Abstract
Here, we report the case of an acute promyelocytic leukemia (APL) patient who—although negative for FLT3 mutations at diagnosis—developed isolated FLT3 tyrosine kinase II domain (FLT3-TKD)-positive meningeal relapse, which, in retrospect, could be traced back to a minute bone marrow subclone present at first diagnosis. Initially, the 48-yr-old female diagnosed with high-risk APL had achieved complete molecular remission after standard treatment with all-trans retinoic acid (ATRA) and chemotherapy according to the AIDA (ATRA plus idarubicin) protocol. Thirteen months after the start of ATRA maintenance, the patient suffered clinically overt meningeal relapse along with minute molecular traces of PML/RARA (promyelocytic leukemia/retinoic acid receptor alpha) in the bone marrow. Following treatment with arsenic trioxide and ATRA in combination with intrathecal cytarabine and methotrexate, the patient achieved a complete molecular remission in both cerebrospinal fluid (CSF) and bone marrow, which currently lasts for 2 yr after completion of therapy. Whole-exome sequencing and subsequent ultradeep targeted resequencing revealed a heterozygous FLT3-TKD mutation in CSF leukemic cells (p.D835Y, c.2503G>T, 1000/1961 reads [51%]), which was undetectable in the concurrent bone marrow sample. Interestingly, the FLT3-TKD mutated meningeal clone originated from a small bone marrow subclone present in a variant allele frequency of 0.4% (6/1553 reads) at initial diagnosis. This case highlights the concept of clonal evolution with a subclone harboring an additional mutation being selected as the “fittest” and leading to meningeal relapse. It also further supports earlier suggestions that FLT3 mutations may play a role for migration and clonal expansion in the CSF sanctuary site.
Keywords: leukemia
INTRODUCTION
Acute promyelocytic leukemia (APL) is a distinct subset of acute myeloid leukemia (AML), associated with the chromosomal translocation t(15;17)(q22;q12), resulting in PML-RARA gene fusion and chimeric protein, which interferes with the maturation process of myeloid cells (Grimwade et al. 2010). Clinical trials report long-term disease-free survival in ∼85% of patients treated with all-trans retinoic acid (ATRA) and anthracycline-based chemotherapy (Lo-Coco et al. 2016). However, ∼10% of patients will suffer relapse. Recently, the therapeutic spectrum has been widened by the introduction of arsenic trioxide (ATO), which in combination with ATRA produced 2-yr event-free survival rates of 97% in patients with low-to-intermediate-risk APL (Lo-Coco et al. 2013).
The Sanz score, which is based on peripheral leukocyte and platelet counts at presentation, forms the mainstay for risk stratification in APL (Sanz et al. 2000). Other factors that have been associated with an adverse outcome include CD56 expression and a promoter polymorphism of the FAS (also known as CD95) gene (Montesinos et al. 2011; Sunter et al. 2012).
Internal tandem duplications of the FLT3 gene (FLT3-ITD) and point mutations of the FLT3 tyrosine kinase II domain (FLT3-TKD), typically at Asp 835, are detected in 15%–40% and 10%–20% of APL cases, respectively (Noguera et al. 2002; Arrigoni et al. 2003; Au et al. 2004; Callens et al. 2005; Gale et al. 2005; Kuchenbauer et al. 2005; Chillon et al. 2010; Barragan et al. 2011; Schnittger et al. 2011; Welch et al. 2012; Shen et al. 2015). In APL, FLT3-ITD mutations are associated with leukocytosis at presentation but did not independently correlate with response to treatment or survival (Noguera et al. 2002; Au et al. 2004; Gale et al. 2005; Kutny et al. 2012; Breccia et al. 2013; Poire et al. 2014; Shen et al. 2015). No distinct clinical phenotype or consistent impact on prognosis has been identified for FLT3-TKD mutations in APL so far, perhaps because of their relative rarity (Shih et al. 2003; Barragan et al. 2011; Schnittger et al. 2011; Kutny et al. 2012).
Similar to other types of AML, extramedullary manifestations are rare in APL. However, extramedullary relapses have been reported with increasing frequency during the last 20 years, corresponding to the time when ATRA, which penetrates the central nervous system (CNS) poorly, became the mainstay of APL treatment (Byrd et al. 1995; Bakst et al. 2011). Approximately 3%–5% of APL patients will experience extramedullary relapse, most commonly in the CNS, with CNS relapse being either isolated or associated with bone marrow relapse (de Botton et al. 2006; Albano and Specchia 2011; Wang et al. 2014). Although high white blood cell counts at diagnosis and prior CNS hemorrhage are established risk factors for CNS relapse, the significance of increased age, differentiation syndrome, CD56 expression, and FLT3-ITD positivity remains unclear (Montesinos et al. 2009; Tashiro et al. 2011; Colovic et al. 2012).
