Abstract
The filamentous fungus Trichoderma reesei is a widely used strain for cellulolytic enzyme production. A hypercellulolytic T. reesei variant SN1 was identified in this study and found to be different from the well-known cellulase producers QM9414 and RUT-C30. The cellulose-degrading enzymes of T. reesei SN1 show higher endoglucanase (EG) activity but lower β-glucosidase (BGL) activity than those of the others. A uracil auxotroph strain, SP4, was constructed by pyr4 deletion in SN1 to improve transformation efficiency. The BGL1-encoding gene bgl1 under the control of a modified cbh1 promoter was overexpressed in SP4. A transformant, SPB2, with four additional copies of bgl1 exhibited a 17.1-fold increase in BGL activity and a 30.0% increase in filter paper activity. Saccharification of corncob residues with crude enzyme showed that the glucose yield of SPB2 is 65.0% higher than that of SP4. These results reveal the feasibility of strain improvement through the development of an efficient genetic transformation platform to construct a balanced cellulase system for biomass conversion.
Keywords: Trichoderma reesei, uracil auxotrophy, β-glucosidase, biomass conversion, pretreated corncob residues
Introduction
Lignocellulosic biomass, which includes agricultural crop and forest residues, is the most abundant renewable resource on earth (Singhania et al., 2013). It is considered a promising alternative to fossil fuel because it can be converted to second-generation biofuels, such as bioethanol (Gusakov, 2013). However, the relatively high cost of cellulase preparation for cellulose deconstruction is a major obstacle that limits the large-scale commercial production of bioethanol with lignocellulosic biomass as the substrate (Wu et al., 2011). Most of the current cellulase preparations involve the use of filamentous fungi, such as Trichoderma reesei, but the ability to produce cellulase through the use of a wild strain is not always fit for industrial requirements (Weiss et al., 2013). Therefore, different well-known hypercellulolytic mutants, such as T. reesei QM9414 and RUT-C30, have been developed through classical mutation (Montenecourt and Eveleigh, 1977, 1979). Compared with random mutation, genetic modification provides a rational strategy for strain improvement. However, the main limitation of this strategy is the selection of a proper marker for genetic transformation. Diverse selection mechanisms, including auxotroph complementation (Penttilä et al., 1987), antibiotic resistance (Bailey et al., 2002), and nitrogen utilization (Te’o et al., 2002), have been applied to filamentous fungi. Auxotrophic markers, especially the mutation of the orotidine 5′-monophosphate decarboxylase gene (pyr4/G or ura3) that leads to uracil auxotrophy, are widely applied in fungal transformation because of their high efficiency and ease in obtaining transformants (Smith et al., 1991). Moreover, a pyr4 blaster cassette containing a direct repeat sequence can be recycled by screening with 5′-FOA as the counter selection for multiple gene operations (Hartl and Seiboth, 2005). Evidently, construction of a uracil auxotroph strain derived from a cellulase hyperproducing mutant could facilitate genetic modification and strain improvement.
Trichoderma reesei is one of the most popular industrial filamentous fungi utilized for cellulase production. Its cellulase system cannot be present in optimal ratios for the deconstruction of lignocellulosic substrates, because it was originally isolated from decayed canvas (Mandels and Reese, 1957; Gomes et al., 1992). The well-known T. reesei mutants, including QM9414 and RUT-C30, were generated through random mutation with only cellulose as the screening substrate. Hence, their cellulase preparations are unfeasible for large-scale production and conversion of lignocellulosic materials into bioethanol. Given their capacity to secrete high-level cellulases, T. reesei strains still occupy the dominant position as a suitable starting point for the construction of engineered strains used to produce highly active and economically feasible cellulases for the bioethanol industry (Rosgaard et al., 2006). The T. reesei cellulase system is composed of endoglucanase (EG) and cellobiohydrolase (CBH), which jointly hydrolyze cellulose to produce cellooligosaccharide and cellobiose (Teugjas and Väljamäe, 2013). These products are further degraded into glucose by β-glucosidase (BGL) to complete the rate-limiting step of cellulose hydrolysis (Sørensen et al., 2013). However, the low amount of secreted BGL (i.e., BGL1) usually leads to incomplete cellobiose hydrolysis, which in turn inhibits the product of the T. reesei enzyme system (Rosgaard et al., 2006). Hence, various strategies, such as exogenous addition of BGLs (Duenas et al., 1995), co-cultivation of high-BGL activity strains with T. reesei (Gutierrez-Correa et al., 1999), and genetic transformation of T. reesei with homologous and heterologous bgl genes, have been proposed to supplement BGL activity (Zhang et al., 2010; Ma et al., 2011; Treebupachatsakul et al., 2015).
In our previous work, a hypercellulolytic variant SN1 was isolated on Carboxy Methyl Cellulose (CMC) agar plates and also used for cellulase production. Here, the strain SN1 was identified as T. reesei and characterized with high endoglucanase activity. For strain improvement, a uracil auxotroph strain was constructed by deleting the pyr4 gene via homologous recombination in SN1. The β-glucosidase gene was further overexpressed by using the strong engineered cbh1 promoter to optimize the cellulase system for the saccharification of different pretreated corncob residues.
Materials and Methods
Strains, Plasmids, and Culture Conditions
Trichoderma reesei QM9414 (ATCC 26921), which is a general cellulase producer, and RUT-C30 (ATCC 56765), which is another cellulase high-producing strain that is less sensitive to glucose repression, were utilized as the control strains for the comparison of cellulase production. The strain SN1 is a hypercellulolytic variant isolated from our laboratory (Song et al., 2016) and used as the initial host for strain improvement. Escherichia coli DH5a (TransGen, Beijing, China) was used for vector construction and propagation. The pMD18-T cloning vector (Takara, Otsu, Japan) was purchased for TA cloning. The pTHB vector was used for bgl1 overexpression; this vector contained a T. reesei bgl1 expression cassette under the control of a modified four-copy cbh1 promoter, as described by Zhang et al. (2010). The plasmid pAB4-1 contained the Aspergillus niger pyrG gene as a selection marker to encode orotidine-5′-phosphate decarboxylase (Hartl and Seiboth, 2005). All strains were grown and maintained on a potato dextrose agar plate (PDA) containing 20.0 g/L D-glucose and 20.0 g/L agar for 5–7 days at 30°C. The conidia were harvested, and 106 conidia were inoculated in a 500 mL flask containing 150 mL minimal medium (MM; Penttilä et al., 1987). MM with 300 μg/mL hygromycin B and 1.5 mg/mL 5′-FOA (Sigma, USA) was applied as the selective medium to screen the uracil auxotroph transformants (Singh et al., 2015). An esculin plate containing 3 g/L of esculin, 10 g/L of sodium carboxymethy cellulose (CMC–Na), 0.5 g/L of ferric citrate, and 20 g/L agar was utilized to screen the strains showing β-glucosidase (BGL) activity. A CMC plate containing 10 g/L of CMC–Na, 1 g of yeast extract, and 20 g/L agar was utilized to screen the strains showing cellulase (EG) activity.
