Here, Signer et al. investigated the mechanism underlying the limited rate of protein synthesis in hematopoietic stem cells (HSCs). The authors found that adult HSCs had more hypophosphorylated eukaryotic initiation factor 4E-binding protein1 (4E-BP1) and 4E-BP2 as compared with most other hematopoietic progenitors, providing new insight into the mechanism by which HSCs attenuate protein synthesis.
Keywords: 4E-BP, protein synthesis, stem cell
Abstract
Adult stem cells must limit their rate of protein synthesis, but the underlying mechanisms remain largely unexplored. Differences in protein synthesis among hematopoietic stem cells (HSCs) and progenitor cells did not correlate with differences in proteasome activity, total RNA content, mRNA content, or cell division rate. However, adult HSCs had more hypophosphorylated eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and 4E-BP2 as compared with most other hematopoietic progenitors. Deficiency for 4E-BP1 and 4E-BP2 significantly increased global protein synthesis in HSCs, but not in other hematopoietic progenitors, and impaired their reconstituting activity, identifying a mechanism that promotes HSC maintenance by attenuating protein synthesis.
Adult hematopoietic stem cell (HSC) maintenance depends on these cells making less protein per hour as compared with other hematopoietic progenitors (Signer et al. 2014). This does not reflect merely HSC quiescence, as dividing HSCs also make less protein per hour as compared with other hematopoietic progenitors. Several other adult stem cells also exhibit lower rates of protein synthesis as compared with progenitors in the same tissues, including neural stem cells (Llorens-Bobadilla et al. 2015), skeletal muscle stem cells (Zismanov et al. 2016), hair follicle stem cells (Blanco et al. 2016), and Drosophila germline stem cells (Sanchez et al. 2016). Like HSCs, each of these stem cells is depleted by genetic changes that increase their rate of protein synthesis, suggesting that this is a widely shared property of adult stem cells. However, there is limited insight into the mechanisms that suppress protein synthesis in stem cells.
Ribosome biogenesis limits protein synthesis in some stem cells. Quiescent neural stem cells express lower levels of ribosomal subunits and synthesize protein at a lower rate as compared with activated neural stem cells (Llorens-Bobadilla et al. 2015). Increased ribosome biogenesis in Drosophila germline stem cells increases protein synthesis, promoting differentiation and the loss of stem cells (Sanchez et al. 2016). Replication stress in HSCs from old mice transcriptionally silences ribosome genes, reducing ribosome biogenesis and perhaps impairing HSC function during aging (Flach et al. 2014). Runx1-deficient HSCs have impaired ribosome biogenesis, lower protein synthesis, and altered function (Cai et al. 2015). Defects in ribosome biogenesis impair HSC function (Le Bouteiller et al. 2013).
Phosphorylated eukaryotic initiation factor 2α (eIF2α) inhibits translation initiation and is part of the mechanism that limits protein synthesis in some stem cells. eIF2α can be phosphorylated under steady-state circumstances, although phosphorylation is increased by a wide range of stresses, including unfolded protein responses (Wek et al. 2006). Protein synthesis in skeletal muscle stem cells is limited by high levels of phosphorylated eIF2α, which promotes quiescence and stem cell maintenance (Zismanov et al. 2016). However, eIF2α function may vary among stem cells, as HSCs express relatively low levels of phosphorylated eIF2α (Signer et al. 2014), and activation of the unfolded protein response can be deleterious to individual HSCs (van Galen et al. 2014).
mTORC1 signaling promotes protein synthesis through phosphorylation and activation of ribosomal protein S6 kinase 1 (Brown et al. 1995) and phosphorylation and inhibition of eIF4E-binding proteins (4E-BPs) (Beretta et al. 1996; Brunn et al. 1997). Three genes (Eif4ebp1, Eif4ebp2, and Eif4ebp3) encode 4E-BPs that negatively regulate translation by binding the cap-binding protein eIF4E, inhibiting eIF4G binding, and preventing eIF4F complex assembly (eIF4E–eIF4G–eIF4A). Phosphorylation of 4E-BPs by mTORC1 weakens their binding to eIF4E, promoting eIF4F assembly and cap-dependent translation (Pause et al. 1994; Gingras et al. 1999).
