In this study, Yang et al. show that mice lacking Xist RNA, which is needed for X-chromosome inactivation, can develop normally and survive to term; however, Xist-null mice die before adulthood. This study provides new insight into Xist-independent mechanisms of dosage compensation and demonstrates that small deviations from X-autosomal balance can have profound effects on overall fitness.
Keywords: genome balance, inverse effect, X inactivation, Xist, dosage compensation, knockout mouse, transcriptomics
Abstract
X-chromosome inactivation (XCI) compensates for differences in X-chromosome number between male and female mammals. XCI is orchestrated by Xist RNA, whose expression in early development leads to transcriptional silencing of one X chromosome in the female. Knockout studies have established a requirement for Xist with inviability of female embryos that inherit an Xist deletion from the father. Here, we report that female mice lacking Xist RNA can, surprisingly, develop and survive to term. Xist-null females are born at lower frequency and are smaller at birth, but organogenesis is mostly normal. Transcriptomic analysis indicates significant overexpression of hundreds of X-linked genes across multiple tissues. Therefore, Xist-null mice can develop to term in spite of a deficiency of dosage compensation. However, the degree of X-autosomal dosage imbalance was less than anticipated (1.14-fold to 1.36-fold). Thus, partial dosage compensation can be achieved without Xist, supporting the idea of inherent genome balance. Nevertheless, to date, none of the mutant mice has survived beyond weaning stage. Sudden death is associated with failure of postnatal organ maturation. Our data suggest Xist-independent mechanisms of dosage compensation and demonstrate that small deviations from X-autosomal balance can have profound effects on overall fitness.
In mammals, equal dosage of X-linked genes between the sexes is ensured through the process of X-chromosome inactivation (XCI). XCI has been extensively studied using mouse models and occurs in two waves during early embryogenesis in females (Starmer and Magnuson 2009; Lee 2011; Disteche 2012; Maduro et al. 2016). Imprinted XCI is observed in preimplantation embryos and involves silencing of the paternal X chromosome (XP) in all cells of the early embryo (Takagi and Sasaki 1975; West et al. 1977; Takagi et al. 1978). As development proceeds, the silent XP is perpetuated by all cells of the extraembryonic tissues (e.g., cells of the trophectoderm and primitive endoderm, which eventually give rise to the placenta and yolk sac). However, for the embryo proper, the inactive XP is maintained only until the early blastocyst stage, when cells of the inner cell mass (ICM) briefly reactivate the XP (Mak et al. 2004) before a silent X chromosome is reinstated. In this second round of silencing, XCI occurs randomly on either the XP or XM (maternal X). In mice, random XCI occurs in the embryo proper between embryonic day 5.5 (E5.5) and E6.5 and gives rise to the mosaic pattern of XP or XM inactivation that characterizes somatic cells of the adult female (Lyon 1961; Gardner and Lyon 1971; Monk and Harper 1979).
Previous studies have established that XCI is mediated by the long noncoding Xist RNA (Borsani et al. 1991; Brockdorff et al. 1991, 1992). Xist RNA is transcribed exclusively from the future inactive X chromosome, coats the chromosome (Clemson et al. 1996; Engreitz et al. 2013; Simon et al. 2013), and triggers formation of silent chromatin that is stably inherited through cell divisions (Starmer and Magnuson 2009; Lee 2011; Disteche 2012; Maduro et al. 2016). In the preimplantation embryo and the extraembryonic lineages, Xist is expressed only from the XP, in keeping with imprinted XP inactivation. Female embryos that inherit a paternal Xist deletion die shortly after implantation as a consequence of dosage imbalance in extraembryonic tissue (Marahrens et al. 1997). In contrast, female embryos that inherit a maternal Xist deletion develop normally for two reasons: First, the maternal Xist allele is not expressed during normal development in the early embryo. Second, in the embryo proper, the random XCI process becomes nonrandom such that the X chromosome with an intact copy of Xist is always an inactive X chromosome (Lyon et al. 1964; Takagi 1980; Penny et al. 1996; Marahrens et al. 1997; Senner et al. 2011). These findings also demonstrate that Xist is essential for not only imprinted XCI but also random XCI. In an embryonic stem (ES) cell model, knocking out Xist on one X chromosome results in a skewed pattern of XCI, with the intact X always adopting the inactive X chromosome state (Penny et al. 1996). Similarly, a heterozygous deletion of Xist in vivo results in a nonrandom XCI pattern in all cells of the embryo proper and the adult soma (Hoki et al. 2009, 2011).
In spite of these classic studies, the impact of losing both Xist alleles on the developing embryo has never been addressed, as the loss of imprinted XCI causes embryonic lethality before effects on random XCI can be addressed. Of specific interest is whether deviation from a 1:1 X to autosome (X:A) dosage balance has consequences for the developing soma. One recent study showed that conditionally deleting Xist in the blood compartment resulted in a partial inactive X-chromosome reactivation and the development of highly lethal blood cancers with full penetrance, demonstrating that Xist is a potent suppressor of hematologic cancer in adult mice (Yildirim et al. 2013). On the other hand, in the ex vivo context, cells derived from the ICM, such as mouse ES cells (Lin et al. 2007; Schulz et al. 2014) and embryonal carcinoma (EC) cells (Martin et al. 1978), appear to tolerate some degree of X-linked hypertranscription. Furthermore, because ES cells do not express Xist and consequently carry two active X chromosomes, they are not dosage-compensated and have ∼60% more X-linked gene expression relative to their differentiated male counterparts (Nguyen and Disteche 2006; Lin et al. 2007; Deng et al. 2011; Kharchenko et al. 2011; Yildirim et al. 2012). This overexpression may explain why, although female mouse ES and EC cells can be maintained indefinitely ex vivo, they have a tendency to lose one X chromosome with prolonged culture. Thus, it is currently believed that full X:A dosage compensation is essential for embryonic development.
