANF and BNP [37]. Given the beneficial effects of increasing ANF expression in the heart, we also filtered compounds for their ability to concomitantly reduce cell size and increase ANF levels in NRVMs. These compounds, termed Class II hits, were further categorized based on whether they altered the sub-cellular localization of ANF protein within NRVMs. We hypothesize that some Class II hits, such as colchicine and vindesine, which are microtubule poisons, increase intracellular levels of ANF by disrupting trafficking and secretion of the natriuretic peptide. In contrast, ANF localization was normal in NRVMs treated with other Class II hits, such as biperiden and conessine, suggesting direct effects of these small molecules on ANF synthesis and/or turnover.
5. Conclusions
Drug discovery typically involves the identification of potential drug targets and validation of a role for these targets in disease. Target discovery and validation often takes many years, thereby delaying the start of small molecule screening efforts, chemical optimization of hit compounds, and in vivo testing to confirm that manipulation of the target will provide therapeutic benefit without unacceptable side effects. The work presented here provides an example of an alternative approach, in which the starting point is a “phenotype-based” high-throughput screen. Hits from this unbiased screen can be used as tools to identify novel cellular regulators of cardiac hypertrophy that have been recalcitrant to classical discovery efforts. In addition, optimized analogues of hit compounds can be employed to validate targets using animal models of pathological cardiac hypertrophy. Finally, existing drugs that emerged as anti-hypertrophic hits (e.g. artesunate) could be tested for potential as “repurposed” drugs for cardiac hypertrophy and heart failure, thereby streamlining efforts to translate preclinical discoveries to clinical testing in humans.
Acknowledgments
We thank B. Ferguson for NRVM preparation, R. Fickes and J. Spiltoir for compound administration, and M. Bristow for valuable discussions. This work was supported by grants from the Butcher Foundation and a Bioscience Discovery Evaluation Grant from the University of Colorado Denver Technology Transfer Office (CU3167H). T.A.M. was supported by the NIH (HL116848, HL127240 and AG043822) and the American Heart Association (Grant-in-Aid, 14510001). M.S.S. was funded by T32 training grants and an F32 fellowship from the NIH (5T32HL007822, 5T32HL007171 and F32HL126354).
References
- 1.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira DA, Williams JA. Origin and evolution of high throughput screening. Br J Pharmacol. 2007;152:53–61. doi: 10.1038/sj.bjp.0707373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devereux RB, Wachtell K, Gerdts E, Boman K, Nieminen MS, Papademetriou V, et al. Prognostic significance of left ventricular mass change during treatment of hypertension. JAMA. 2004;292:2350–2356. doi: 10.1001/jama.292.19.2350. [DOI] [PubMed] [Google Scholar]
- 6.Gardin JM, Lauer MS. Left ventricular hypertrophy: the next treatable, silent killer? JAMA. 2004;292:2396–2398. doi: 10.1001/jama.292.19.2396. [DOI] [PubMed] [Google Scholar]
- 7.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 8.Braunwald E. Research advances in heart failure: a compendium. Circ Res. 2013;113:633–645. doi: 10.1161/CIRCRESAHA.113.302254. [DOI] [PubMed] [Google Scholar]
- 9.McKinsey TA, Kass DA. Small-molecule therapies for cardiac hypertrophy: moving beneath the cell surface. Nat Rev Drug Discov. 2007;6:617–635. doi: 10.1038/nrd2193. [DOI] [PubMed] [Google Scholar]
- 10.Long CS, Henrich CJ, Simpson PC. A growth factor for cardiac myocytes is produced by cardiac nonmyocytes. Cell Regul. 1991;2:1081–1095. doi: 10.1091/mbc.2.12.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiltoir JI, Stratton MS, Cavasin MA, Demos-Davies K, Reid BG, Qi J, et al. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J Mol Cell Cardiol. 2013;63:175–179. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 13.Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 14.Hess KM, Goad JA, Arguin PM. Intravenous artesunate for the treatment of severe malaria. Ann Pharmacother. 2010;44:1250–1258. doi: 10.1345/aph.1M732. [DOI] [PubMed] [Google Scholar]
- 15.Desai AS, McMurray JJ, Packer M, Swedberg K, Rouleau JL, Chen F, et al. Effect of the angiotensin-receptor-neprilysin inhibitor LCZ696 compared with enalapril on mode of death in heart failure patients. Eur Heart J. 2015;36:1990–1997. doi: 10.1093/eurheartj/ehv186. [DOI] [PubMed] [Google Scholar]
- 16.McMurray JJ, Packer M, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, et al. Angiotensin-neprilysin inhibition versus enalapril in heart failure. N Engl J Med. 2014;371:993–1004. doi: 10.1056/NEJMoa1409077. [DOI] [PubMed] [Google Scholar]