RESULTS
Clinical Presentation and Treatment Outcome
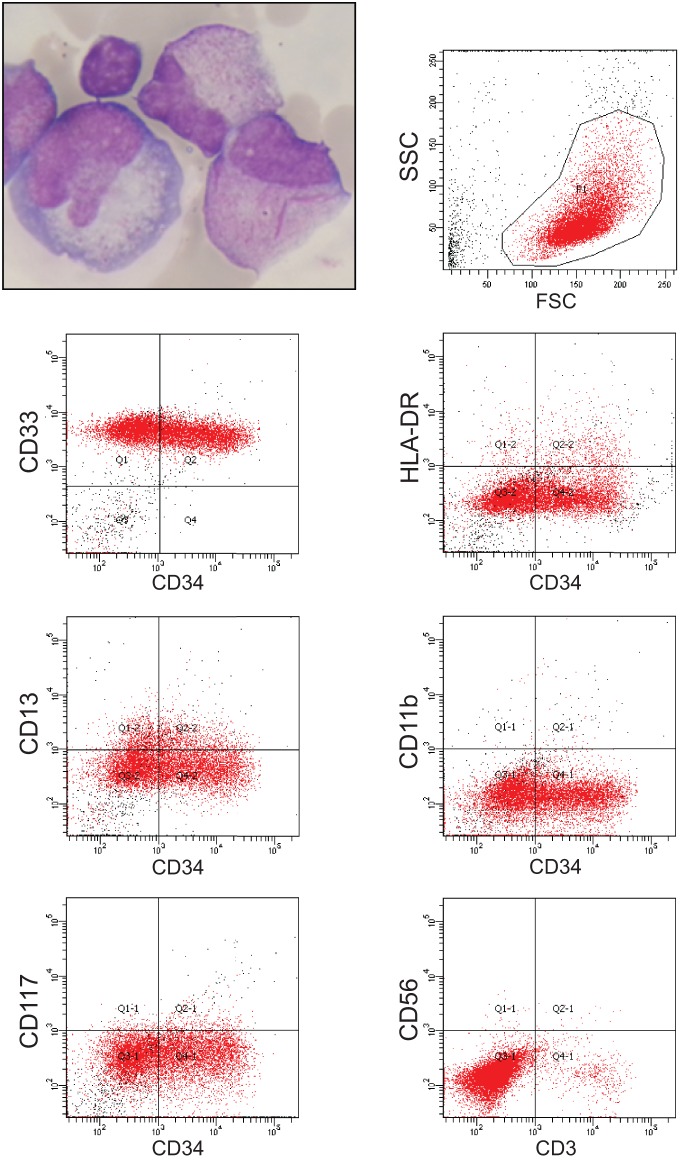
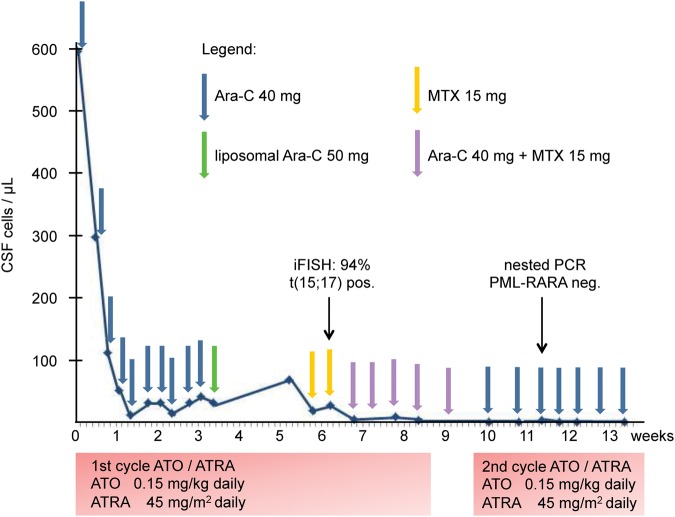
The 48-yr-old female patient was diagnosed with high-risk APL in January 2012. After treatment according to the high-risk arm of the AIDA (ATRA plus idarubicin) protocol with ATRA and chemotherapy (Lo-Coco et al. 2010), complete molecular remission was achieved by the end of consolidation chemotherapy. Subsequently, maintenance therapy with ATRA was administered. In August 2013, 13 mo after initiation of maintenance, the patient developed meningeal relapse with pleocytosis of 597/µL in the cerebrospinal fluid (CSF) (Fig. 1), which caused headaches and paresthesia of the arms. Meningeal relapse was confirmed by immunophenotyping (CD33+, CD13+/−, CD34+/−, CD11b−, HLA-DR−, CD117−, CD56−; Fig. 1), interphase fluorescence in situ hybridization, PML-RARα (promyelocytic leukemia/retinoic acid receptor-α) reverse transcription–polymerase chain reaction (RT-PCR), and brain magnetic resonance imaging. At the time of meningeal relapse, the bone marrow continued to be in complete hematologic remission, although minimal residual disease (MRD) testing was positive for PML-RARA by nested RT-PCR without quantifiable load. The patient was treated with intrathecal applications of cytarabine and methotrexate and received ATO/ATRA according to the APL0406 protocol (Lo-Coco et al. 2013), with the first cycle given over 2 mo and intervals between consolidation cycles shortened to 1 wk to increase dose density. At Day 26 of ATO/ATRA induction, elemental arsenic levels in peripheral blood and CSF were 114 µg/L and 13 µg/L as measured by inductively coupled plasma mass spectrometry. Following neutrophilic differentiation of meningeal APL blasts by Day 42, CSF cell counts normalized by week 8 and complete molecular remission was achieved by week 11 (Fig. 2). The patient continued on ATO/ATRA until February 2014 and on intrathecal chemotherapy until April 2014. For consolidation, the patient was recommended autologous hematopoietic stem cell transplantation with busulfan-based conditioning but opted for a watch-and-wait strategy instead. As of March 2016, with 2 yr of follow-up after the end of salvage treatment, the patient continues to be in complete molecular remission in bone marrow and CSF alike.