Here, we set out to investigate the effects of dosage imbalance on the developing embryo. We asked whether it is possible to create female mice lacking Xist RNA throughout the body. To our surprise, it is. Although X-linked gene expression is significantly increased, the overall increase is far less than expected of two active X chromosomes. These data thereby provide support for inherent genome balance and “inverse effects” as historically observed in plants and flies (Stenberg et al. 2009; Birchler 2013; Sun et al. 2013). However, they also demonstrate that small deviations from X-autosomal equivalence can have a major impact on survival and fitness.
Results
Female mice lacking Xist RNA undergo embryogenesis and are viable to term
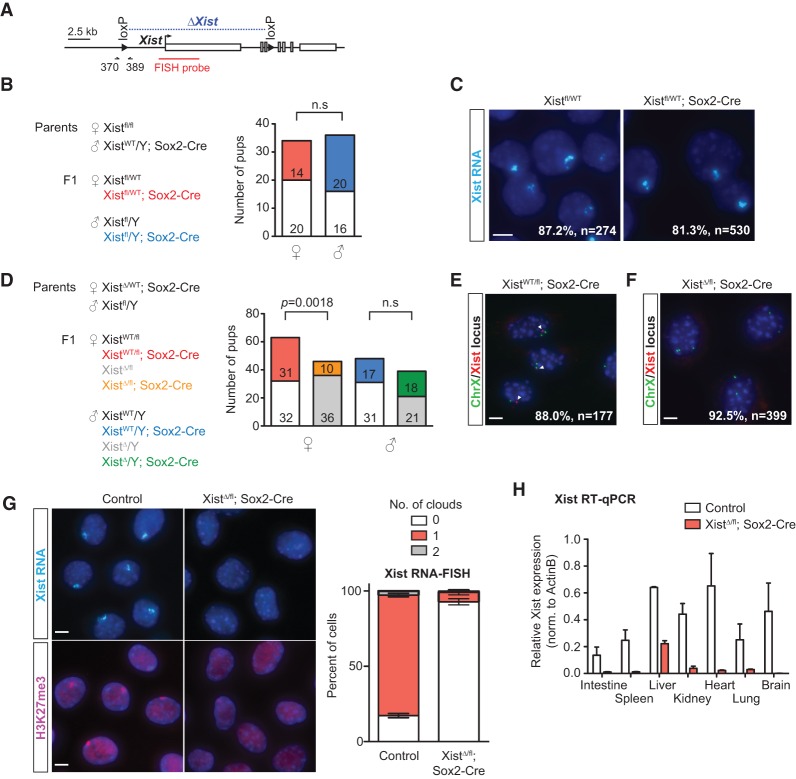
To create a whole-body Xist-null mouse, we used a conditional allele for Xist that would delete the promoter and first three exons upon exposure to Cre recombinase (Fig. 1A; Csankovszki et al. 1999). To bypass the requirement for Xist during imprinted XCI in placental lineages (Marahrens et al. 1997), we used a Sox2 promoter-driven Cre recombinase (Hayashi et al. 2002) in order to generate mice deficient for Xist only in the epiblast lineage. Previous work showed that Sox2-Cre expression is observed at the blastocyst stage in the ICM but not the trophectoderm or primitive endoderm (subjected to imprinted XCI), thereby enabling us to decouple the effects of Xist on random versus imprinted XCI. Heterozygous Xist mutants (Xistfl/WT; Sox2-Cre) were born at normal Mendelian ratios (χ2, P = 0.6724) (Fig. 1B). The viability of these animals was expected in light of previous work showing skewed XCI patterns in female cells carrying a single Xist knockout allele (Penny et al. 1996; Marahrens et al. 1997). Indeed, RNA-FISH (fluorescence in situ hybridization) analysis in mouse embryonic fibroblasts (MEFs) showed that >80% of cells in both heterozygous mutants and wild-type controls harbored an Xist cloud (Fig. 1C), consistent with the occurrence of skewed inactivation favoring the wild-type X chromosome. Hence, these heterozygous animals will be used as the “control” group.
Figure 1.
Female mice lacking Xist RNA survive to term. (A) Map of the Xist conditional allele used in this study. The region of Xist deletion is indicated in blue. Arrows mark the location of the 370/389 primer set (used for detection of XistWT and Xistfl). The DNA-FISH probe for the Xist locus is indicated in red. (B, left) Schematic for generating heterozygous Xist deletants. (Right) Genotype data for cross; the number of pups for each genotype is listed. All genotypes in both sexes were derived at normal Mendelian ratios. χ2 test, P = 0.67. (n.s.) Not significant. (C) Representative images for Xist RNA-FISH from control (Xistfl/WT) and heterozygous mutant (Xistfl/WT; Sox2-Cre) E14.5 MEFs. Percentages denote cells with one Xist cloud and represent the average of at least two animals per genotype. (n) Number of nuclei counted. Bar, 5 µm. (D, left) Schematic for generating homozygous Xist deletants. (Right) Genotype data for cross, number of pups of each genotype obtained is listed. Homozygous Xist deletants were derived at significantly lower frequency (χ2 test). (n.s.) Not significant. (E) Representative images for Xist DNA-FISH in XistWT/fl; Sox2-Cre TTFs. The percentage denotes cells with one Xist focus (red) and two chromosome X foci (green) and represents the average of two animals. The location of the Xist probe is indicated in A. Arrowheads point to Xist signal. (n) Number of nuclei counted. Bar, 5 µm. (F) Representative images for Xist DNA-FISH in XistΔ/fl; Sox2-Cre TTFs. The percentage denotes cells with no Xist focus (red) and two chromosome X foci (green) and represents the average of three animals. The location of the Xist probe is indicated in A. (n) Number of nuclei counted. Bar, 5 µm. (G, left) Representative images for Xist RNA-FISH (top) and H3K27me3 immunofluorescence (bottom) in control (XistWT/fl; Sox2-Cre) and XistΔ/fl; Sox2-Cre TTFs. (Right) Quantification for RNA-FISH. At least 100 nuclei from each animal were counted. Data represent mean ± SEM of at least two animals per genotype. Bar, 5 µm. (H) RT-qPCR for Xist levels in different tissues. Data represent mean ± SEM for two controls and three Xist-null mutants.