4E-BP levels vary among tissues. Hematopoietic cells express abundant 4E-BP1 and 4E-BP2 but not 4E-BP3 (Tsukiyama-Kohara et al. 2001). 4E-BP-mediated translation inhibition does not generally affect global translation rates but particularly impairs the translation of certain subsets of mRNAs (Morita et al. 2013; Tahmasebi et al. 2014). Conditional Pten deletion increases mTORC1 signaling in HSCs (Lee et al. 2010), increasing 4E-BP phosphorylation (Magee et al. 2012), elevating protein synthesis, and promoting HSC depletion (Yilmaz et al. 2006; Signer et al. 2014). It is not clear whether 4E-BPs regulate HSC function, although knockdown of 4E-BP2 (Hartman et al. 2013) or eIF4E1 (Yang et al. 2014) in neural progenitors increases protein synthesis and promotes premature neuronal differentiation.
The synthesis of O-propargyl-puromycin (OP-Puro) has facilitated the quantification of protein synthesis in individual cells in vivo (Liu et al. 2012). OP-Puro enters the acceptor site of ribosomes and is incorporated into nascent polypeptide chains. The amount of protein synthesis per hour in individual cells in vivo can then be quantified based on OP-Puro incorporation (Liu et al. 2012; Signer et al. 2014). In this study, we examined potential mechanisms that might influence differences in protein synthesis between HSCs and restricted hematopoietic progenitors in adult mouse bone marrow.
Results and Discussion
HSCs do not have high proteasome activity
We administered OP-Puro to mice, killed them 1 h later, and compared OP-Puro incorporation by subpopulations of bone marrow cells by flow cytometry. Consistent with our prior study (Signer et al. 2014), we observed significantly less OP-Puro incorporation into CD150+CD48−Lineage− Sca-1+c-kit+ (CD150+CD48−LSK) HSCs (Kiel et al. 2005) and CD150−CD48−LSK multipotent progenitor cells (MPPs) (Oguro et al. 2013) as compared with restricted hematopoietic progenitors (Fig. 1A).
Figure 1.
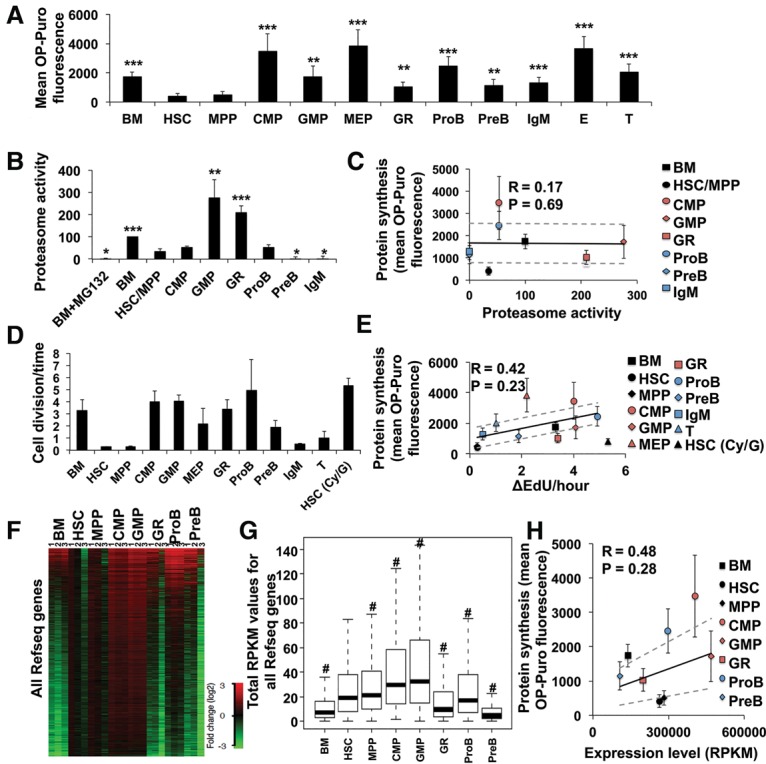
Protein synthesis rates in HSCs and progenitor cells show little correlation with proteasome activity, frequency of cell division, or RNA content. (A) OP-Puro incorporation by HSCs and progenitor cells in vivo. n = 8 mice in eight experiments. (B) Proteasome activity in HSCs and progenitor cells normalized to unfractionated bone marrow cells. n = 3 experiments. (C) Mean protein synthesis rate (from A) plotted against relative proteasome activity (from B). (D) The frequency of cell division for each population based on the rate of 5-ethynyl-2′-deoxyuridine (EdU) incorporation in vivo (see Supplemental Fig. S1B–M for primary data). n = 5 mice in two experiments. (E) Mean protein synthesis (from A) plotted against the rate of cell division (from D). (F) Gene expression values for all mouse RefSeq genes in each cell population (in total reads per kilobase of exonic length per million mapped reads [RPKM]). Log2 fold change of gene expression in each sample is shown relative to the average values in HSCs. (G) Total RPKMs of all mouse RefSeq genes in each cell population. Two-sided paired Wilcoxon rank-sum test was used for pair-wise comparisons between HSCs and each cell type. (#) P < 2.2 × 10−16. (H) Mean protein synthesis (from A) plotted against median total RPKMs (from G). Data represent mean ± SD unless indicated otherwise. The statistical significance of differences relative to HSCs in A, B, and D were assessed using a repeated-measures one-way analysis of variance (ANOVA) followed by Dunnett's test for multiple comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Regression analyses in C, E, and F were performed excluding HSCs, which were plotted independently. Ninety-five percent confidence intervals (dashed lines) and Pearson's correlation coefficients (R) are shown.