Morphology and Molecular Identification
The fungal strains were grown on PDA plates or solid MM with 2% glucose, 2% glycerol, or 2% lactose as the carbon sources. Photographs of the colonies were obtained with a SONY DSC-HX400 camera for morphology comparison. Microscopic images were captured with a Nikon Eclipse80i light microscope (Nikon, Japan). The fungal genomic DNA was isolated according to the method of Penttilä et al. (1987). For phylogenetic identification of the variant strain SN1, 18S and internally transcribed spacer (ITS) rDNA gene fragments were amplified through polymerase chain reaction (PCR) with the universal primer pairs 18S-F1/18S-R1 and ITS-F1/ITS-R1 (Table 1; White et al., 1990), respectively. The SN1 genomic DNA was utilized as the PCR template. The amplified products were purified, sequenced, and compared with the sequences of fungal rDNA 18S and ITS from the NCBI database through the use of the BLAST algorithm. Multiple sequence alignment was conducted with ClustalX. Maximum composite likelihood method analysis was conducted with the MEGA 5 software.
Table 1.
Primers used in this study.
| Primers | Sequences (5′–3′) | Employment |
| 18S-F1 | TTCCAGCTCCAATAGCGTAT | Strain identification |
| 18S-R1 | CAGACAAATCACTCCACCAAC | Strain identification |
| ITS-F1 | TCCGTAGGTGAACCTGCGG | Strain identification |
| ITS-R1 | TCCTCCGCTTATTGATATGC | Strain identification |
| pyr4-UF1 | AGTGTTTGATGCTCACGCTC | Mutant construction |
| pyr4-UR1 | GGAGATGTTGCTGAAGTCGA GGCGAGGGAGTTGCTTTA | Mutant construction |
| pyr4-DF1 | ATGAGTCGTTTACCCAGAATAAG AAAGGCATTTAGCAAGA | Mutant construction |
| pyr4-DR1 | TGAACAGTAAGGTGTCAGCA | Mutant construction |
| pyr4-UF2 | TGATGCTCACGCTCGGAT | Mutant construction |
| pyr4-DR2 | TCGTCTCGTTCAGCTCGTAATC | Mutant construction |
| Ypyr4-UF1 | ACACAACCTACTGAGCAGAACC | Mutant construction |
| Yhph-F2 | GTCTGGACCGATGGCTGTG | Mutant construction |
| Hph-F1 | CGACTTCAGCAACATCTCC | Mutant construction |
| Hph-R1 | ATTCTGGGTAAACGACTCAT | Mutant construction |
| Yhph-F1 | GCAAAGTGCCGATAAACA | Mutant construction |
| Yhph-R1 | GCGAAGGAGAATGTGAAG | Mutant construction |
| pyrG-S | CTTCCTAATACCGCCTAGTCAT | Mutant construction |
| pyrG-A | AGCCGCTGGTCAATGTTATC | Mutant construction |
| YpyrG-F1 | ATCAACACCATGTCCTCCAA | Mutant construction |
| YpyrG-R1 | ACACGAATCCCATAACGAAG | Mutant construction |
| Y1 | GCCAGGGATGCTTGAGTGTA | Mutant construction |
| Y2 | CCCAGCCACAGGACCAAGTATG | Mutant construction |
| pyr4-probe-F | GCCATCCTCCTTCTTCCTCT | Probe |
| pyr4-probe-R | TGATACACACAAGTCTGCCAGAT | Probe |
| bgl-probe-F | TTGAGCCCAATCAGAAATGCGT | Probe |
| bgl-probe-R | CCCAGCCACAGGACCAAGTATG | Probe |
Construction of the pyr4 Deletion Strain
The Δpyr4::hph cassette for pyr4 gene disruption was constructed with the double-joint PCR method (Yu et al., 2004). The cassette carried the E. coli hygromycin B phosphotransferase (hph) gene as the selection marker between the 5′- and 3′-flanking regions of pyr4. Specifically, the pyr4 5′- and 3′-flanking sequences were amplified with two primer pairs, pyr4-1500-UF1/pyr4-72-UR1 and pyr4-60-DF1/pyr4-1540-DR1 (Table 1), respectively, with the SN1 genomic DNA as the template. The hygromycin B phosphotransferase gene (hph) was amplified with hph-F1/hph-R1 primers (Table 1). The amplicons were then mixed in 1:3:1 molar ratio of 5′-flanking region:hph:3′-flanking region and jointed together during another round of PCR without primers. The resulting PCR product was used directly as a template for the third round of PCR to construct the pyr4 disruption cassette with the nest primers pyr4-1400-UF2/pyr4-1488-UR2 (Table 1). The pyr4 disruption cassette was transformed into SN1 protoplasts afterward through the method described by Penttilä et al. (1987). The target transformants were screened on MM containing 300 μg/mL hygromycin B and 1.5 mg/mL 5′-FOA. The purified candidate transformants were identified through PCR and Southern blot with the genomic DNA as the template to verify if the pyr4 locus was replaced by the deletion cassette.
Overexpression of the β-Glucosidase bgl1 Gene in T. reesei SP4
The vectors pTHB and pAB4-1 were co-transformed into protoplasts of T. reesei SN1 through the above mentioned method. The transformants were directly screened on MM. The candidates were then verified through PCR by using primers Y1 and Y2 (Table 1), as described by Zhang et al. (2010). Esculin agar plates were prepared and utilized to confirm the bgl1 overexpression strain.
Southern Blot Analysis
The probe of pyr4 was a fragment amplified through PCR by using primers pyr4-probe-F and pyr4-probe-R to detect the pyr4 gene. The XbaI/HindIII-digested genomic DNA was hybridized by the pyr4 probe. The probe of bgl1 was a fragment amplified through PCR by using primer bgl1-probe-F/bgl1-probe-R to detect the bgl1 gene. The EcoRI-digested genomic DNA was hybridized by the bgl1 probe. The probe-hybridized DNA fragment was detected with the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Diagnostics, Mannheim, Germany).
Shake Flask Cultivation for Cellulase Production
The seed cultures for cellulase production were prepared in MM after incubation at 200 rpm and 30°C for 36 h in a 300 mL flask. Then 15 mL of the activated cells was transferred to 150 mL of the cellulase production medium (CPM) in a 500 mL flask. The CPM composition was as follows (g/L): 20 Microcrystalline cellulose, 2.5 (NH4)2SO4, 5 KH2PO4, 0.6 MgSO4.7H2O, 1 CaCl2.2H2O, and 20 corn steep liquor.
Cellulase Activity Assay and Zymogram Analysis of β-Glucosidase Activity
The filter paper (FP), EG, CBH, and BGL activities of T. reesei were measured as described before (Ghose, 1987) by using Whatman No. 1 FP (Whatman, UK), CMC–Na (Sigma, USA), p-nitrophenyl-β-D-cellobioside (pNPC; Sigma, USA), and p-nitrophenyl-β-D-glucopyranoside (PNPG; Sigma, USA) as the substrates, respectively. One unit of enzyme activity was defined as the amount of enzymes releasing 1 mole of reducing sugar (or p-nitrophenol in the CBH and BGL assays) per minute under the assay conditions. Renaturing SDS–PAGE electrophoresis was performed in 12% polyacrylamide separating gel containing 0.1% SDS with 0.3% CMC–Na as the substrate; this procedure was performed in a Mini-PROTEAN tetra electrophoresis cell (Bio-Rad Laboratories, Milan, Italy).