We then attempted to generate homozygous Xist mutants by crossing Xistfl/WT; Sox2-Cre females to Xistfl/Y males (Fig. 1D). To our surprise, we observed live-born pups of the XistΔ/fl; Sox2-Cre genotype, which should in principle have become Xist−/− upon Cre-mediated excision of the floxed allele around the time of implantation. Litter sizes did not deviate significantly from normal (average litter size of 7.5 pups, with 3.4 females per litter). We isolated primary tail-tip fibroblasts (TTFs) from XistΔ/fl; Sox2-Cre neonates and performed DNA-FISH using an Xist probe that mapped within the region targeted for deletion. Xist signal was undetectable in at least 92.5% of TTFs (Fig. 1F). In contrast, as expected, DNA-FISH in control XistWT/fl; Sox2-Cre TTFs detected the presence of one Xist focus in 88% of cells (Fig. 1E). Xist RNA-FISH analysis confirmed the absence of the Xist cloud along with loss of the H3K27me3 modification, a classic epigenetic mark associated with the inactive X chromosome (Fig. 1G; Plath et al. 2003). We also performed RT-qPCR for multiple organs from homozygous mutants and control animals. Xist RNA was almost undetectable in most tissues, including the gut, spleen, kidney, heart, lung, and brain (Fig. 1H). Residual Xist expression was detected in the livers of all three homozygous mutants but at significantly reduced levels. Thus, we created female mice lacking Xist RNA (referred to here as “mutant”).
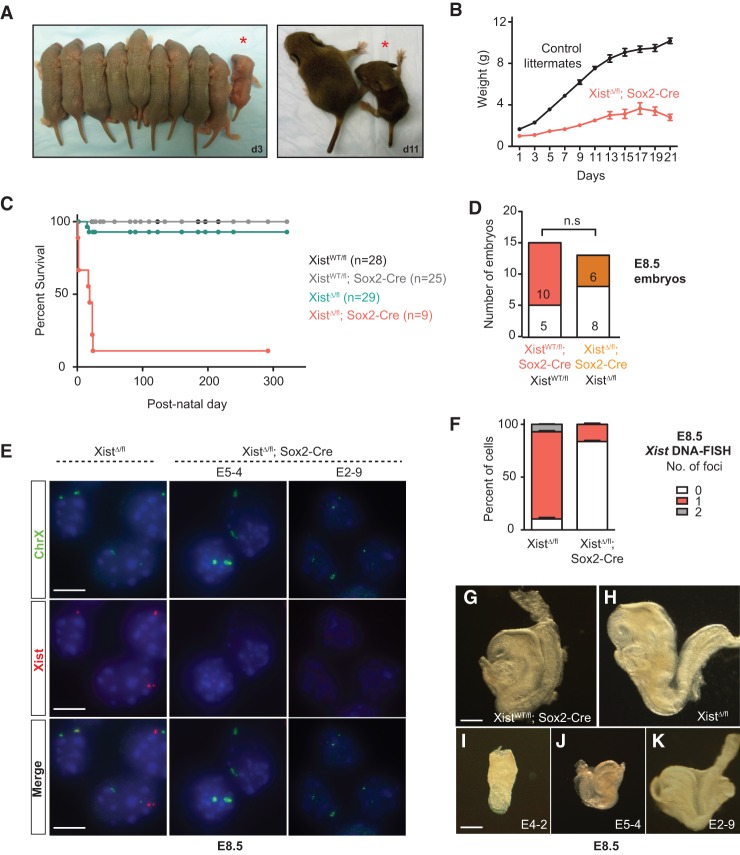
Although mutant females were born at regular intervals, their births occurred at only ∼25% of the expected Mendelian frequency (χ2, P = 0.0018) (Fig. 1D). All mutant pups were runted at birth (Fig. 2A), with an average body weight 60% that of female littermates (Fig. 2B). These mice showed persistent growth retardation and failed to thrive such that, at postnatal day 21 (P21), their average body weight was ∼30% that of female littermates (Fig. 2B). With one exception, all Xist-null females died by weaning age, with a median survival of 18 d ± 10.4 d (Fig. 2C). Reducing competition for food by removing male littermates did prolong survival of the Xist-null females, suggesting that a failure to compete for resources might have contributed to their early demise. Nevertheless, these resource-rich mutants still died within the first month of life, with the longest survivor persisting to 24 d. Thus, both inherent physiological defects and external competition contributed to decreased overall fitness.
Figure 2.
Xist-null female mice have decreased fitness. (A) Female littermates from homozygous crosses at P3 (d3, left) and P11 (d11, right). Homozygous Xist mutants are indicated by asterisks. (B) Body weight of Xist mutants (n = 3) and female littermate controls (n = 10). Error bars indicate mean ± SEM. (C) Kaplan-Meier survival curve for homozygous Xist mutants and female littermate controls. (D) Genotype data from homozygous cross at E8.5. The observed number of embryos is listed. (n.s.) Not significant. (E) Representative images for Xist and chromosome X DNA-FISH to quantify deletion efficiency at E8.5. Data for two Xist-null mutants are shown. Bar, 10 µm. (F) Quantification for DNA-FISH shown in E. Fifty to 100 nuclei from each animal were counted, depending on the size of the embryo. Data represent mean ± SEM of two XistΔ/fl and three XistΔ/fl; Sox2-Cre embryos. (G–K) E8.5 embryos of various genotypes. E4-2, E5-4, and E2-9 are XistΔ/fl; Sox2-Cre embryos showing different degrees of developmental defect and/or runting. Bar, 250 µm.