To test whether HSCs have high proteasome activity like embryonic stem cells (Vilchez et al. 2013), we compared the proteasome activity of HSCs versus restricted progenitors. We sorted 30,000 unfractionated bone marrow cells, CD48−LSK cells (HSCs/MPPs), CD34+CD16/32lowCD127−Sca-1−LK common myeloid progenitors (CMPs) (Akashi et al. 2000), CD34+CD16/32highCD127− Sca-1−LK granulocyte macrophage progenitors (GMPs) (Akashi et al. 2000), Gr-1+ myeloid, IgM−CD43+B220+ pro-B, IgM−CD43−B220+ pre-B (Hardy et al. 1991), and B220+IgM+ B cells. Proteasome activity in HSCs/MPPs was similar to CMPs and pro-B cells but significantly lower than unfractionated bone marrow cells, GMPs, and Gr-1+ cells (Fig. 1B; Supplemental Fig. S1A). Overall, proteasome activity showed little correlation with OP-Puro incorporation (R = 0.17, P = 0.69) (Fig. 1C).
Little correlation between protein synthesis and cell division or RNA content
We administered the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) to mice and measured its incorporation by HSCs and restricted progenitors in the bone marrow after 2, 6, and 12–24 h to determine the frequency of cell division in each population (Fig. 1D). We observed a reasonably linear increase in the fraction of EdU+ cells over time in each population (Supplemental Fig. S1B–M). We observed a modest correlation between protein synthesis rates and cell division (R = 0.42, P = 0.23) (Fig. 1E). However, when we drove HSCs into cycle by treating them with cyclophosphamide and GCSF, we observed only a limited increase in protein synthesis (Fig. 1E). Therefore, the rate of protein synthesis in HSCs was not determined primarily by the rate at which they divide.
To test whether protein synthesis rates reflected total RNA content (largely ribosomal RNA), we quantified total RNA by nanofluidic electrophoresis using a BioAnalyzer. HSCs had amounts of total RNA similar to those of CMPs and pro-B cells and significantly more total RNA per cell than Gr-1+, pre-B, and unfractionated bone marrow cells (Supplemental Fig. S1N), each of which had higher protein synthesis rates than HSCs (Fig. 1A). Therefore, we observed little correlation between protein synthesis rates and total RNA content per cell (R = 0.32, P = 0.44) (Supplemental Fig. S1O).
We quantified the amount of mRNA per cell by RNA sequencing (RNA-seq) after adding RNA standards that allow normalization of transcript numbers to cell number (Fig. 1F; Loven et al. 2012). On average, HSCs had significantly more mRNA per cell as compared with pre-B cells, unfractionated bone marrow, and Gr-1+ cells but less mRNA per cell than CMPs, GMPs, and pro-B cells (Fig. 1G), all of which had higher protein synthesis rates than HSCs (Fig. 1A). Therefore, we observed little correlation between protein synthesis rates and mRNA content in hematopoietic cells (R = 0.48, P = 0.28) (Fig. 1H).