Saccharification of the Pretreated Corncob Residues
Acid-pretreated and delignined corncob residues were used as substrates in the saccharification process; their composition has been described by Zhao et al. (2008) (Supplementary Table S1). The cellulase crude complexes for the saccharification of the pretreated corncob residues were placed in 100 mL flasks containing 5% (w/v) substrate. The pH value and temperature were adjusted to 4.8 (with 50 mM citric acid buffer) and 50°C, respectively. Enzyme loading was adjusted to the same FP activity (10 FPU/g substrates). Glucose production was detected with an SBA-40C biological sensor analyzer (BISAS, Shandong, China) after incubation for 24 or 48 h. Cellulose conversion was calculated as follows:
Results
Molecular and Morphological Identification of Cellulolytic Strain SN1
The cellulase production variant SN1 was identified with partial 18S rDNA and ITS sequences to characterize its phylogenetic relationships. Both the 18S rDNA gene and ITS sequences of SN1 showed 100% identity to those of T. reesei (Supplementary Figure S1). Hence, the strain SN1 was defined as a variant of T. reesei.
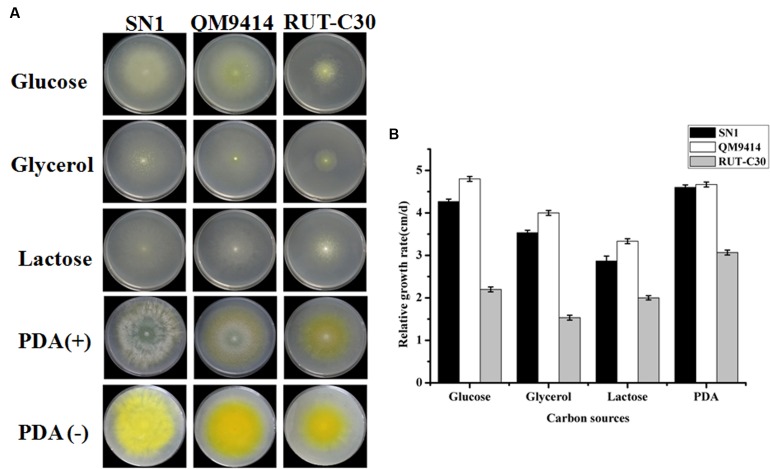
Trichoderma reesei is a cellulolytic filamentous fungus originally isolated in the Solomon Islands during World War II. T. reesei QM9414 and RUT-C30 are among the most well-known high cellulase-producing mutants after 30 years of strain improvements (Peterson and Nevalainen, 2012). They are utilized as control strains in the current study for comparison with SN1 to analyze growth in different carbon sources. All strains were cultured on plates containing 2% glucose, 2% glycerol, and 2% lactose and on PDA plates for 4 days. As shown in Figure 1A, SN1 exhibited a colony morphology similar to that of QM9414 and RUT-C30 on glucose, glycerol, and lactose but showed increased radial growth and reduced pigment production on the PDA plates. The growth rate of SN1was comparable to that of QM9414 but higher than that of RUT-C30 on all the growth media tested in this study (Figure 1B). Microscopic observations revealed that SN1 had a spore morphology similar to that of the others but possessed a less-branched mycelium (Supplementary Figure S2). In summary, the strain SN1 is a novel T. reesei variant with high EG activity.
FIGURE 1.
Growth of Trichoderma reesei SN1, QM9414 and RUT-C30 on different carbon sources. (A) Strains were grown on plates containing minimal medium supplemented with 2% glucose, 2% glycerol, 2% lactose, or PDA at 30°C for 3 days. (B) The growth was represented by the relative growth rate which was measured as the colony diameter increase per centimeter per day.
Cellulase Production and Its Cellulolytic Potential in Biomass Saccharification by T. reesei SN1
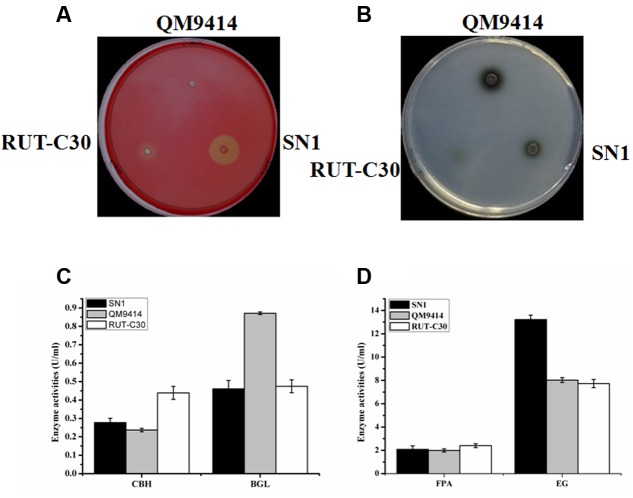
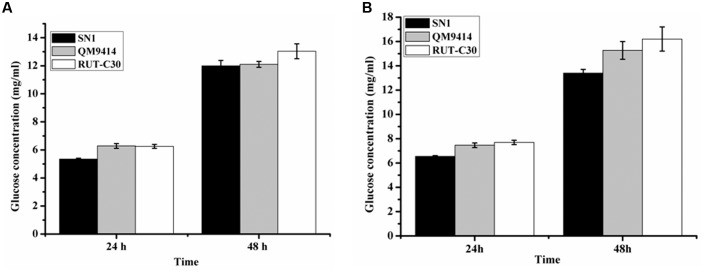
To investigate the capacity to secrete cellulase, the fungal strains were cultured on media containing CMC as the substrate, which is generally used to detect EG activity. The media plate was dyed with 1% congo red and decolored with 1 M NaCl after 3 days of cultivation. The cellulolytic halo around the SN1 colony was much larger than those of RUT-C30 and QM9414 (Figure 2A). This result indicates that T. reesei SN1 probably secreted high-level EG. Another medium containing CMC and esculin as the substrate was applied to test the ability to secrete β-glucosidase (Figure 2B). The size of the black halo around the SN1 colony was smaller than that of QM9414, suggesting that T. reesei SN1 secreted a relatively low level of BGL. T. reesei SN1, QM9414 and RUT-C30 were fermented in CPM to further verify cellulase production. The culture supernatants were also used for the cellulase activity assay. As shown in Figure 2C, T. reesei SN1 produced a total cellulase activity (FPA) comparable to that of QM9414 but exhibited a notably higher (nearly two times) EG activity than QM9414 and RUT-C30. Comparison of CBH and BGL activities showed that SN1 also produced a CBH activity similar to that of QM9414 but had a much reduced (almost half) BGL activity (Figure 2D). These results confirm that T. reesei SN1 is a novel hypercellulytic strain that produces prominent EG activity but minimal BGL activity. These results prompted us to examine the saccharification potential of the SN1 enzyme to convert cellulosic materials. The experimental design involved the enzymatic hydrolysis of two types of pretreated corncob residues. This hydrolysis was performed at 50°C and pH of 4.8 for 48 h. In the saccharification of the acid-pretreated corncob residues, the amount of glucose released using the SN1enzyme at 24 h was only 5.3 mg/mL (i.e., 15.2% cellulose conversion), which was lower than the values for QM9414 (6.3 mg/mL corresponding to 17.9% cellulose conversion) and RUT-C30 (6.4 mg/mL corresponding to 17.9% cellulose conversion). The glucose releases in SN1 (12.0 mg/mL corresponding to 34.4% cellulose conversion) and QM9414 (12.1 mg/mL corresponding to 34.8% cellulose conversion) were similar after an additional 24 h of enzymatic hydrolysis (i.e., after a total enzymatic reaction of 48 h). However, they were still lower than that in RUT-C30 (13.3 mg/mL corresponding to 38.4% cellulose conversion; Figure 3A). When the delignined corncob residues were used as the substrate, the final glucose release in SN1 (13.4 mg/mL corresponding to 36.7% cellulose conversion) after a 48 h reaction was still lower than those in QM9414 (15.3 mg/mL corresponding to 42.0% cellulose conversion) and RUT-C30 (16.2 mg/mL corresponding to 44.4% cellulose conversion; Figure 3B). Therefore, T. reesei SN1 exhibited outstanding EG activity. However, its saccharification efficiency for cellulosic materials was obviously lower than those of QM9414 and RUT-C30 probably because of the insufficiency of β-glucosidase in the SN1enzyme system.