The reduced number of homozygous females at birth and the progressive loss prior to weaning are consistent with a failure of XCI in the pregastrulation embryo. Indeed, analysis of E8.5 embryos showed that effects began to manifest during this early post-implantation stage. Unlike in neonates, embryos of all genotypes were recovered at approximately equal frequencies at E8.5 (χ2, P = 0.5653) (Fig. 2D). However, embryos of the XistΔ/fl; Sox2-Cre genotype already appeared different from XistWT/fl; Sox2-Cre and XistΔ/fl control littermates (Fig. 2G–K). Interestingly, whereas XistΔ/fl; Sox2-Cre embryos often showed a developmental delay, there was a range of severity, with some embryos appearing only slightly runted, while others showed severe retardation (Fig. 2I–K). Regardless of phenotype, Sox2-Cre efficiently deleted Xist in all embryos of the XistΔ/fl; Sox2-Cre genotype, leaving no detectable cells with two intact Xist alleles and rare cells with one intact allele in each embryo (Fig. 2E,F). By PCR, the intact allele could be detected only faintly, if at all, in null mutant females at E8.5 (Supplemental Fig. S1).
Thus, we conclude that a significant fraction of female mice lacking Xist RNA is viable to term. This viability of the Xist-null animals was unexpected for several reasons. First, XCI is tightly coupled to cell differentiation both in vivo in the epiblast and ex vivo in ES cells (Monk and Harper 1979; Penny et al. 1996). Furthermore, on the basis of numerous in vivo genetic studies over the past 20 years, it is widely believed that proper dosage compensation is essential for developmental progression and the ultimate survival of female embryos (Marahrens et al. 1997; Hoki et al. 2009, 2011). Indeed, mice lacking Xist expression due to either a deletion of Xist itself (Marahrens et al. 1997; Kalantry et al. 2009; Namekawa et al. 2010) or mutations of essential regulators all die shortly after implantation (Lee 2000, 2002; Hoki et al. 2009, 2011; Shin et al. 2010). Deleting effectors of the dosage compensation machinery in other organisms often results in early embryonic lethality as well. For example, female roundworms lacking proteins of the dosage compensation machinery die in early embryonic development, and male fruit flies lacking proteins of the male-specific lethal (MSL) complex or the essential roX RNAs perish during early embryogenesis due to impaired dosage compensation (Cline and Meyer 1996). Finally, even female mice that are conditionally deleted for Xist only in the blood lineage die from highly lethal blood cancers with full penetrance (Yildirim et al. 2013). Therefore, the ability of female mice to withstand a near-total loss of Xist across all organs was indeed highly surprising.
The lone survivor is a mosaic of XX and XO cells
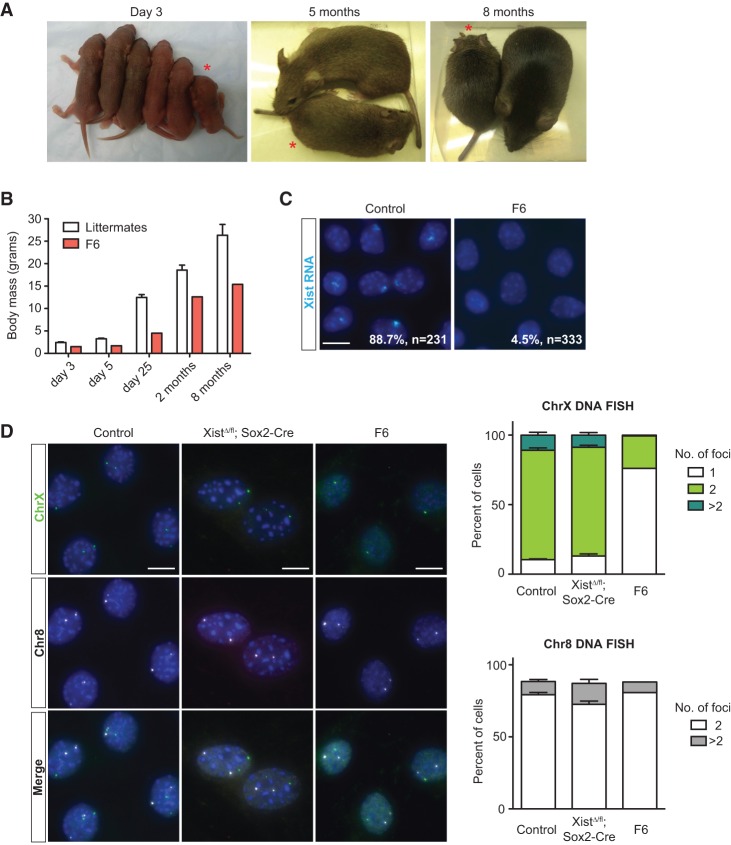
One mutant female (F6) survived to adulthood and remained viable at 1 yr of age (Figs. 2C, 3A). Like other Xist-null females, this animal was also runted at birth but gained substantial weight during the first 2 mo, although she consistently remained smaller than female littermates (Fig. 3B). Xist RNA-FISH performed on TTFs confirmed the absence of Xist clouds in >95% of cells (Fig. 3C). However, Xist DNA-FISH revealed that 76% of the diploid fibroblasts carried only one X chromosome (Fig. 3D). We suggest that the high percentage of “XO” cells in female F6 rescued the lethality associated with biallelic deletion of Xist, as, indeed, XO cells would not require X-linked dosage compensation. In contrast, mutant animals that perished were not evidently XO females (Fig. 3D). The ability of XO cells to rescue postnatal lethality supports the idea that the death of Xist-null mutants resulted from XCI-related effects and dosage imbalance.