Figure 1.
Clinical manifestations of meningeal APL relapse with Giemsa staining (upper left panel) and fluorescence-activated cell sorting analysis of CD33, CD34, HLA-DR, CD13, CD11b, CD117, CD56, and CD3 expression from cerebrospinal fluid blasts at relapse. Cells were gated in the side scatter (SSC)/forward scatter (FSC) analysis as shown in the upper right panel.
Figure 2.
Treatment and course of cerebrospinal fluid (CSF) cell counts during 3 mo following relapse. The normalization of the CSF cell count sets in with delay. Ara-C, cytosine arabinoside; MTX, methotrexate; PCR, polymerase chain reaction; PML, promyelocytic leukemia; RARA, retinoic acid receptor-α; iFISH, interphase fluorescence in situ hybridization; ATO, arsenic trioxide; ATRA, all-trans retinoic acid.
Genomic Analysis
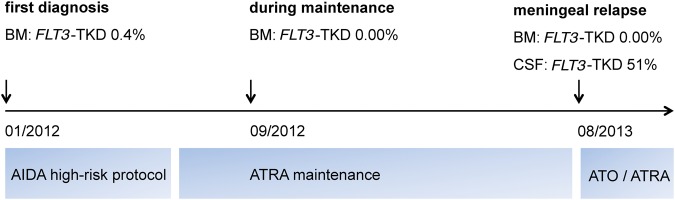
To identify molecular drivers of meningeal relapse, we performed whole-exome sequencing of CSF-derived leukemic cells within NCT MASTER (Molecularly Aided Stratification for Tumor Eradication Research), a clinical sequencing program for advanced-stage cancer across all histologies, which identified a FLT3-TKD mutation (p.D835Y, c.2503G>T) (Table 1). Ultradeep targeted resequencing revealed the heterozygous presence of this mutation, which had not been found at first diagnosis, in virtually all CSF cells (51%, 1000/1961 reads). Of note, the mutation was undetectable (0/1666 reads) in the corresponding MRD-positive bone marrow sample drawn concurrently at meningeal relapse. Next, ultradeep targeted sequencing was performed to trace back the meningeal relapse clone to bone marrow samples obtained during the course of the disease. Interestingly, a small subclone comprising 0.4% (6/1553 reads) of bone marrow cells at first diagnosis already harbored the FLT3-TKD mutation, whereas it was undetectable in the bone marrow at all other time points—that is, during ATRA maintenance (0/874 reads), at meningeal relapse (0/1666 reads), and after the first cycle of ATO/ATRA (0/1198 reads) (Fig. 3).
Table 1.
Somatic mutations in CSF-derived leukemic cells identified by whole-exome sequencing
| Chromosome | Position | Gene | RefSeq accession | Exon | Nucleotide change | VAF | Protein change |
|---|---|---|---|---|---|---|---|
| SNVs | |||||||
| 1q22 | 155,931,985 | ARHGEF2 | NM_001162383 | 10 | c.G1130T | 0.6 | p.R377L |
| 3q25.32 | 158,449,993 | RARRES1 | NM_002888 | 1 | c.A212G | 0.33 | p.N71S |
| 5q31.3 | 140,215,358 | PCDHA7 | NM_018910 | 1 | c.G1390A | 0.35 | p.V464M |
| 9q21.2 | 79,320,582 | PRUNE2 | NM_015225 | 8 | c.T6608A | 0.43 | p.L2203X |
| 9q33.1 | 118,974,010 | PAPPA | NM_002581 | 4 | c.G1717A | 0.32 | p.V573I |
| 11p15.1 | 21,135,209 | NELL1 | NM_006157 | 13 | c.C1375T | 0.57 | p.R459C |
| 13q12.2 | 28,592,642 | FLT3 | NM_004119 | 20 | c.G2503T | 0.4 | p.D835Y |
| 13q14.3 | 52,678,680 | NEK5 | NM_199289 | 9 | c.G563T | 0.38 | p.W188L |
| 14q23.2 | 62,194,324 | HIF1A | NM_001243084 | 6 | c.T796G | 0.36 | p.F266V |
| 16q24.2 | 87,678,376 | JPH3 | NM_020655 | 2 | c.G895A | 0.55 | p.G299S |
| 19p13.2 | 10,091,528 | COL5A3 | NM_015719 | 34 | c.G2513A | 0.5 | p.R838H |
| 20p11.23 | 18,474,662 | RBBP9 | NM_006606 | 3 | c.T188C | 0.49 | p.L63P |
| Xq24 | 117,676,802 | DOCK11 | NM_144658 | 2 | c.T217C | 0.43 | p.S73P |
| Indels | |||||||
| 8p22 | 17,067,557 | ZDHHC2 | NM_016353 | 9 | c.848dupT | 0.5 | p.I283fs |
| 8q24.21 | 128,750,873 | MYC | NM_002467 | 2 | c.411_412insGACCTTCTG | 0.34 | p.E137delinsEDLL |
| 14q32.32 | 103,592,943 | TNFAIP2 | NM_006291 | 1 | c.150_152del | 0.12 | p.50_51del |
| 15q26.1 | 90,378,824 | AP3S2 | NM_001199058 | 10 | c.1107delC | 0.49 | p.I369fs |
CSF, cerebrospinal fluid; SNVs, single-nucleotide variants; Indels, insertion/deletion mutations; VAF, variant allele frequency.