FIGURE 2.
A comparison of cellulase production in T. reesei SN1, QM9414 and RUT-C30. (A) The endoglucanase production of T. reesei SN1, QM9414 and RUT-C30 on the condition of 1% CMC-Na with 0.5% triton at 30°C for 2 days. (B) Detection of β-glucosidase activity on the CMC-esculin plate in T. reesei SN1, QM9414 and RUT-C30 at 30°C for 24 h. (C) The FPA and EG activities of the T. reesei SN1, QM9414 and RUT-C30, were measured after being induced in cellulase fermentation medium for 5 days. (D) The CBH and BGL activities of the T. reesei SN1, QM9414 and RUT-C30, were measured as the above mentioned. All results are represented as the mean of three independent experiments; error bars express the standard deviation.
FIGURE 3.
Saccharification of different pretreated concob residues by T. reesei SN1, QM9414 and RUT-C30. (A) Saccharification of acid pretreated corncob residues with equal FPA activity. (B) Saccharification of delignined corncob residues. Data are represented as the mean of three independent experiments; error bars express the standard deviations.
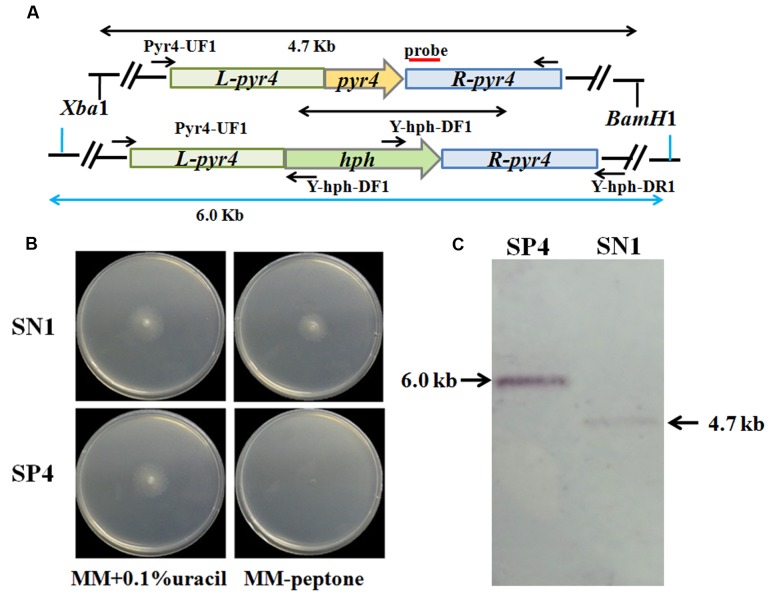
Generation of a pyr4 Disruption Strain from T. reesei SN1
The uracil auxotroph strain maximizes high-efficiency genetic transformation and can be used for multiple gene manipulations depending on the pyr4 marker gene that can be bidirectionally selected (Seidl and Seiboth, 2010). Thus, disruption of the pyr4 gene in T. reesei SN1 to construct the uracil auxotroph strain will contribute to strain improvement through genetic modification. In this study, a Δpyr4::hph cassette was constructed and utilized to disrupt the pyr4 gene in T. reesei SN1 (Figure 4A). The generated uracil auxotroph strain SP4 was confirmed through plate screening. As shown in Figure 4B, SP4 could normally grow on MM containing 0.1% uracil but could not grow on MM without uracil. The XbaI/HindIII-digested genomic DNA was hybridized with the pyr4 probe for further Southern blot assay. It yielded a 6.0 kb fragment for the strain SP4 and a 4.7 kb fragment for the parent strain SN1 (Figure 4C). These results suggest that the pyr4 gene was successfully knocked out in SP4. Furthermore, comparison of the cellulase production of SP4 and SN1 showed that the cellulase activities of the SP4 crude enzyme preparations were almost similar to those of SN1 (data not shown).
FIGURE 4.
Deletion of pyr4 gene in T. reesei SN1. (A) Schematic representation of the pyr4 locus in the Δpyr4 and parent strains. Relative positions of the XbaI/HindIII enzyme restriction sites are noted. Probe used for Southern analysis is shown as red box. (B) Phenotypic assay on MM supplemented with uracil (left) and MM without uracil (right) at 30°C for 2 days. (C) Southern blot of the genome digested with XbaI/HindIII. A fragment of 4.7 kb is present in the parent strain, the 6.0 kb band is shown in Δpyr4 strains.
Overexpression of bgl1 under the Control of the Artificial Four-Copy cbh1 Promoter in T. reesei SP4
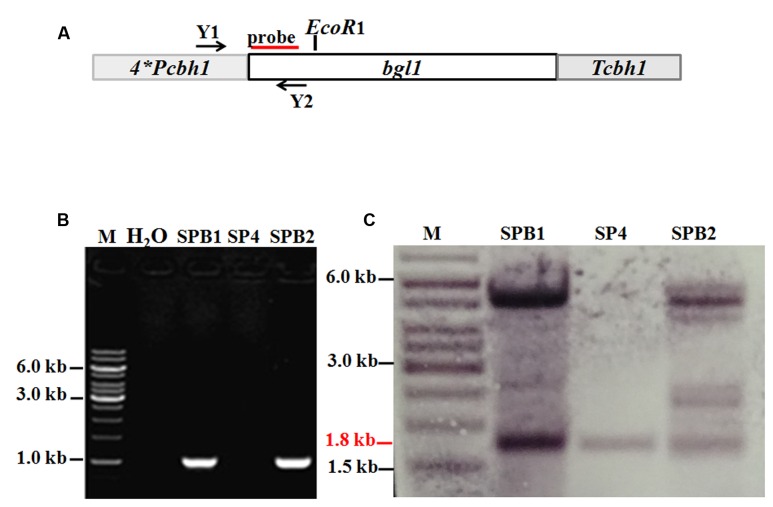
To overcome the lack of BGL in the SP4 enzyme system, the pTHB plasmid containing the bgl1 gene under the control of a strong modified cbh1 promoter, as described by Zhang et al. (2010) (Figure 5A), was co-transformed with the A. niger pyrG-containing plasmid pAB4-1 into T. reesei SP4. The 126 positive transformants were all further screened on esculin plates to test the BGL secretion (data not shown). This esculin method has been widely used to screen strains showing BGL activity (Lee et al., 2014). Two of the transformants, namely, SPB1 and SPB2, showed much larger black zones around the colony than the parental strain SP4 (Figures 6A,B). They were then selected and verified through PCR amplification of the bgl1 gene using the primers Y1 and Y2 (Figure 5B). For further analysis by Southern blot, the EcoRI-digested genomic DNA was hybridized by the bgl1 probe and yielded a 1.8 kb fragment for the parent strain SP4 (Figure 5C). An additional 5.0 kb band was observed for SPB1, and four additional bands (2.3, 4.5, 5.0, and 5.5 kb) were found for SPB2. These results suggest that one and four bgl1 copies were successfully integrated into the genomes of SPB1 and SPB2, respectively.