Figure 3.
The surviving Xist-null female is a mosaic of XX and XO cells. (A) Xist mutant F6 and female littermates at P3 (day 3) and 5 mo and 8 mo of age. F6 is indicated by an asterisk. (B) Body weight of F6 and female littermate controls (n = 5) up to 8 mo of age. Data represent mean ± SEM. (C) Representative images for Xist RNA-FISH in TTFs of controls (XistWT/fl; Sox2-Cre) and F6 females. Percentages denote cells with one Xist cloud and, for controls, represent the average of two animals. (n) Number of nuclei counted. Bar, 10 µm. (D, left) Representative images for DNA-FISH using probes mapping to chromosome X and chromosome 8 in controls (XistWT/fl; Sox2-Cre), XistΔ/fl; Sox2-Cre animals, and female F6. (Right) Quantification for chromosome X (top) and chromosome 8 (bottom) DNA-FISH. At least 100 nuclei from each animal were counted. For chromosome X analysis, cells with more than two chromosome 8 foci were omitted. Data represent mean ± SEM. Controls, n = 3; XistΔ/fl; Sox2-Cre, n = 7. Bar, 10 µm.
Partial dosage compensation occurs in female mice lacking Xist RNA
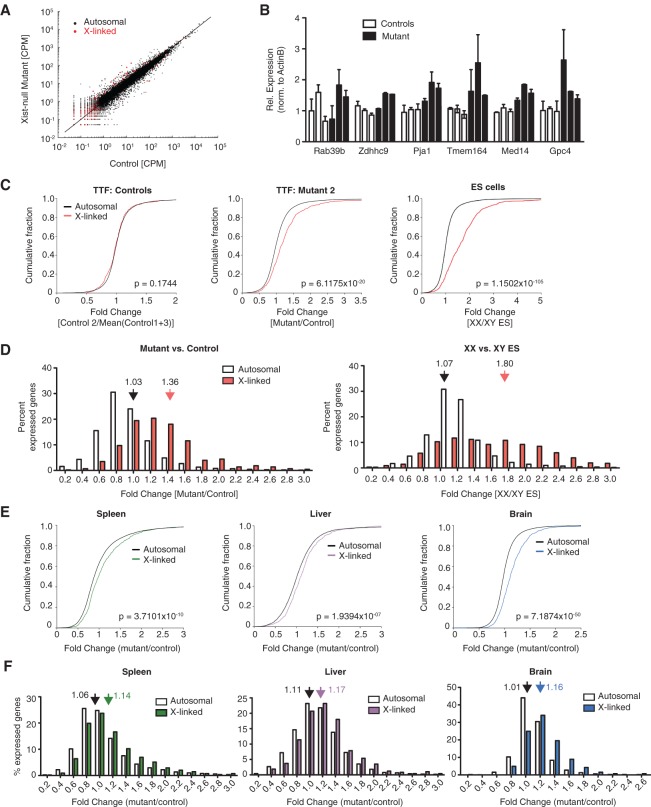
Given the ability of Xist-null mutants to survive to term with apparently normal embryogenesis, we investigated whether dosage compensation might have occurred. To this end, we performed RNA sequencing (RNA-seq) for primary TTFs derived from three mutants and three control animals. Scatter plots of gene expression (read counts per million [CPM]) showed that, whereas autosomal gene expression in mutants did not deviate from that in controls, X-linked gene expression showed a net positive change, as evidenced by the offset of red counts from the diagonal (Fig. 4A; Supplemental Fig. S2A). This observation suggested that the X chromosome was overexpressed in mutants and was supported by the orthogonal RT-qPCR approach that sampled a panel of X-linked genes from three mutants versus controls (Fig. 4B). Cumulative distribution plots of fold changes from RNA-seq revealed a statistically significant rightward shift for X-linked genes relative to autosomal genes (P < 0.05, Wilcoxon rank sum test) (Fig. 4C; Supplemental Fig. S2B). This was not observed for X-linked genes when comparing gene expression data from any one control with the average of two other controls. Probability density plots further argued for increased X-linked gene expression in mutant TTFs (filtered for CPM ≥ 1) (Supplemental Fig. S2C).
Figure 4.
Partial dosage compensation in Xist-null female mice. (A) Scatter plot for normalized read counts of all X-linked (red) and autosomal (black) genes in primary TTFs derived from control (XistWT/fl; Sox2-Cre) versus Xist-null mutant (XistΔ/fl; Sox2-Cre) mice. (B) RT-qPCR validation of RNA-seq data. Expression was normalized to β-actin. Data represent mean ± SEM of two replicate experiments. Data for six animals (three controls and three Xist-null mutants) are shown individually. (C) Cumulative distribution plots for fold changes in X-linked (red) and autosomal (black) genes in a control TTF line (relative to the average of other controls) (left), Xist-null TTF line (relative to the average of all controls) (middle), and female undifferentiated ES cells (relative to male ES cells) (right). Only genes with CPM ≥ 1 were considered. P-values given by Wilcoxon's rank sum test. (D, left) Distribution of X-linked (red) versus autosomal (white) fold changes in Xist-null TTFs relative to control cells. To minimize the effects due to noise, only genes with CPM ≥ 1 were considered. Fold changes are binned in steps of 0.2 (i.e., label of 0.2 on the X-axis includes all genes with fold changes between 0.0 and 0.2). Average fold changes of X-linked or autosomal genes are indicated. (Right) Distribution of X-linked (red) versus autosomal (white) fold changes in XX female ES cells relative to XY male ES cells. Average fold changes of X-linked or autosomal genes are indicated. (E) Cumulative distribution plots for fold changes in X-linked and autosomal genes (black) for the spleen (left), liver (middle), and brain (right) of the P1 Xist-null mutant relative to the average of age-matched control females. n = 2. Only genes with CPM ≥ 1 were considered. P-values were given by Wilcoxon's rank sum test. (F) Distribution of X-linked versus autosomal fold changes for the spleen (left), liver (middle), and brain (right) of the Xist-null mutant from D relative to age-matched control females. n = 2. Only genes with CPM ≥ 1 were considered. Average fold changes of X-linked or autosomal genes are indicated.