HSCs have relatively high levels of hypophosphorylated 4E-BPs
We assessed the levels of total, phosphorylated, and nonphosphorylated 4E-BP1 and 4E-BP2 in 30,000 sorted cells from each population (Fig. 2A). Control experiments using peripheral blood from 4E-BP1−/− (Tsukiyama-Kohara et al. 2001), 4E-BP2−/− (Banko et al. 2005), and 4E-BP1−/−; 4E-BP2−/− mice (Le Bacquer et al. 2007) showed that the upper band was 4E-BP1, and the lower band was 4E-BP2 (Supplemental Fig. S2A,B). HSCs/MPPs expressed relatively high levels of total 4E-BP1 and 4E-BP2 as compared with most hematopoietic progenitors—significantly higher than Gr-1+, pro-B, pre-B, and IgM+ B cells (Fig. 2A–C). Levels of nonphosphorylated 4E-BP1 and 4E-BP2 were also significantly higher in HSCs/MPPs as compared with Gr-1+, pro-B, pre-B, and IgM+ B cells (Fig. 2D,E). The ratio of phosphorylated to total 4E-BP1/2 tended to be lower in HSCs as compared with restricted progenitors (Supplemental Fig. S2C,D). Differences in β-actin levels among cell populations represented differences in the amount of β-actin per cell, as lysates from equal numbers of cells were used in these experiments (Fig. 2A). Nonetheless, even when levels of total and nonphosphorylated 4E-BP1 and 4E-BP2 were normalized to differences in β-actin, they tended to remain higher in HSCs/MPPs as compared with Gr-1+, pro-B, pre-B, and IgM+ B cells (although not all of the differences were statistically significant) (Supplemental Fig. S2E–H).
Figure 2.
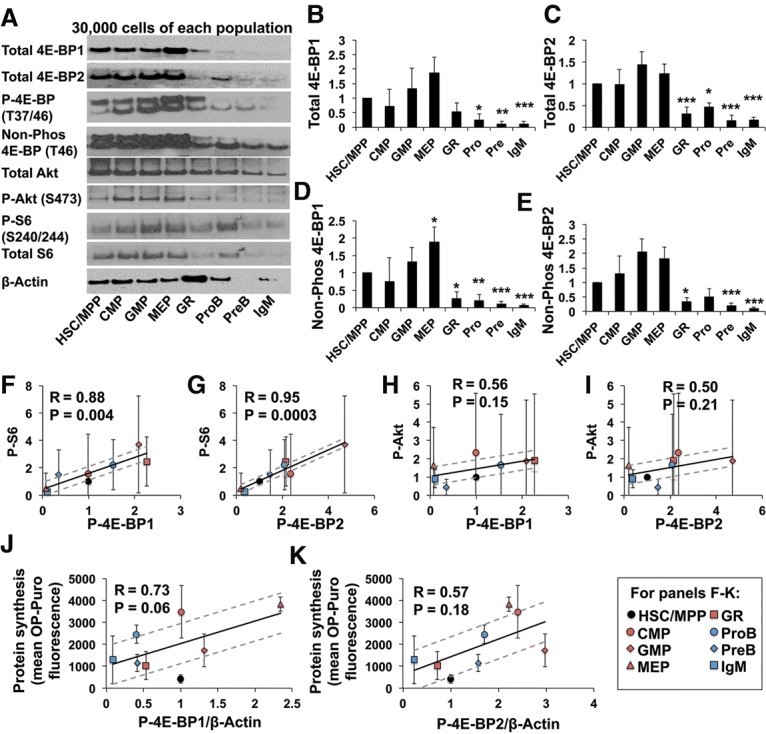
HSCs tend to have high levels of hypophosphorylated 4E-BPs relative to most other hematopoietic progenitors. (A) Western blot of 3 × 104 cells from each cell population (one of four representative blots). Band intensity was normalized by cell number for each population. For phosphorylated and nonphosphorylated 4E-BP, the upper band is 4E-BP1, and the lower band is 4E-BP2 (see Supplemental Fig. S2A,B). (B–E) Quantification of band intensity for total 4E-BP1 (B), total 4E-BP2 (C), nonphosphorylated 4E-BP1 (D), and nonphosphorylated 4E-BP2 (E). (F,G) Mean intensity of phosphorylated S6 plotted against phosphorylated 4E-BP1 (F) or phosphorylated 4E-BP2 (G) levels for each cell population. (H,I) Mean intensity of phosphorylated Akt plotted against phosphorylated 4E-BP1 (H) or phosphorylated 4E-BP2 (I). (J,K) Mean protein synthesis (from Fig. 1A) plotted against phosphorylated 4E-BP1 (J) or 4E-BP2 (K) levels normalized to β-Actin. The regressions exclude HSCs/MPPs, which were plotted independently. Data in B–K represent mean ± SD from four experiments, including the one in A. The statistical significance of differences between HSCs/MPPs and other cell populations in B–E were assessed with one-way ANOVAs followed by Dunnett's test for multiple comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. For F–K, Pearson's correlation coefficient (R) and 95% confidence intervals (dashed lines) are shown.