FIGURE 5.
Overexpression of bgl1 gene in the Δpyr4 mutant. (A) An expression cassette containing the bgl1 gene is controlled by the T. reesei modified cbh1 promoter. Relative positions of the EcoRI enzyme restriction sites are noted. Probe used for Southern analysis is shown as red box. (B) PCR assay of the BGL overexpression transformants. Both the transformants SPB1 and SPB2 show a 1.0 kb DNA fragments panning using the primers Y1 and Y2, M: DNA marker. (C) Southern blot of the genome digested with EcoR1. The 1.8 kb (red line) wild-type band is present in all the strains. The additional bands indicate the bgl1-overexpression cassette was successfully integrated into the chromosome.
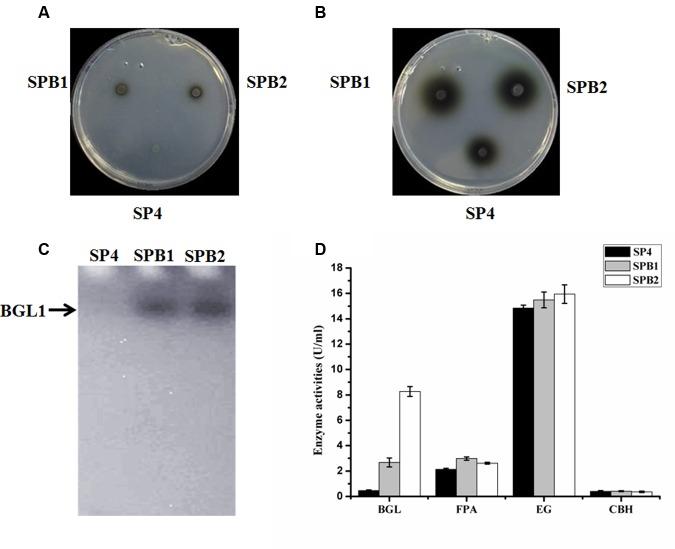
FIGURE 6.
Phenotypic and cellulase production assay of bgl1 overexpression transformants. (A) The black halo was observed on the transformants SPB1 and SPB2 at the 12 h. (B) The black halos were observed on the transformants SPB1 and SPB2 at the 24 h. (C) Detection of β-glucosidase activity from the culture-free supernatant of T. reesei SN1, SPB1and SPB2 performed on 12% SDS–PAGE containing 0.5% esculin as substrate. (D) BGL, FPA, EG, CBH activities were measured after being induced in cellulase fermentation medium for 5 days. Data are represented as the mean of three independent experiments; error bars express the standard deviations.
Characterization of Cellulase Production by the bgl1-Overexpression Strains
The bgl1-overexpression strains were cultivated in a CPM for 5 days. The culture supernatants were collected for the cellulase activity assay. First, renaturing SDS-PAGE analysis on 12% polyacrylamide gel containing 0.3% esculin as a substrate was applied to detect the BGL secreted by the fungal strains. Figure 6C shows a clear band in the secreted proteins of transformant SPB1 or SPB2 but not in SP4. This result indicates that the two bgl1-overexpression strains are very likely to secrete a much higher level of BGL than the parental strain. In accordance with this result, the BGL activities of SPB1 and SPB2 measured with pNPG as a substrate were 4.7 and 17.1 times higher than that of SP4, respectively (Figure 6D). FP activity (FPA) is generally utilized to evaluate the total cellulase activities. The FPA of SPB1 and SPB2 showed 39.7 and 22.6% increase compared with that of the parent strain SP4 (Figure 6D). However, the EG and CBH activities of the two transformants were not significantly changed, besides a relatively lower CBH activity (10.0% decrease) in SPB2 (Figure 6D). Interestingly, the highest β-glucosidase activity (8.3 U/mL) was found in SPB2; the value was 3.2-fold higher than that of SPB2 (2.6 U/mL). However, the FPA in SPB2 (2.6 U/mL) was lower than that in SPB1 (3.0 U/mL). This difference in FPA in the two transformants may have been caused by the negative effect of the additional four copies of cbh1 promoters in the SPB2. It was reported that multiple copies of promoters might result in the titration effect of transcriptional regulators for cellulase gene expression (Rahman et al., 2009; Zhang et al., 2010).
Saccharification of the Corncob Substrates by Cellulase Preparations from T. reesei SPB1 and SPB2
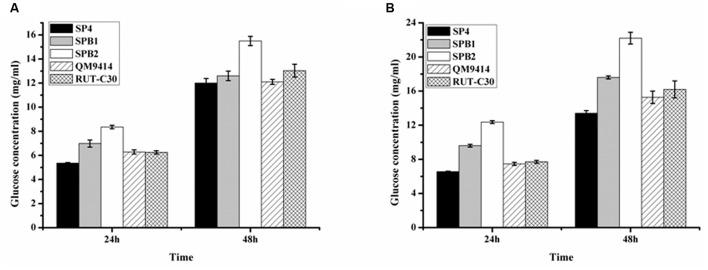
The ability of the cellulase preparations produced by T. reesei SPB1 and SPB2 to hydrolyze the pretreated corncob residues was investigated. In the saccharification of the acid-pretreated corncob residues, the glucose release (12.6 mg/mL corresponding to 36.2% cellulose conversion) using the SPB1 enzyme was slightly higher than that for SP4 (12.1 mg/mL corresponding to 34.4% cellulose conversion) but lower than that for RUT-C30 (13.3 mg/mL corresponding to 38.4% cellulose conversion) after a total enzymatic reaction of 48 h (Figure 7A). The SPB2 enzyme exhibited the highest glucose release (15.5 mg/mL corresponding to 44.6% cellulose conversion). This result indicates that the high BGL activity in the SPB2 enzyme contributed to the efficient hydrolysis of the acid-pretreated corncob residues. The final glucose yields of SPB1 (17.6 mg/mL corresponding to 48.2% cellulose conversion) and SPB2 (22.2 mg/mL corresponding to 61.0% cellulose conversion) after 48 h reaction were higher than those of SP4 (13.4 mg/mL corresponding to 36.7% cellulose conversion) and RUT-C30 (16.2 mg/mL corresponding to 44.4% cellulose conversion) when the delignined corncob residues were used as a substrate (Figure 7B).Therefore, both bgl1-overexpression strains (SPB1and SPB2) exhibited better performance than the parental strain SP4 regardless of whether acid-pretreated or delignined corncob residues were used as the substrate. These results suggest that BGL activity has a notable positive correlation with cellulose conversion and indicates that β-glucosidase plays a key role in the enzymatic hydrolysis of corncob residues.
FIGURE 7.