We then analyzed gene expression changes by binning fold change values in increments of 0.2 (Fig. 4D). A substantial right shift in X-gene expression was also evident (average fold change 1.36) (Fig. 4D, left panel), whereas this was not observed for autosomal genes (average fold change 1.03) (Fig. 4D, left panel). Interestingly, although ∼12.5% of expressed X-linked genes were up-regulated by ≥1.8-fold, most changes fell between 1.0-fold and 1.8-fold. In fact, the average fold change was only 1.36—a value considerably below the doubling of X-linked expression expected of cells with two active X chromosomes (Fig. 4D, right panel, Supplemental Fig. S2C). However, our empiric analyses showed that this theoretical doubling does not occur even in undifferentiated female ES cells that carry two active X chromosomes. In actuality, X-linked expression in female ES cells was, on average, 1.8-fold that of the single active X chromosome in male cells (Fig. 4D, right panel), a value in close agreement with previous studies (Nguyen and Disteche 2006; Lin et al. 2007; Deng et al. 2011; Kharchenko et al. 2011; Yildirim et al. 2012).
With the exception of F6 (who is partially XO), none of the mutant females survived beyond P24. Even in a non-nutrient-limiting environment, the Xist-null mice succumbed to sudden death. We examined this lethality more closely by sampling three different tissues (spleen, liver, and brain) from mutant and control females at P1 for RNA-seq. In agreement with data from fibroblasts, comparison of X versus autosomal gene expression changes showed significantly elevated X-linked gene expression in all three tissues (P < 0.05, Wilcoxon rank sum test) (Fig. 4E,F). As with TTFs, the degree of up-regulation was modest in most X-linked genes. There was some variability in the severity of imbalance, with brain tissue demonstrating the most pronounced changes, while the liver showed the least deviation from control samples (presumably due to residual Xist-expressing cells) (Fig. 1H). The tempered effect in the liver suggests that this tissue may be less tolerant of Xist loss and select for progenitors that retain at least one copy of Xist. Taken together, our data demonstrate that female mice lacking Xist show partial dosage compensation.
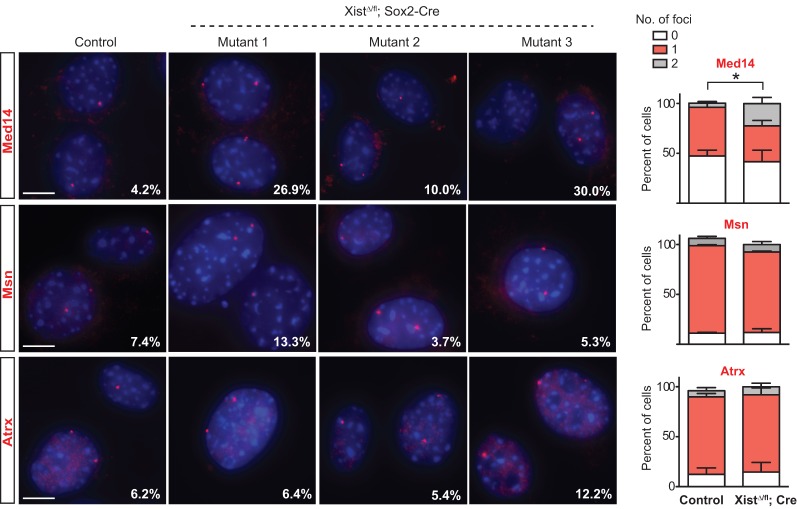
In principle, X-linked up-regulation could result from increased expression from a single X or partial expression from both Xs. To distinguish between these possibilities, we performed RNA-FISH for three X-linked genes—Med14, Msn, and Atrx—in mutant and control TTFs (Fig. 5). Med14, a gene identified to be up-regulated in the mutant TTF lines, showed significantly increased biallelic expression from 4.2% of control cells to up to 30% of mutant cells (P = 0.044) (Fig. 5). We also noted an increase in biallelic Msn expression (up to 13.3%) in the mutant TTF line, whose RNA-seq showed up-regulated bulk Msn RNA relative to controls (7.4%). This was not observed in mutant TTFs that did not demonstrate increased bulk Msn levels (mutants 2 and 3). Similarly, expression of Atrx RNA remained mostly monoallelic, correlating with minimal changes in Atrx levels observed by RNA-seq analysis. These results demonstrate a correlation between X-linked up-regulation and occurrence of biallelic expression and also imply the presence of an Xist-independent mechanism of dosage compensation in mice.
Figure 5.
RNA-FISH analysis of up-regulated X-linked genes. Representative RNA-FISH images for Med14, Msn, and Atrx. Data for three Xist-null TTF lines are shown. Percentages denote cells with two foci. At least 100 nuclei were counted per sample. RNA-FISH results are quantified in the graphs for each gene. The percentage of cells with two Med14 foci is significantly increased in Xist-null TTFs. t-test, P = 0.044. Data represent mean ± SEM. n = 3 per genotype. Bar, 10 µm.