Figure 3.
FLT3-TKD subclone architecture based on ultradeep sequencing results, inserted into the time line of the disease course. A heterozygous FLT3-TKD mutation was present in virtually all cerebrospinal fluid (CSF) cells at meningeal relapse. Interestingly, the mutation could be traced back to a tiny FLT3-TKD clone present in the bone marrow sample from first diagnosis. BM, bone marrow; AIDA, ATRA plus idarubicin; ATRA, all-trans retinoic acid; ATO, arsenic trioxide.
DISCUSSION
From a clinical perspective, this case corroborates the effectiveness of ATO as a relapse treatment and, because of its permeability across the blood–brain barrier (Au et al. 2006, 2008), as a treatment for meningeal disease. ATO has been described to exert dose-dependent dual effects on APL cells, inducing preferentially apoptosis at high concentrations and differentiation at low concentrations (Chen et al. 1997). ATO significantly penetrates into the CSF, leading to CSF arsenic concentrations of ∼20% of the respective plasma levels, with considerable variation between individual patients (Au et al. 2006, 2008; Helwig et al. 2007). In our patient, the CSF arsenic concentration reached only 11% of the respective plasma concentration at day 26 of induction treatment, explaining the delayed differentiation effect that became detectable only after several weeks and cautioning us not to stop ATO-based protocols prematurely because of a perceived lack of efficacy.
From a molecular perspective, this case highlights the concept of genetic heterogeneity and selection of the “fittest” subclone as a driving mechanism of leukemia progression and relapse. This concept has recently been illustrated by whole-genome sequencing on paired diagnostic and relapse samples from patients with normal karyotype AML, establishing that relapse clones can originate from a minor subclone at initial diagnosis (Ding et al. 2012). Our case corroborates and extends this concept of clonal evolution as pathogenetic mechanism in AML.
Similar to normal karyotype AML, both FLT3-ITD and FLT3-TKD mutations were recurrently found as cooperating mutations in APL (Au et al. 2004; Callens et al. 2005; Gale et al. 2005; Kuchenbauer et al. 2005; Chillon et al. 2010; Barragan et al. 2011; Schnittger et al. 2011; Welch et al. 2012; Shen et al. 2015). Several FLT3-mutant APL cases with extramedullary relapse manifestations have been published (Housman et al. 2010; Tashiro et al. 2011; Colovic et al. 2012; Zou et al. 2013; Gill et al. 2015; Liu et al. 2015). In the majority of these cases, FLT3 mutational analysis has only been performed in bone marrow or peripheral blood samples and either at initial diagnosis or relapse, respectively. Interestingly, however, in one patient, a FLT3-ITD mutation was detected at the time of combined CNS and bone marrow relapse, whereas the bone marrow at initial APL diagnosis was FLT3-ITD-negative (Tashiro et al. 2011). Additionally, a case of meningeal involvement in second relapse of APL was reported recently, which harbored a FLT3-ITD mutation, whereas the concurrently drawn bone marrow sample was FLT3-ITD-negative (Gill et al. 2015). Analogous to our case, the FLT3-ITD-positive meningeal clone could be traced back to a minor, although not quantified, subclone in the bone marrow at first relapse.
Recently, it was shown that, analogous to normal hematopoietic cells, the level of C-X-C chemokine receptor 4 (CXCR4) expression on AML cells correlates with stromal cell-derived factor-1 (SDF1, encoded by CXCL12)-induced chemotaxis and poor prognosis (Rombouts et al. 2004). Importantly, CXCR4 expression was significantly higher in FLT3-ITD mutated when compared with FLT3 wild-type AML, and cells expressing constitutively active FLT3-ITD displayed dramatically enhanced migration toward SDF1/CXCL12 (Fukuda et al. 2005). In addition, SDF1/CXCL12 is highly expressed on the vasculature of the blood–brain barrier where it regulates the entry of leukocytes into the CNS (Williams et al. 2014). In conclusion, it appears that FLT3 mutations favor migration and seeding of APL cells to the CSF sanctuary. The mechanisms of clonal selection in this case are closely intertwined with the biology of this privileged CSF site.