Saccharification of different pretreated concob residues by the BGL-overexpression strains. (A) Saccharification of acid pretreated corncob residues by T. reesei SP4 and the BGL-overexpresion strains SPB1 and SPB2 with equal FPA activity. (B) Saccharification of delignined corncob residues T. reesei SP4 and the BGL-overexpresion strains SPB1 and SPB2 with equal FPA activity. Data are represented as the mean of three independent experiments; error bars express the standard deviations.
Discussion
The application of cellulolytic microorganisms to cellulosic biomass bioconversion is considered a potential sustainable approach to develop novel bioproducts, such as biofuels (Premalatha et al., 2015). Diverse cellulolytic microorganisms, including fungi and bacteria, have been isolated from various environments (Jyotsna et al., 2010; Raddadi et al., 2013; Dantur et al., 2015; Premalatha et al., 2015). However, the ability to produce cellulose-degrading enzymes by using wild strains is not always fit for industrial requirements (Weiss et al., 2013). Hence, much effort has been exerted to mutate or engineer potential strains for application in the cellulase industry. In this study, a hypercellulolytic fungus, SN1, was identified as a T. reesei variant according to morphological characteristics and ribosomal DNA sequences.
Trichoderma reesei was initially isolated from decayed canvas during World War II and is currently one of the most well-known industrial fungi used for cellulase production (Seidl et al., 2008). Several mutants with high cellulase activity, such as NG-14, QM9414, and RUT-C30, have been obtained after more than half a century of strain improvement. Among these mutants, QM9414 and RUT-C30 are the most widely used in cellulase production and as model strains for basic research on cellulase expression (Stricker et al., 2006; Gyalai-Korpos et al., 2011). Therefore, the SN1 strain was compared with these two mutants for hyphal growth and cellulase production in this study. The growth rate of SN1 was comparable to that of QM9414 but was faster than that of RUT-C30. SN1 produced remarkably reduced pigment and sporulated earlier (Figure 1). RUT-C30 can produce approximately 1.4 times the total cellulase activity of QM9414 but only 50% of the β-glucosidase activity of QM9414 (Peterson and Nevalainen, 2012). In this study, SN1 showed a total cellulase activity comparable to that of QM9414 and a notably higher (nearly two times) EG activity than both QM9414 and RUT-C30 (Figure 2). Considering that our lab studied T. reesei for years, and this strain is quite different from QM9414 (carbon catabolite-repressed, containing the full-length cre1 gene) and RUT-C30 (carbon catabolite-derepressed, containing the truncated cre1 gene) whether on morphological phenotype or cellulase production, so we further tested whether the SN1 strain was carbon catabolite-derepressed and found that it was carbon catabolite-repressed and contained a complete carbon catabolite repressor gene cre1 (data not shown). Therefore, we speculated that SN1 was a novel T. reesei variant, which was most probably spontaneously mutated from QM9414 due to the cellulose substrates used as the continued screening pressure. Many studies have shown that EGs can be widely used in industrial activities, such as pulp beatability (Kamaya, 1996), deinking (Gübitz et al., 1998), and bio-polishing (Li et al., 2011). The high-level endoglucanase production showed that SN1 is a potential strain for industrial cellulase production.
However, despite the feature of SN1 cellulases, the respective activities in the cellulase products appeared to be present in suboptimal ratios for lignocellulose degradation. Hence, improvement of the cellulolytic capacity of T. reesei SN1 is required. This enhancement is often accomplished through genetic engineering. Auxotrophy selection markers, such as uracil auxotrophy, have been proven highly efficient for T. reesei genetic transformation (Hartl and Seiboth, 2005; Steiger, 2013). T. reesei TU6 is an auxotrophic strain widely used for its high transformation efficiency (Gruber et al., 1990). However, TU6 was obtained through random γ-radiation mutagenesis from QM9414, resulting in slow growth, decreased conidiation, and reduced cellulase production. Therefore, a new uracil auxotrophic strain, SP4, was constructed in this study through targeted gene deletion from a hypercellulolytic strain (SN1). No discernible difference in growth, conidiation, or cellular morphology was observed between SP4 and its parental strain SN1 (data not shown). Furthermore, SP4 produced a high cellulase level and could be genetically transformed with high efficiency because of the uracil auxotroph. Therefore, T. reesei SP4 provides a platform strain suitable for genetic manipulation to improve cellulolytic efficiency in biomass conversion.
The major challenges in the bioconversion of lignocellulosic biomass include obtaining the optimum amount of BGL to complete the final step of cellulose hydrolysis (Singhania et al., 2013; Pryor and Nahar, 2015). Low β-glucosidase activity results in incomplete cellobiose hydrolysis, which further leads to product inhibition in the cellulolytic enzymes. Construction of engineering strains with high BGL activity is the preferred strategy to address this problem. Indeed, studies have shown that overexpression of BGL-encoding genes in T. reesei could result in improved BGL activity; this overexpression was mostly accomplished through genetic transformation with hygromycin B resistance (Liu et al., 2013). Wang and Xia (2011) constructed a T. reesei strain with a BGL activity of 5.3 U/mL by using the heterologous expression of the A. nigerβ-glucosidase (cellobiase) gene. Ma et al. (2011) reported that heterologous expression of the Penicillium decumbentsβ-glucosidase gene via Agrobacterium-mediated transformation in T. reesei leads to an eightfold increase in BGL activity. In another study, Zhang et al. (2010) utilized a modified cbh1 promoter to overexpress the native bgl1 gene in T. reesei and obtained a pyrithiamine-resistant strain with 5.7-fold higher BGL activity. In this study, the same bgl1 expression cassette was transformed into SP4 with uracil auxotrophy as a selection marker. This uracil auxotroph transformation system showed high efficiency. One transformant, SPB2, exhibiting a 17.1-fold higher BGL activity with additional four copies of bgl1 gene was selected from 126 positive transformants. The ratio of BGL activity to FPA in SPB2 reached 2.8, which was much higher than that in the parental strain (0.2). Chen et al. (2008) reported that cellobiose would be efficiently degraded into glucose if the ratio of BGL activity to FPA is more than 0.3. Thus, the enzyme system produced by the bgl1-overexpression strains constructed in this study can be directly applied to the saccharification of lignocellulosic materials without adding extra BGL.
The SPB2 enzyme containing the highest BGL activity displayed excellent performance in the saccharification of both corncob substrates (Figure 7). The SPB1 enzyme was more effective than SP4 in the saccharification of the delignined corncob residues, but both had a similar effect on the hydrolysis of the acid-pretreated corncob residues. A possible reason for the difference maybe that a small amount of β-glucosidase in the enzyme complex was adsorbed on to the lignin component in the acid-pretreated corncob residues, which is in accordance with the postulation of Haven and Jørgensen (2013). The enhancement effect of excessive BGL on the saccharification of cellulosic materials (shown by the SPB2 enzyme) has been previously reported (Peterson and Nevalainen, 2012). Such results can be explained by the possibility that a large amount of β-glucosidase would reach the saturated adsorption of lignin and further degrade cellobiose, thus helping alleviate product inhibition and facilitate cellulose conversion.