X up-regulation tends to occur in regions with normally high Xist density
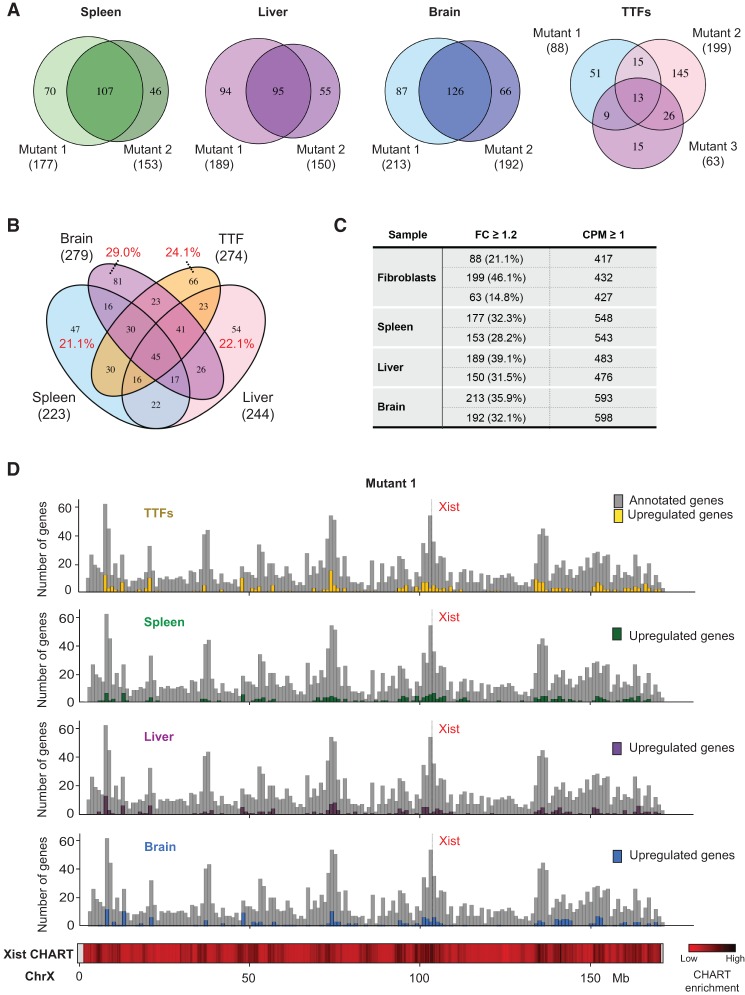
We asked whether commonalities existed among X-linked genes that were up-regulated (≥1.2-fold, CPM ≥ 1) across the four cell/tissue types profiled. In each tissue type, hundreds of X-linked genes were up-regulated (Fig. 6C; Supplemental Tables S1, S2). There was extensive overlap between biological replicates and between tissue types (Fig. 6A,B). Gene ontology (GO) analysis for up-regulated genes revealed consistent enrichment (adjusted P < 0.05) of factors implicated in chromatin regulation, chromatin modification, and chromatin/chromosome organization for TTFs, the liver, and the brain (Supplemental Fig. S3). The cluster of transcription elongation factor A-like genes (Tceal5, Tceal6, Tceal7, and Wbp5) on the X chromosome was enriched in the brain. Interestingly, genes known to escape X inactivation were also up-regulated. At the genome-wide level, differential expression analysis identified 266, 290, and 98 differentially expressed genes (DEGs) from the spleen, liver, and brain, respectively (false discovery rate < 0.05) (Supplemental Table S3A–C). GO analysis for DEGs did not identify significant enrichment for any processes in the liver and brain. However, DEGs in the spleen were highly enriched for proteases (Supplemental Table S3D).
Figure 6.
Up-regulated X-linked genes show significant overlap across tissues and correlate with high-density Xist-binding sites observed in normal cells. (A) Venn diagrams showing overlap in up-regulated (fold change [FC] ≥ 1.2) X-linked genes between two (for tissues) or three (for fibroblasts) biological replicates. The total number of up-regulated genes for each sample is indicated in parentheses. (B) Venn diagram showing overlap in up-regulated (fold change ≥ 1.2) X-linked genes across the spleen, liver, heart, and fibroblasts. The total number of up-regulated genes for each cell/tissue type is indicated in parentheses. The percentages refer to the fraction of up-regulated genes specific to one cell/tissue type. (C) The number of up-regulated X-linked genes in each cell/tissue type, along with the total number of expressed genes in the corresponding sample. Replicates are shown individually. (D) Histogram (in 1-Mb bins) showing the number of up-regulated genes and their locations along the X chromosome. Gene density is plotted in gray. Heat map for Xist CHART data performed for day 3 differentiating female ES cells shown (Simon et al. 2013).
The overlap in up-regulated X-linked genes across different cell/tissue types suggested that some chromosomal domains may be more prone to dosage imbalance in the absence of Xist. To test this idea, we divided the X chromosome into 1-Mb bins and computed the frequency of gene up-regulation in each 1-Mb segment (Fig. 6D; Supplemental Fig. S4). Up-regulated genes were found throughout the chromosome, and their occurrence was correlated with gene density (r = 0.53–0.68) (Supplemental Fig. S5). Up-regulated genes were also generally correlated with regions with high-level Xist RNA binding, as determined by correlation analysis with published CHART-seq data sets (Simon et al. 2013). Thus, up-regulated genes fell within or near regions targeted by Xist RNA in female cells undergoing XCI.