APL cases with meningeal involvement are rare, thereby precluding more systematic analyses on the role of specific mutations for extramedullary disease manifestations. Nevertheless, the above cases, together with the data reported here, strongly suggest a role for FLT3 mutations in establishment and expansion of APL CNS manifestations.
METHODS
Isolation of DNA
Tissue samples were provided in accordance with the regulations of the National Center for Tumor Diseases (NCT) Heidelberg Tissue Bank and the approval of the Ethics Committee of Heidelberg University. DNA from APL cells and DNA from the normal blood sample were isolated using the AllPrep DNA/RNA/Protein Mini Kit (QIAGEN), followed by quality control and quantification using a Qubit 2.0 Fluorometer (Life Technologies), a 2200 TapeStation system (Agilent), and a 2100 Bioanalyzer system (Agilent).
Whole-Exome Sequencing
High-throughput sequencing and data analysis were performed as described (Kordes et al. 2016). In brief, exome capturing was performed using SureSelect Human All Exon V5+UTRs in-solution capture reagents (Agilent). Briefly, 1.5 µg genomic DNA were fragmented to 150–200 bp (paired-end) insert size with a Covaris S2 device, and 250 ng of Illumina adapter-containing libraries were hybridized with exome baits at 65°C for 16 h. Paired-end sequencing (101 bp) was performed with a HiSeq 2500 instrument (Illumina) in rapid mode.
Mapping and Analysis of Whole-Exome Sequencing Data
Reads were mapped to the 1000 Genomes Phase 2 assembly of the human reference genome (NCBI build 37.1) using BWA (version 0.6.2) with default parameters and maximum insert size set to 1000 bp (Li and Durbin 2009). BAM files were sorted with SAMtools (version 0.1.19) (Li et al. 2009), and duplicates were marked with Picard tools (version 1.90). Average target coverage was 116.13× for the CSF leukemia and 113.41× for the bone marrow control. In both, >80% of the targets had a coverage of at least 40× (see Table 2).
Table 2.
Sequencing coverage for the leukemic CSF cells and the bone marrow control
| Sample | Percentage of reads aligned | Average read coverage | Number of CCDS r15 sites with ≥10-fold coverage | Percentage of CCDS r15 with ≥10-fold coverage | Percentage of CCDS r15 with ≥20-fold coverage | Percentage of FLT3 sites (2958 bases) with ≥10-fold coverage |
|---|---|---|---|---|---|---|
| Bone marrow | 99.65% | 113.41 | 46,754,560 | 94.78% | 89.84% | 98.72% |
| CSF blasts | 99.64% | 116.13 | 46,758,736 | 94.79% | 89.84% | 97.73% |
The 49 Mbp of consensus coding sequence (CCDS release 15, ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/archive/15/CCDS.current.txt dated August 3, 2014) is used as the reference for protein-coding sequence coverage. The CCDS release was downloaded and converted into a BED file containing the exon coordinates with a custom Perl script. Only genes with ccds_status “Public” were considered. Using SAMtools version 0.1.19 (Li et al. 2009) view and coverageBed v2.16.2 (Quinlan and Hall 2010), the read number per site was determined for nonduplicate reads with a minimal mapping quality of 1.
CSF, cerebrospinal fluid; CCDS, Consensus Coding DNA Sequence.
For the detection of single-nucleotide variants, we applied our in-house analysis pipeline based on SAMtools mpileup and BCFtools with parameter adjustments to allow for calling of somatic variants with heuristic filtering as previously described (Jones et al. 2012, 2013; Yaktapour et al. 2014). After annotation with RefSeq (version September 2013) using ANNOVAR (Wang et al. 2010), somatic, nonsilent coding variants of high confidence were selected.
Ultradeep Targeted Resequencing
For the FLT3-D835Y-TKD mutation, ultradeep targeted resequencing was performed.
Library Preparation and Semiconductor Sequencing
For library preparation, the multiplex PCR-based Ion Torrent AmpliSeq technology (Life Technologies) with the Cancer HotSpot Panel v2 (IonTorrent/Thermo Fisher Scientific) was used. Amplicon library preparation was performed with the Ion AmpliSeq Library Kit v2.0 using ∼10 ng of DNA. Briefly, the DNA was mixed with the primer pool, containing all primers for generating the 207 amplicons and the AmpliSeq HiFi Master Mix and transferred to a PCR cycler (Bio-Rad). After the end of the PCR, primer end sequences were partially digested using FuPa reagent, followed by the ligation of barcoded sequencing adapters (Ion Xpress Barcode Adapters, Life Technologies). The final library was purified using AMPure XP magnetic beads (Beckman Coulter) and quantified using qPCR (Ion Library Quantitation Kit, Thermo Fisher Scientific) on a StepOne qPCR machine (Thermo Fisher Scientific). The individual libraries were diluted to a final concentration of 100 pM and eight to 10 libraries were pooled and processed to library amplification on Ion Spheres using Ion PGM Template OT2 200 Kit. Unenriched libraries were quality-controlled using Ion Sphere quality control measurement on a QuBit instrument. After library enrichment (Ion OneTouch ES), the library was processed for sequencing using the Ion Torrent 200 bp sequencing v2 chemistry and the barcoded libraries were loaded onto a chip. Our way of pooling eight samples on a 318 chip resulted in a mean coverage of 3000-fold per amplicon.