Conclusion
A novel hypercellulolytic T. reesei variant, SN1, was characterized. A uracil auxotrophic strain, SP4, was constructed through gene deletion to achieve high-efficiency genetic transformation. Further overexpression of the bgl1 gene in SP4 significantly enhanced the BGL activity in the cellulolytic enzyme complex and was proven to be highly effective in the saccharification of the pretreated corncob residues. These results reveal the feasibility of strain improvement from a mutant industrial strain via ab initio genetic manipulation. This strategy appears to be practicable as an essential step in the construction of genetically engineered strains for the production of commercially viable enzyme preparations for biomass conversion.
Author Contributions
YuQ, YH, YiQ, and YZ designed and coordinated the study and wrote the manuscript. YuQ and LZ carried out the experiments and analyzed the results. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31370135), Fundamental Research Funds of Shandong University (No. 2015CJ005), and Agricultural Science and Technology Achievement Transformation Fund of Shandong Province (2014 No. 45).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01349
FIGURE S1 | Phylogenetic tree of strain SN1 based on the 18S rDNA gene sequence and internally transcribed spacer (ITS) region. (A) 18S rDNA sequences. The 18s rDNA gene sequences from Trichoderma reesei (XM_006965206.1), T. sp (AB923813.1), T. harzianum (KP133166.1), T. viride (AF218788), Penicillium oxalicun (JQ349066.1), Aspergillus niger (AM270052.1), Cladonia arbuscula (KT148631.1). (B) ITS region sequences, the ITS region sequences from T. reesei (KT336514.1), T. harzianum (KR232487.1), T. viride (DQ442274.1), T. saturnisporum (KF294853.1), T. asperelloides (KT426897.1), T. pseudokoningii (AF140046), P. oxalicun (KT189516.1), A. niger (KM613139.1).
FIGURE S2 | Mycelium morphology of SN1, QM9414, and RUT-C30. All strains were cultivated 2–5 days on PDA plates (glucose) to analyze the microscopic morphology of mycelium and spore. The images were captured using DIC. Scale bars are listed 20 μm in the figure.
References
- Bailey M., Askolin S., Hörhammer N., Tenkanen M., Linder M., Penttilä M., et al. (2002). Process technological effects of deletion and amplification of hydrophobins I and II in transformants of Trichoderma reesei. Appl. Microbiol. Biotechnol. 58 721–727. 10.1007/s00253-002-0966-z [DOI] [PubMed] [Google Scholar]
- Chen M., Zhao J., Xia L. (2008). Enzymatic hydrolysis of maize straw polysaccharides for the production of reducing sugars. Carbohydr. Polym. 71 411–415. 10.1016/j.carbpol.2007.06.011 [DOI] [Google Scholar]
- Dantur K. I., Enrique R., Welin B., Castagnaro A. P. (2015). Isolation of cellulolytic bacteria from the intestine of diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 5 15 10.1186/s13568-015-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas R., Tengerdy R. P., Gutierrez-Correa M. (1995). Cellulase production by mixed fungi in solid-substrate fermentation of bagasse. World J. Microbiol. Biotechnol. 11 333–337. 10.1007/BF00367112 [DOI] [PubMed] [Google Scholar]
- Ghose T. K. (1987). Measurement of cellulase activities. Pure Appl. Chem. 59 257–268. 10.1351/pac198759020257 [DOI] [Google Scholar]
- Gomes I., Gomes J., Steiner W., Esterbauer H. (1992). Production of cellulase and xylanase by a wild strain of Trichoderma viride. Appl. Microbiol. Biotechnol. 36 701–707. 10.1007/BF00183253 [DOI] [Google Scholar]
- Gruber F., Visser J., Kubicek C. P., De Graaff L. H. (1990). The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18 71–76. 10.1007/BF00321118 [DOI] [PubMed] [Google Scholar]
- Gübitz G. M., Mansfield S. D., Böhm D., Saddler J. N. (1998). Effect of endoglucanases and hemicellulases in magnetic and flotation deinking of xerographic and laser-printed papers. J. Biotechnol. 65 209–215. 10.1016/S0168-1656(98)00130-8 [DOI] [Google Scholar]
- Gusakov A. V. (2013). Cellulases and hemicellulases in the 21st century race for cellulosic ethanol. Biofuels 4 567–569. 10.4155/bfs.13.55 [DOI] [Google Scholar]
- Gutierrez-Correa M., Portal L., Moreno P., Tengerdy R. P. (1999). Mixed culture solid substrate fermentation of Trichoderma reesei with Aspergillus niger on sugar cane bagasse. Bioresour. Technol. 68 173–178. 10.1016/S0960-8524(98)00139-4 [DOI] [Google Scholar]
- Gyalai-Korpos M., Mangel R., Alvira P., Dienes D., Ballesteros M., Réczey K. (2011). Cellulase production using different streams of wheat grain-and wheat straw-based ethanol processes. J. Ind. Microbiol. Biotechnol. 38 791–802. 10.1007/s10295-010-0811-9 [DOI] [PubMed] [Google Scholar]
- Hartl L., Seiboth B. (2005). Sequential gene deletions in Hypocrea jecorina using a single blaster cassette. Curr. Genet. 48 204–211. 10.1007/s00294-005-0011-8 [DOI] [PubMed] [Google Scholar]
- Haven M. Ø., Jørgensen H. (2013). Adsorption of β-glucosidases in two commercial preparations onto pretreated biomass and lignin. Biotechnol. Biofuels 6 165 10.1186/1754-6834-6-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyotsna K. P. P., Vijayalakshmi K., Prasanna N. D., Shaheen S. K. (2010). Isolation and characterization of cellulose producing Lysinibacillus sphaericus MTCC No 9468 from gut of Eisenia Foetida. Bioscan 6 325–327. [Google Scholar]
- Kamaya Y. (1996). Role of endoglucanase in enzymatic modification of bleached kraft pulp. J. Ferm. Bioeng. 82 549–553. 10.1016/S0922-338X(97)81250-0 [DOI] [Google Scholar]
- Lee S., Lee Y. H., Park J. M., Bai D. H., Jang J. K., Park Y. S. (2014). Bioconversion of ginsenosides from red ginseng extract using candida allociferrii JNO301 isolated from Meju. Mycobiology 42 368–375. 10.5941/MYCO.2014.42.4.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tang C., Shi H., Wu M. (2011). Cloning and optimized expression of a neutral endoglucanase gene (ncel5A) from Volvariella volvacea WX32 in Pichia pastoris. J. Biosci. Bioeng. 111 537–540. 10.1016/j.jbiosc.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Liu G., Qin Y., Li Z., Qu Y. (2013). Development of highly efficient, low-cost lignocellulolytic enzyme systems in the post-genomic era. Biotechnol. Adv. 31 962–975. 10.1016/j.biotechadv.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang J., Zou G., Wang C., Zhou Z. (2011). Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzyme Microb. Technol. 49 366–371. 10.1016/j.enzmictec.2011.06.013 [DOI] [PubMed] [Google Scholar]
- Mandels M., Reese E. T. (1957). Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 73 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenecourt B. S., Eveleigh D. E. (1977). Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl. Environ. Microbiol. 34 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenecourt B. S., Eveleigh D. E. (1979). Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 181 289–301. 10.1021/ba-1979-0181.ch014 [DOI] [Google Scholar]