Levels of phosphorylated S6 strongly correlated with levels of phosphorylated 4E-BP1 (R = 0.88, P = 0.004) (Fig. 2F) or phosphorylated 4E-BP2 (R = 0.95, P = 0.0003) (Fig. 2G). However, correlations between phosphorylated Akt and phosphorylated 4E-BP1 (R = 0.56, P = 0.15) (Fig. 2H) or phosphorylated 4E-BP2 (R = 0.50, P = 0.21) (Fig. 2I) were weaker. These data suggest that differences in 4E-BP phosphorylation among HSCs and progenitor cells likely reflect differences in mTORC1 signaling. Levels of phosphorylated 4E-BP1 (R = 0.73, P = 0.06) (Fig. 2J) and phosphorylated 4E-BP2 (R = 0.57, P = 0.18) (Fig. 2K) normalized to β-actin exhibited stronger correlations with protein synthesis rates than any other parameter that we tested. Levels of phosphorylated 4E-BP1 (R = 0.40, P = 0.37) (Supplemental Fig. S2I) and phosphorylated 4E-BP2 (R = 0.24, P = 0.6) (Supplemental Fig. S2J) that were normalized based on cell numbers did not exhibit as strong a correlation with protein synthesis.
To test whether 4E-BPs limit the rate of protein synthesis in HSCs, we examined OP-Puro incorporation by HSCs and progenitors in 4E-BP1−/−, 4E-BP2−/−, and 4E-BP1−/−; 4E-BP2−/− mice. 4E-BP1−/− or 4E-BP2−/− HSCs did not exhibit significant changes in OP-Puro incorporation compared with wild-type HSCs (Fig. 3A). However, 4E-BP1−/−; 4E-BP2−/− HSCs exhibited a significant increase in protein synthesis as compared with wild-type HSCs (23%, P < 0.05) (Fig. 3A). The magnitude of this increase was close to that observed after Pten deletion, which leads to HSC depletion (Signer et al. 2014). We did not detect a statistically significant effect of 4E-BP1 deficiency, 4E-BP2 deficiency, or combined 4E-BP1;4E-BP2 deficiency on the rate of protein synthesis in other hematopoietic progenitors (Fig. 3B). HSCs thus appear to be more sensitive to the effects of 4E-BP1/2 on global protein synthesis.
Figure 3.
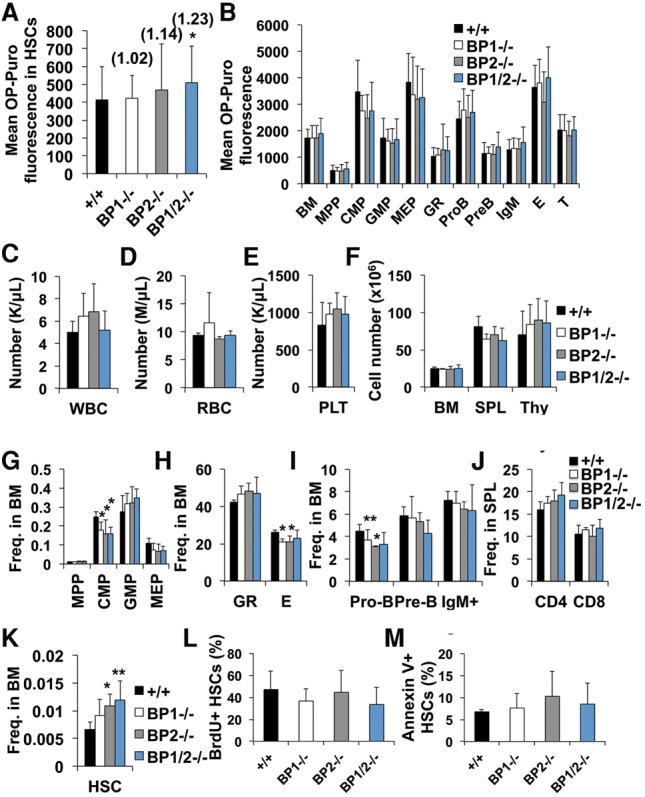
4E-BP1/2-deficient mice have HSCs with increased protein synthesis but largely normal hematopoiesis. (A) Mean OP-Puro incorporation by HSCs from 4E-BP1−/−, 4E-BP2−/−, 4E-BP1−/−;4E-BP2−/−, or wild-type mice. n = 9 mice per genotype in nine experiments. The value relative to wild type is shown above each bar. (B) Mean OP-Puro incorporation into each restricted progenitor population from the same mice as in A. (C–E) Number of white blood cells (WBC), red blood cells (RBC), and platelets (PLT) in the peripheral blood. (F) Cell number in the bone marrow (BM; one femur and one tibia), spleen (SPL), and thymus (Thy). (G–I) Frequencies of MPPs, CMPs, GMPs, and megakaryocyte erythroid-restricted progenitors (MEPs) (G); Gr-1+ myeloid and Ter119+ erythroid cells (H); and pro-B, pre-B, and IgM+ B cells (I) in the bone marrow. (J) Frequencies of CD4+ and CD8+ T cells in the spleen. (K) Frequency of HSCs in the bone marrow. n = 6 mice per genotype in three experiments in C–K. (L) Frequency of BrdU+ HSCs after a 72-h pulse of BrdU in vivo. (M) Frequency of annexin V+ HSCs. n = 5–6 mice per genotype in four experiments in L and M. All data represent mean ± SD. The statistical significance of differences relative to wild type was assessed using one-way ANOVAs (repeated-measures in A and B) followed by Dunnett's test for multiple comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
4E-BP1−/−;4E-BP2−/− HSCs have impaired regenerative activity
4E-BP1−/−, 4E-BP2−/−, and 4E-BP1−/−;4E-BP2−/− compound mutant mice all exhibited relatively normal hematopoiesis (Fig. 3C–J). HSC frequency appeared to be higher in 4E-BP2−/− (65% ± 32%, P < 0.05) (Fig. 3K) and 4E-BP1−/−; 4E-BP2−/− (80% ± 53%, P < 0.01) mutant mice (Fig. 3K) as compared with controls. This modest increase in HSC frequency was not associated with significant changes in the frequency of dividing (Fig. 3L) or dying (Fig. 3M) HSCs. This suggests that the increase in HSC frequency may have been caused by a lower rate of differentiation by 4E-BP1/2-deficient HSCs. We also observed modest but significant declines in the frequencies of CMPs, Ter119+ erythroid progenitors, and pro-B cells in the bone marrow of some 4E-BP-deficient mice as compared with controls (Fig. 3G–I).
To test the effect of 4E-BP deficiency on HSC function, we performed competitive reconstitution assays in irradiated mice. Bone marrow cells (5 × 105 cells) from 4E-BP1−/−, 4E-BP2−/−, 4E-BP1−/−;4E-BP2−/−, or wild-type mice (all CD45.2+) were transplanted with equal numbers of wild-type recipient bone marrow cells (CD45.1+) into irradiated mice (CD45.1+). Recipients of 4E-BP-deficient bone marrow cells had significantly higher levels of donor-derived hematopoietic cells in their blood 4–16 wk after transplantation (Fig. 4A–D). To test whether this increased reconstitution reflected increased HSC frequency in the bone marrow of 4E-BP-deficient donors (Fig. 3K) or increased function of 4E-BP-deficient HSCs, we competitively transplanted 10 CD150+CD48−LSK HSCs from 4E-BP1−/−;4E-BP2−/− or wild-type donors into irradiated recipient mice along with recipient bone marrow cells. 4E-BP-deficient HSCs gave normal levels of donor cell reconstitution (Fig. 4E–H). The higher level of donor cell reconstitution by 4E-BP-deficient bone marrow cells thus reflects higher HSC frequency in the donor bone marrow rather than increased reconstituting activity by 4E-BP-deficient HSCs.
Figure 4.