Variant Calling and Annotation
Data analysis was performed using the Ion Torrent Suite Software (version 4.4). After base calling, the reads were aligned against the human genome (hg19) using the TMAP algorithm within the Torrent Suite. Variant calling was performed with the variant caller plugin within the Torrent Suite Software and the IonReporter package using a corresponding BED file containing the coordinates of the amplified regions. Only variants with an allele frequency >5% and minimum coverage >100 reads were taken into account. Variant annotation was performed using ANNOVAR (Wang et al. 2010). Annotations included information about nucleotide and amino acid changes of RefSeq annotated genes, COSMIC and dbSNP entries, and detection of possible splice site mutations. For data interpretation and verification, the aligned reads were visualized using the IGV browser (Broad Institute) (Robinson et al. 2011).
ADDITIONAL INFORMATION
Data Deposition and Access
The raw analytical data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/datasets), accession number EGAS00001001848.
Ethics Statement
APL cells and a matched normal blood sample were obtained following written informed consent under an institutional review board–approved protocol covering all aspects relevant to clinical cancer genome sequencing. This study was conducted in accordance with the Declaration of Helsinki. The patient also granted written consent to the publication of her case in Molecular Case Studies.
Author Contributions
T.B. and A.K. designed the research. T.B., S.F., W.W., B.H., M.H., and V.E. performed the research and analyzed the data. A.D.H. contributed to clinical data collection. C.T. contributed to sample preparation. T.B. and A.K. wrote the manuscript. S.F. and W.W. edited the manuscript. All authors made significant contributions to, reviewed, and approved the final version of the manuscript.
Funding
This work was supported by grants H021 and H001 from the German Cancer Research Center (DKFZ)-Heldelberg Center for Personalized Oncology (HIPO) (S.F., A.K.).
Competing Interest Statement
T.B. and A.K. have received travel grants from Teva, the manufacturer of arsenic trioxide (ATO).
REFERENCES
- Albano F, Specchia G. 2011. Extramedullary disease in acute promyelocytic leukemia: two-in-one disease. Mediterr J Hematol Infect Dis 3: e2011066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni P, Beretta C, Silvestri D, Rossi V, Rizzari C, Valsecchi MG, Cazzaniga G, Biondi A. 2003. FLT3 internal tandem duplication in childhood acute myeloid leukaemia: association with hyperleukocytosis in acute promyelocytic leukaemia. Br J Haematol 120: 89–92. [DOI] [PubMed] [Google Scholar]
- Au WY, Fung A, Chim CS, Lie AK, Liang R, Ma ES, Chan CH, Wong KF, Kwong YL. 2004. FLT-3 aberrations in acute promyelocytic leukaemia: clinicopathological associations and prognostic impact. Br J Haematol 125: 463–469. [DOI] [PubMed] [Google Scholar]
- Au WY, Tam S, Fong BM, Kwong YL. 2006. Elemental arsenic entered the cerebrospinal fluid during oral arsenic trioxide treatment of meningeal relapse of acute promyelocytic leukemia. Blood 107: 3012–3013. [DOI] [PubMed] [Google Scholar]
- Au WY, Tam S, Fong BM, Kwong YL. 2008. Determinants of cerebrospinal fluid arsenic concentration in patients with acute promyelocytic leukemia on oral arsenic trioxide therapy. Blood 112: 3587–3590. [DOI] [PubMed] [Google Scholar]
- Bakst RL, Tallman MS, Douer D, Yahalom J. 2011. How I treat extramedullary acute myeloid leukemia. Blood 118: 3785–3793. [DOI] [PubMed] [Google Scholar]
- Barragan E, Montesinos P, Camos M, González M, Calasanz MJ, Román-Gómez J, Gómez-Casares MT, Ayala R, López J, Fuster Ó, et al. 2011. Prognostic value of FLT3 mutations in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline monochemotherapy. Haematologica 96: 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breccia M, Loglisci G, Loglisci MG, Ricci R, Diverio D, Latagliata R, Foà R, Lo-Coco F. 2013. FLT3-ITD confers poor prognosis in patients with acute promyelocytic leukemia treated with AIDA protocols: long-term follow-up analysis. Haematologica 98: e161–e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Edenfield WJ, Shields DJ, Dawson NA. 1995. Extramedullary myeloid cell tumors in acute nonlymphocytic leukemia: a clinical review. J Clin Oncol 13: 1800–1816. [DOI] [PubMed] [Google Scholar]