- 17.Packer M, McMurray JJ, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, et al. Angiotensin receptor neprilysin inhibition compared with enalapril on the risk of clinical progression in surviving patients with heart failure. Circulation. 2015;131:54–61. doi: 10.1161/CIRCULATIONAHA.114.013748. [DOI] [PubMed] [Google Scholar]
- 18.Eichhorn EJ, Bristow MR. Medical therapy can improve the biological properties of the chronically failing heart. A new era in the treatment of heart failure. Circulation. 1996;94:2285–2296. doi: 10.1161/01.cir.94.9.2285. [DOI] [PubMed] [Google Scholar]
- 19.Jentzsch C, Leierseder S, Loyer X, Flohrschutz I, Sassi Y, Hartmann D, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Ryall KA, Bezzerides VJ, Rosenzweig A, Saucerman JJ. Phenotypic screen quantifying differential regulation of cardiac myocyte hypertrophy identifies CITED4 regulation of myocyte elongation. J Mol Cell Cardiol. 2014;72:74–84. doi: 10.1016/j.yjmcc.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, et al. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J Biomol Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 22.Foldes G, Matsa E, Kriston-Vizi J, Leja T, Amisten S, Kolker L, et al. Aberrant α-adrenergic hypertrophic response in cardiomyocytes from human induced pluripotent cells. Stem Cell Rep. 2014;3:905–914. doi: 10.1016/j.stemcr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Force T, Kolaja KL. Cardiotoxicity of kinase inhibitors: the prediction and translation of preclinical models to clinical outcomes. Nat Rev Drug Discov. 2011;10:111–126. doi: 10.1038/nrd3252. [DOI] [PubMed] [Google Scholar]
- 24.Kagiyama S, Eguchi S, Frank GD, Inagami T, Zhang YC, Phillips MI. Angiotensin II-induced cardiac hypertrophy and hypertension are attenuated by epidermal growth factor receptor antisense. Circulation. 2002;106:909–912. doi: 10.1161/01.cir.0000030181.63741.56. [DOI] [PubMed] [Google Scholar]
- 25.McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, et al. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 26.Nakagami H, Jensen KS, Liao JK. A novel pleiotropic effect of statins: prevention of cardiac hypertrophy by cholesterol-independent mechanisms. Ann Med. 2003;35:398–403. doi: 10.1080/07853890310001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ, et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 28.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Wang X, Wang X, Yuan M, Wu G, Hu J, et al. Artemisinin attenuates post-infarct myocardial remodeling by down-regulating the NF-κB pathway. Tohoku J Exp Med. 2012;227:161–170. doi: 10.1620/tjem.227.161. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Z, Sun G, Zhu C, Cheng B, Zhang C, Ma Y, et al. Artemisinin, an anti-malarial agent, inhibits rat cardiac hypertrophy via inhibition of NF-κB signaling. Eur J Pharmacol. 2010;649:277–284. doi: 10.1016/j.ejphar.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Guo Z. Artemisinin anti-malarial drugs in China. Acta Pharm Sin B. 2016;6:115–124. doi: 10.1016/j.apsb.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee IS, Ryu DK, Lim J, Cho S, Kang BY, Choi HJ. Artesunate activates Nrf2 pathway-driven anti-inflammatory potential through ERK signaling in microglial BV2 cells. Neurosci Lett. 2012;509:17–21. doi: 10.1016/j.neulet.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Wang S, Wang Y, Zhou C, Chen G, Shen W, et al. Inhibitory effect of the antimalarial agent artesunate on collagen-induced arthritis in rats through nuclear factor κ B and mitogen-activated protein kinase signaling pathway. Transl Res. 2013;161:89–98. doi: 10.1016/j.trsl.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Cominacini L, Mozzini C, Garbin U, Pasini A, Stranieri C, Solani E, et al. Endoplasmic reticulum stress and Nrf2 signaling in cardiovascular diseases. Free Radic Biol Med. 2015;88:233–242. doi: 10.1016/j.freeradbiomed.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 35.Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. 2010;122:2727–2735. doi: 10.1161/CIRCULATIONAHA.110.942268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagel-Langenickel I, Buttgereit J, Bader M, Langenickel TH. Natriuretic peptide receptor B signaling in the cardiovascular system: protection from cardiac hypertrophy. J Mol Med (Berl) 2007;85:797–810. doi: 10.1007/s00109-007-0183-4. [DOI] [PubMed] [Google Scholar]
- 37.Minguet J, Sutton G, Ferrero C, Gomez T, Bramlage P. LCZ696: a new paradigm for the treatment of heart failure? Expert Opin Pharmacother. 2015;16:435–446. doi: 10.1517/14656566.2015.1000300. [DOI] [PubMed] [Google Scholar]