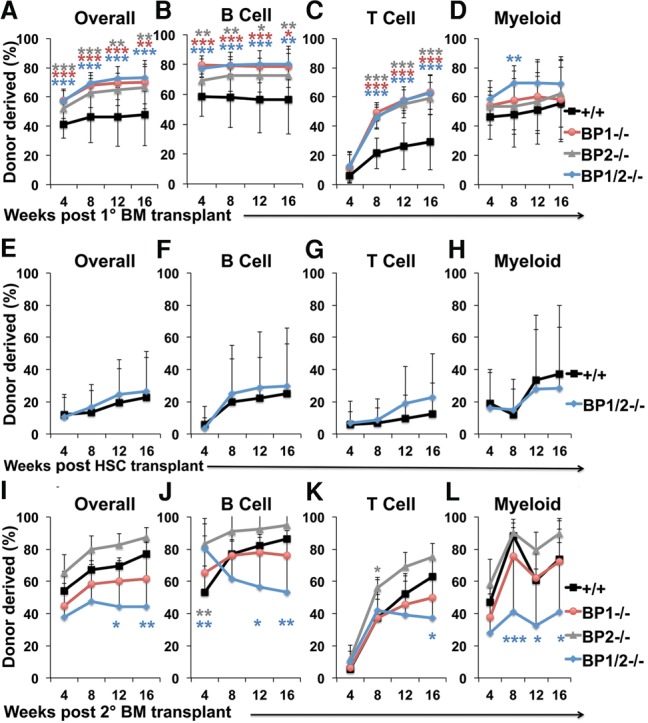
4E-BP1−/−;4E-BP2−/− HSCs have impaired reconstituting potential upon serial transplantation. (A–D) Donor cell engraftment when 5 × 105 bone marrow cells from mice of the indicated genotypes were transplanted along with 5 × 105 recipient bone marrow cells into irradiated mice. Donor cell engraftment in the bone marrow is shown in Supplemental Figure S3A. (E–H) Donor cell engraftment when 10 HSCs from mice of the indicated genotypes were transplanted with 3 × 105 recipient bone marrow cells into irradiated mice. (I–L) Donor cell engraftment after serial transplantation of 3 × 106 bone marrow cells from primary recipients in A–D into secondary recipient mice. n = 8–11 recipients per genotype from two primary donors from wild type and three primary donors from each of the 4E-BP-deficient genotypes. Data represent mean ± SD. The statistical significance of differences relative to wild-type were assessed with one-way ANOVAs followed by Dunnett's test for multiple comparisons in A–D and I–L and a two-tailed Student's t-test in E–H. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Although 4E-BP2−/− and 4E-BP1−/−;4E-BP2−/− mutant mice had increased HSC frequency in their bone marrow as compared with controls (Fig. 3K), the frequency of donor HSCs in primary recipient mice after bone marrow transplantation did not significantly differ between recipients of 4E-BP-deficient cells as compared with control cells (Supplemental Fig. S3B). These data raised the possibility that 4E-BP deficiency may impair HSC self-renewal after transplantation.
To assess HSC self-renewal potential, we performed secondary transplants of 3 × 106 bone marrow cells from primary recipient mice with levels of donor cell reconstitution nearest the median values in each treatment. Deficiency for only 4E-BP1 or 4E-BP2 did not significantly affect donor cell reconstitution in secondary recipient mice as compared with control cells. In contrast, secondary recipients of 4E-BP1−/−;4E-BP2−/− cells had significantly less donor cell reconstitution in all lineages 16 wk after transplantation as compared with secondary recipients of wild-type donor cells (Fig. 4I–L). These data suggest that 4E-BP1−/−;4E-BP2−/− HSCs have reduced self-renewal potential as compared with control HSCs.
4E-BP1 and 4E-BP2 likely regulate the translation of a subset of mRNAs in HSCs. Increased expression of these mRNAs likely increases HSC frequency (Fig. 3K) and reconstituting capacity in the bone marrow of 4E-BP1/2−/− mice (Fig. 4A–D) while reducing HSC self-renewal upon serial transplantation (Fig. 4I–L). Nonetheless, other mechanisms must also limit protein synthesis in HSCs, as protein synthesis rates in 4E-BP1/2−/− HSCs remained significantly lower than in 4E-BP1/2−/− restricted hematopoietic progenitors. Moreover, both GMPs and megakaryocyte erythroid-restricted progenitors (MEPs) had higher levels of hypophosphorylated 4E-BPs relative to HSCs (Fig. 2D,E) and yet still had higher levels of global protein synthesis (Fig. 1A), perhaps because global protein synthesis in these cells is not as sensitive to 4E-BP1/2 as in HSCs (Fig. 3B).