- Callens C, Chevret S, Cayuela JM, Cassinat B, Raffoux E, de Botton S, Thomas X, Guerci A, Fegueux N, Pigneux A, et al. 2005. Prognostic implication of FLT3 and Ras gene mutations in patients with acute promyelocytic leukemia (APL): a retrospective study from the European APL Group. Leukemia 19: 1153–1160. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Shi XG, Tang W, Xiong SM, Zhu J, Cai X, Han ZG, Ni JH, Shi GY, Jia PM, et al. 1997. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): I. As2O3 exerts dose-dependent dual effects on APL cells. Blood 89: 3345–3353. [PubMed] [Google Scholar]
- Chillon MC, Santamaria C, Garcia-Sanz R, Balanzategui A, Sarasquete ME, Alcoceba M, Martin L, Caballero MD, Vidriales MB, Ramos F, et al. 2010. Long FLT3 internal tandem duplications and reduced PML-RARα expression at diagnosis characterize a high-risk subgroup of acute promyelocytic leukemia patients. Haematologica 95: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovic N, Tomin D, Vidovic A, Tosic N, Atkinson HD, Colovic MD. 2012. Central nervous system relapse in CD56+, FLT3/ITD+ promyelocytic leukemia. Med Oncol 29: 260–262. [DOI] [PubMed] [Google Scholar]
- de Botton S, Sanz MA, Chevret S, Dombret H, Martin G, Thomas X, Mediavilla JD, Recher C, Ades L, Quesnel B, et al. 2006. Extramedullary relapse in acute promyelocytic leukemia treated with all-trans retinoic acid and chemotherapy. Leukemia 20: 35–41. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Broxmeyer HE, Pelus LM. 2005. Flt3 ligand and the Flt3 receptor regulate hematopoietic cell migration by modulating the SDF-1α (CXCL12)/CXCR4 axis. Blood 105: 3117–3126. [DOI] [PubMed] [Google Scholar]
- Gale RE, Hills R, Pizzey AR, Kottaridis PD, Swirsky D, Gilkes AF, Nugent E, Mills KI, Wheatley K, Solomon E, et al. 2005. Relationship between FLT3 mutation status, biologic characteristics, and response to targeted therapy in acute promyelocytic leukemia. Blood 106: 3768–3776. [DOI] [PubMed] [Google Scholar]
- Gill H, Ip HW, Pang AW, Sum J, Leung AY, Kwong YL. 2015. FLT3 internal tandem duplication in acute promyelocytic leukemia: central nervous system relapse. Ann Hematol 94: 1049–1051. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Mistry AR, Solomon E, Guidez F. 2010. Acute promyelocytic leukemia: a paradigm for differentiation therapy. Cancer Treat Res 145: 219–235. [DOI] [PubMed] [Google Scholar]
- Helwig A, Klemm M, Schuttig R, Rollig C, Wassilew N, Ehninger G, Illmer T. 2007. Arsenic-induced APL differentiation in cerebrospinal fluid. Leuk Res 31: 703–705. [DOI] [PubMed] [Google Scholar]
- Housman E, Chang P, Lane SW, Blinder R, Galinsky I, Kesari S, Ho VT, Stone RM, Mullally A. 2010. CNS relapse in acute promyelocytic leukemia. J Clin Oncol 28: e409–e411. [DOI] [PubMed] [Google Scholar]
- Jones DTW, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stütz AM, et al. 2012. Dissecting the genomic complexity underlying medulloblastoma. Nature 488: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DTW, Hutter B, Jäger N, Korshunov A, Kool M, Warnatz H-J, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. 2013. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes M, Röring M, Heining C, Braun S, Hutter B, Richter D, Geörg C, Scholl C, Gröschel S, Roth W, et al. 2016. Cooperation of BRAFF595L and HRAS in histiocytic sarcoma provides new insights into oncogenic BRAF signaling. Leukemia 30: 937–946. [DOI] [PubMed] [Google Scholar]
- Kuchenbauer F, Schoch C, Kern W, Hiddemann W, Haferlach T, Schnittger S. 2005. Impact of FLT3 mutations and promyelocytic leukaemia-breakpoint on clinical characteristics and prognosis in acute promyelocytic leukaemia. Br J Haematol 130: 196–202. [DOI] [PubMed] [Google Scholar]
- Kutny MA, Moser BK, Laumann K, Feusner JH, Gamis A, Gregory J, Larson RA, Powell BL, Stock W, Willman CL, et al. 2012. FLT3 mutation status is a predictor of early death in pediatric acute promyelocytic leukemia: a report from the Children's Oncology Group. Pediatr Blood Cancer 59: 662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Zeng K, Wang L, Liu T, Niu T. 2015. Acute promyelocytic leukemia harbouring rare FLT3-TKD and WT1 mutations: a case report. Oncol Lett 10: 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, Breccia M, Gallo E, Rambaldi A, Paoloni F, Fioritoni G, Ferrara F, Specchia G, et al. 2010. Front-line treatment of acute promyelocytic leukemia with AIDA induction followed by risk-adapted consolidation for adults younger than 61 years: results of the AIDA-2000 trial of the GIMEMA Group. Blood 116: 3171–3179. [DOI] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, et al. 2013. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med 369: 111–121. [DOI] [PubMed] [Google Scholar]