- Penttilä M., Nevalainen H., Rättö M., Salminen E., Knowles J. (1987). A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61 155–164. 10.1016/0378-1119(87)90110-7 [DOI] [PubMed] [Google Scholar]
- Peterson R., Nevalainen H. (2012). Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology 158 58–68. 10.1099/mic.0.054031-0 [DOI] [PubMed] [Google Scholar]
- Premalatha N., Gopal N. O., Jose P. A., Anandham R., Kwon S. W. (2015). Optimization of cellulase production by Enhydrobacter sp. ACCA2 and its application in biomass saccharification. Front. Microbiol. 6:1046 10.3389/fmicb.2015.01046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor S. W., Nahar N. (2015). β-glucosidase supplementation during biomass hydrolysis: how low can we go? Biomass Bioenergy 80 298–302. 10.1016/j.biombioe.2015.06.005 [DOI] [Google Scholar]
- Raddadi N., Cherif A., Daffonchio D., Fava F. (2013). Halo-alkalitolerant and thermostable cellulases with improved tolerance to ionic liquids and organic solvents from Paenibacillus tarimensis isolated from the Chott El Fejej, Sahara desert, Tunisia. Bioresour. Technol. 150 121–128. 10.1016/j.biortech.2013.09.089 [DOI] [PubMed] [Google Scholar]
- Rahman Z., Shida Y., Furukawa T., Suzuki Y., Okada H., Ogasawara W., et al. (2009). Application of Trichoderma reesei cellulase and xylanase promoters through homologous recombination for enhanced production of extracellular β-glucosidase I. Biosci. Biotechnol. Biochem. 73 1083–1089. 10.1271/bbb.80852 [DOI] [PubMed] [Google Scholar]
- Rosgaard L., Pedersen S., Cherry J. R., Harris P., Meyer A. S. (2006). Efficiency of new fungal cellulase systems in boosting enzymatic degradation of barley straw lignocellulose. Biotechnol. Prog. 22 493–498. 10.1021/bp050361 [DOI] [PubMed] [Google Scholar]
- Seidl V., Gamauf C., Druzhinina I. S., Seiboth B., Hartl L., Kubicek C. P. (2008). The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics 9:327 10.1186/1471-2164-9-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V., Seiboth B. (2010). Trichoderma reesei: genetic approaches to improving strain efficiency. Biofuels 1 343–354. 10.4155/bfs.10.1 [DOI] [Google Scholar]
- Singh A., Taylor L. E., Vander Wall T. A., Linger J., Himmel M. E., Podkaminer K., et al. (2015). Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol. Adv. 33 142–154. 10.1016/j.biotechadv.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Singhania R. R., Patel A. K., Sukumaran R. K., Larroche C., Pandey A. (2013). Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 127 500–507. 10.1016/j.biortech.2012.09.012 [DOI] [PubMed] [Google Scholar]
- Smith J. L., Bayliss F. T., Ward M. (1991). Sequence of the cloned pyr4 gene of Trichoderma reesei and its use as a homologous selectable marker for transformation. Curr. Genet. 19 27–33. [DOI] [PubMed] [Google Scholar]
- Song W., Han X., Qian Y., Liu G., Yao G., Zhong Y., et al. (2016). Proteomic analysis of the biomass hydrolytic potentials of Penicillium oxalicum lignocellulolytic enzyme system. Biotechnol. Biofuels 9 68 10.1186/s13068-016-0477-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen A., Lübeck M., Lübeck P. S., Ahring B. K. (2013). Fungal beta-glucosidases: a bottleneck in industrial use of lignocellulosic materials. Biomolecules 3 612–631. 10.3390/biom3030612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M. G. (2013). “Molecular tools in Trichoderma genetic studies,” in Trichoderma: Biology and Applications, eds Mukherjee P. K., Horwitz B. A., Singh U. S., Mukherjee M., Schmoll M. (Wallingford: CABI; ), 128–143. [Google Scholar]
- Stricker A. R., Grosstessner-Hain K., Würleitner E., Mach R. L. (2006). Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5 2128–2137. 10.1128/EC.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te’o V. S. J., Bergquist P. L., Nevalainen K. H. (2002). Biolistic transformation of Trichoderma reesei using the Bio-Rad seven barrels Hepta Adaptor system. J. Microbiol. Methods 51 393–399. 10.1016/S0167-7012(02)00126-4 [DOI] [PubMed] [Google Scholar]
- Teugjas H., Väljamäe P. (2013). Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol. Biofuels 6:105 10.1186/1754-6834-6-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treebupachatsakul T., Shioya K., Nakazawa H., Kawaguchi T., Morikawa Y., Shida Y., et al. (2015). Utilization of recombinant Trichoderma reesei expressing Aspergillus aculeatus β-glucosidase I (JN11) for a more economical production of ethanol from lignocellulosic biomass. J. Biosci. Bioeng. 120 657–665. 10.1016/j.jbiosc.2015.04.015 [DOI] [PubMed] [Google Scholar]
- Wang B., Xia L. (2011). High efficient expression of cellobiase gene from Aspergillus niger in the cells of Trichoderma reesei. Bioresour. Technol. 102 4568–4572. 10.1016/j.biortech.2010.12.099 [DOI] [PubMed] [Google Scholar]
- Weiss N., Börjesson J., Pedersen L. S., Meyer A. S. (2013). Enzymatic lignocellulose hydrolysis: improved cellulase productivity by insoluble solids recycling. Biotechnol. Biofuels 6:5 10.1186/1754-6834-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. J. W. T., Taylor J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protoc. 18 315–322. [Google Scholar]
- Wu G., He R., Jia W., Chao Y., Chen S. (2011). Strain improvement and process optimization of Trichoderma reesei RUT- C30 for enhanced cellulase production. Biofuels 2 545–555. 10.4155/bfs.11.124 [DOI] [Google Scholar]
- Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Domínguez Y., Scazzocchio C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41 973–981. 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhong Y., Zhao X., Wang T. (2010). Development of the cellulolytic fungus Trichoderma reesei strain with enhanced β-glucosidase and filter paper activity using strong artifical cellobiohydrolase 1 promoter. Bioresour. Technol. 101 9815–9818. 10.1016/j.biortech.2010.07.078 [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang L., Liu D. (2008). Comparative study on chemical pretreatment methods for improving enzymatic digestibility of crofton weed stem. Bioresour. Technol. 99 3729–3736. 10.1016/j.biortech.2007.07.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Phylogenetic tree of strain SN1 based on the 18S rDNA gene sequence and internally transcribed spacer (ITS) region. (A) 18S rDNA sequences. The 18s rDNA gene sequences from Trichoderma reesei (XM_006965206.1), T. sp (AB923813.1), T. harzianum (KP133166.1), T. viride (AF218788), Penicillium oxalicun (JQ349066.1), Aspergillus niger (AM270052.1), Cladonia arbuscula (KT148631.1). (B) ITS region sequences, the ITS region sequences from T. reesei (KT336514.1), T. harzianum (KR232487.1), T. viride (DQ442274.1), T. saturnisporum (KF294853.1), T. asperelloides (KT426897.1), T. pseudokoningii (AF140046), P. oxalicun (KT189516.1), A. niger (KM613139.1).
FIGURE S2 | Mycelium morphology of SN1, QM9414, and RUT-C30. All strains were cultivated 2–5 days on PDA plates (glucose) to analyze the microscopic morphology of mycelium and spore. The images were captured using DIC. Scale bars are listed 20 μm in the figure.