Materials and methods
Mice
Eif4ebp1−/− (Tsukiyama-Kohara et al. 2001), Eif4ebp2−/− (Banko et al. 2005), and Eif4ebp1−/−;Eif4ebp2−/− (Le Bacquer et al. 2007) mice have been described previously. These mice were backcrossed for at least 10 generations onto a C57BL background. C57BL/Ka-Thy-1.1 (CD45.2) mice were used as wild type throughout this study. C57BL/Ka-Thy-1.2 (CD45.1) mice were used as transplant recipients. Male and female mice between 8 and 14 wk old were used in all studies. Cyclophosphamide and GCSF were administered as described (Signer et al. 2014). All mice were housed in the Animal Resource Center at the University of Texas Southwestern Medical Center, and all protocols were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Proteasome activity assay
Proteasome activity was determined by measuring the rate of hydrolysis of Suc–Leu–Leu–Val–Tyr–7-amino-4-methylcoumarin (AMC). This substrate is specific for the proteasome under the assay conditions used here and is hydrolyzed by all proteasome holoenzymes. The activity reported by this assay represents the total enzymatic capacity of the cellular proteasome and is typically dominated by the 26S proteasome (Sasaki et al. 1984; Demartino and Gillette 2007). From each population, 3 × 104 cells were double-sorted into 96-well black plates. One-hundred microliters of buffer containing 20 mM Tris-HCl (pH 7.6), 1 mM 2-mercaptoethanol, 5 mM MgCl2, 1 mM ATP, and 0.4% NP-40 was added to each well followed immediately by 50 µL of 200 µM Suc–Leu–Leu–Val–Tyr–AMC (Bachem). The fluorescence of free AMC generated by proteasomal hydrolysis of the substrate was monitored continuously (one read per minute for 180 min at 37°C) at 360ex/460em using a BioTek Synergy II plate reader. Controls included reactions conducted in the absence of cells, in the presence of 10 mM MG132 (UBPBio), or using purified bovine 26S proteasome instead of cells. Data are expressed as arbitrary fluorescent units per well.
The rate of cell division
Two milligrams of EdU (Thermo Scientific) in PBS was injected intraperitoneally per mouse every 6 h. Mice were analyzed 2, 6, 12, and 24 h after EdU administration. These short labeling times minimized the extent to which results were affected by the entry of labeled cells into each population through the maturation of upstream progenitors, the exit of labeled cells through differentiation into downstream progeny, or saturation of labels in rapidly dividing cell populations. Bone marrow cells (3 × 106 cells) were stained with antibodies as described in the Supplemental Material. Cells were fixed and permeabilized, and the azide–alkyne cycloaddition was performed as with OP-Puro detection and analyzed by flow cytometry. For cyclophosphamide- and GCSF-treated mice, BrdU was administered in place of EdU and was detected with the BrdU flow kit.
Supplementary Material
Acknowledgments
We thank K. Correll, N. Loof, and the Moody Foundation Flow Cytometry Facility. S.J.M. is a Howard Hughes Medical Institute Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryn and Gene Bishop Distinguished Chair in Pediatric Research, and the director of the Hamon Laboratory for Stem Cells and Cancer. L.Q. is supported by a fellowship from the Howard Hughes Medical Institute. This work was supported by the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases R01 DK100848) and the Cancer Prevention and Research Institute of Texas.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.282756.116.
Freely available online through the Genes & Development Open Access option.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197. [DOI] [PubMed] [Google Scholar]
- Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. 2005. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci 25: 9581–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15: 658–664. [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377: 441–446. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC Jr, Abraham RT. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277: 99–101. [DOI] [PubMed] [Google Scholar]
- Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, Speck NA. 2015. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demartino GN, Gillette TG. 2007. Proteasomes: machines for all reasons. Cell 129: 659–662. [DOI] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512: 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, Bordey A. 2013. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep 5: 433–444. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. 2007. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 117: 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouteiller M, Souilhol C, Beck-Cormier S, Stedman A, Burlen-Defranoux O, Vandormael-Pournin S, Bernex F, Cumano A, Cohen-Tannoudji M. 2013. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med 210: 2351–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. 2010. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell 7: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A. 2012. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci 109: 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. 2015. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17: 329–340. [DOI] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. 2012. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell 11: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711. [DOI] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. 2013. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 13: 102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr, Sonenberg N. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371: 762–767. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. 2016. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Kikuchi T, Yumoto N, Yoshimura N, Murachi T. 1984. Comparative specificity and kinetic studies on porcine calpain I and calpain II with naturally occurring peptides and synthetic fluorogenic substrates. J Biol Chem 259: 12489–12494. [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, et al. 2014. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14: 606–616. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, et al. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med 7: 1128–1132. [DOI] [PubMed] [Google Scholar]
- van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, Xie S, Laurenti E, Hermans K, Eppert K, et al. 2014. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510: 268–272. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Lutz M, Merkwirth C, Morantte I, Tse C, Spencer B, Page L, Masliah E, Berggren WT, et al. 2013. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell 12: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11. [DOI] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, Miller FD. 2014. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron 84: 723–739. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441: 475–482. [DOI] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. 2016. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18: 79–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