- Lo-Coco F, Cicconi L, Breccia M. 2016. Current standard treatment of adult acute promyelocytic leukaemia. Br J Haematol 172: 841–854. [DOI] [PubMed] [Google Scholar]
- Montesinos P, Diaz-Mediavilla J, Deben G, Prates V, Tormo M, Rubio V, Pérez I, Fernández I, Viguria M, Rayón C, et al. 2009. Central nervous system involvement at first relapse in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline monochemotherapy without intrathecal prophylaxis. Haematologica 94: 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos P, Rayon C, Vellenga E, Brunet S, Gonzalez J, Gonzalez M, Holowiecka A, Esteve J, Bergua J, González JD, et al. 2011. Clinical significance of CD56 expression in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline-based regimens. Blood 117: 1799–1805. [DOI] [PubMed] [Google Scholar]
- Noguera NI, Breccia M, Divona M, Diverio D, Costa V, De Santis S, Avvisati G, Pinazzi MB, Petti MC, Mandelli F, et al. 2002. Alterations of the FLT3 gene in acute promyelocytic leukemia: association with diagnostic characteristics and analysis of clinical outcome in patients treated with the Italian AIDA protocol. Leukemia 16: 2185–2189. [DOI] [PubMed] [Google Scholar]
- Poire X, Moser BK, Gallagher RE, Laumann K, Bloomfield CD, Powell BL, Koval G, Gulati K, Holowka N, Larson RA, et al. 2014. Arsenic trioxide in front-line therapy of acute promyelocytic leukemia (C9710): prognostic significance of FLT3 mutations and complex karyotype. Leuk Lymphoma 55: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts EJ, Pavic B, Löwenberg B, Ploemacher RE. 2004. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult myeloid leukemia. Blood 104: 550–557. [DOI] [PubMed] [Google Scholar]
- Sanz MA, Lo-Coco F, Martin G, Avvisati G, Rayon C, Barbui T, Díaz-Mediavilla J, Fioritoni G, González JD, Liso V, et al. 2000. Definition of relapse risk and role of nonanthracycline drugs for consolidation in patients with acute promyelocytic leukemia: a joint study of the PETHEMA and GIMEMA cooperative groups. Blood 96: 1247–1253. [PubMed] [Google Scholar]
- Schnittger S, Bacher U, Haferlach C, Kern W, Alpermann T, Haferlach T. 2011. Clinical impact of FLT3 mutation load in acute promyelocytic leukemia with T(15;17)/PML-RARA. Haematologica 96: 1799–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Fu YK, Zhu YM, Lou YJ, Gu ZH, Shi JY, Chen B, Chen C, Zhu HH, Hu J, et al. 2015. Mutations of epigenetic modifier genes as a poor prognostic factor in acute promyelocytic leukemia under treatment with all-trans retinoic acid and arsenic trioxide. EBioMedicine 2: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih LY, Kuo MC, Liang DC, Huang CF, Lin TL, Wu JH, Wang PN, Dunn P, Lai CL. 2003. Internal tandem duplication and Asp835 mutations of the FMS-like tyrosine kinase 3 (FLT3) gene in acute promyelocytic leukemia. Cancer 98: 1206–1216. [DOI] [PubMed] [Google Scholar]
- Sunter NJ, Scott K, Hills R, Grimwade D, Taylor S, Worrillow LJ, Fordham SE, Forster VJ, Jackson G, Bomken S, et al. 2012. A functional variant in the core promoter of the CD95 cell death receptor gene predicts prognosis in acute promyelocytic leukemia. Blood 119: 196–205. [DOI] [PubMed] [Google Scholar]
- Tashiro H, Shirasaki R, Oka Y, Sugao T, Mizutani-Nogushi M, Yamamoto T, Akiyama N, Kawasugi K, Shirafuji N. 2011. FLT3 internal tandem duplication is associated with a high relapse rate and central nervous system involvement in acute promyelocytic leukemia cases: single institutional analysis. Eur J Haematol 86: 272–273. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao F, Li J, Li L, Li Y, Shi C, Lan W, Li D, Zhao H, Zhang Y, et al. 2014. Arsenic trioxide and mannitol for the treatment of acute promyelocytic leukemia relapse in the central nervous system. Blood 124: 1998–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Holman DW, Klein RS. 2014. Chemokines in the balance: maintenance of homeostasis and protection at CNS barriers. Front Cell Neurosci 8: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaktapour N, Meiss F, Mastroianni J, Zenz T, Andrlova H, Mathew NR, Claus R, Hutter B, Fröhling S, Brors B, et al. 2014. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest 124: 5074–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou XL, Zeng K, Xie LP, Wang L, Chen M, Liu T, Niu T. 2013. Acute promyelocytic leukemia with Flt3-TKD and WT1 mutations relapsing in a testicle and followed by systemic relapse. Acta Haematol 130: 223–229. [DOI] [PubMed] [Google Scholar]