Maturation defects of the spleen and heart
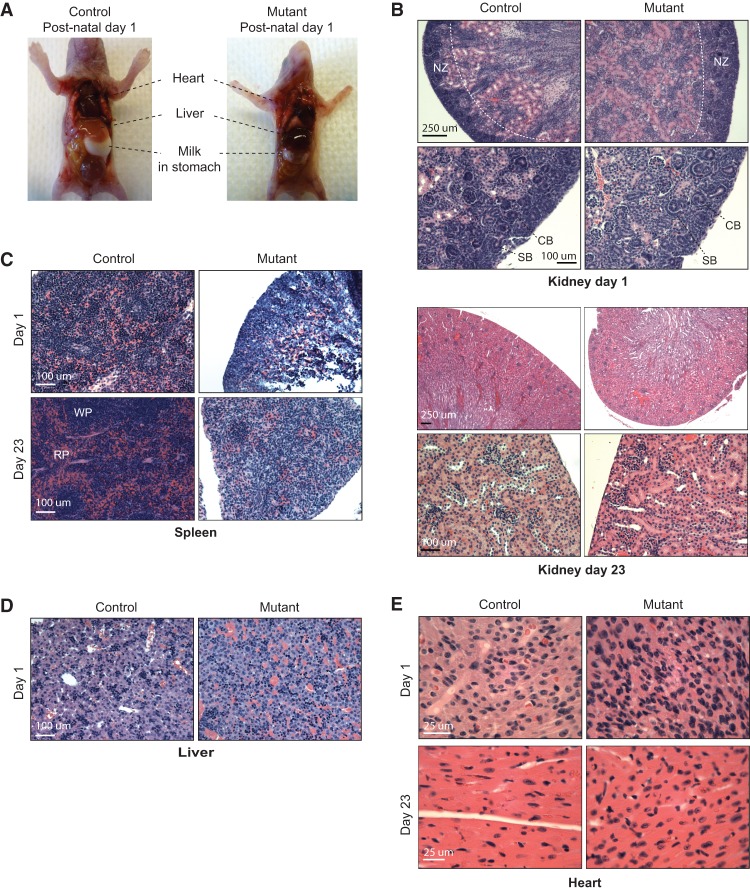
Finally, to investigate proximal causes of death, we analyzed tissues from moribund pups and age-matched controls at P1 and P23. Mutants appeared grossly normal except for a general reduction in organ size, in keeping with the smaller stature of these animals (Fig. 7A; data not shown). This indicated that body patterning and organogenesis were largely normal in spite of impaired dosage compensation. Hematoxylin and eosin (H&E) staining of the kidney, lung, and brain did not reveal any remarkable differences between Xist-null females and controls (Supplemental Fig. S6A,B). In mice, postnatal developmental staging could be performed using the kidney, where the nephrogenic zone (consisting of immature glomeruli and nephrons undergoing proliferation) at P1 normally gives way to mature cortex and fully formed glomeruli by P23 (Little et al. 2007; Rinkevich et al. 2014; Romagnani et al. 2015). This postnatal transition occurred in the kidneys of both mutant and control females (Fig. 7B). Thus, Xist-deficient kidneys could in fact progress normally on a postnatal developmental time scale.
Figure 7.
Xist-null mutants are grossly normal but show developmental defects of the spleen and heart. (A) Xist-null mutant and female control littermates at P1. (B) H&E stainings for kidneys from one mutant and one control animal at P1 (top panel) and P23 (bottom panel). For each animal, H&E stainings are shown at two magnifications. Bars: lower magnification (4× or 10× objectives), 250 µm; higher magnification (20× objectives), 100 µm. (NZ) Nephrogenic zone; (CB) comma-shaped body; (SB) S-shaped body. (C) H&E stainings for spleens from one mutant and one control animal at P1 (day 1) and P23 (day 23). Bars (20× objective), 100 µm. (RP) Red pulp; (WP) white pulp. (D) H&E stainings for livers from one mutant and one control animal at P1 (day 1). Bars (20× objective), 100 µm. (E) H&E stainings for cardiac muscle from one mutant and one control animal at P1 (day 1) and P23 (day 23). Bars (40× objective), 25 µm.
However, in spite of the capacity to progress postnatally, mutant females displayed abnormalities in several organs. First, the spleen failed to show the development of distinctive red and white pulp zones that was observed in controls on day 23 (Fig. 7C). In fact, between days 1 and 23, mutant spleens remained the same size grossly and were disproportionately small. There was also a lack of histological organization (Fig. 7C, note the lack of red and white pulp). The splenic phenotype is consistent with hematologic abnormalities observed in mouse models that either overexpress Xist or conditionally delete Xist in the blood compartment (Savarese et al. 2006; Yildirim et al. 2013) and further supports a critical role of Xist expression and dosage compensation for hematopoietic cells.
We also noticed abnormalities in the liver, with a darkening suggestive of hepatic congestion (Fig. 7A). Histologic analysis confirmed a hepatopathy consistent with venous congestion (Fig. 7D). Such venous congestion is often associated with right-sided cardiac failure. Interestingly, histologic examination of the heart revealed overall features of developmental delay at both day 1 and day 23 (Fig. 7E). During normal mouse development, the perinatal heart usually increases in size through hypertrophy of individual myocytes as the cytoplasm undergoes a dramatic enlargement (Naqvi et al. 2014; Alkass et al. 2015; Soonpaa et al. 2015). However, we observed overall smaller sizes of the heart that correspond to histologically immature cardiomyocytes in the mutant animals. Cells demonstrated a much higher nuclear to cytoplasmic ratio and had correspondingly more densely packed nuclei typical of prenatal embryonic hearts (Fig. 7E). Mutant cardiomyocytes therefore demonstrated features of an earlier development state. Aberrant development of cardiac tissue might thus have been inadequate to support the increasing circulatory demands of the neonates. We propose that aberrant postnatal development in the cardiac and hematologic compartments at least partially explains the observed sudden death of Xist-deficient females.
Discussion
Work by Takagi and Abe (1990) previously showed that the persistent presence of two active X chromosomes is detrimental to early mouse development. The nature of their mouse model (involving a truncated X chromosome), however, made it difficult to distinguish between the effects of partial X monosomy during imprinted XCI versus increased dosage of X-linked genes during random XCI. Xist-null ES cells have been reported to demonstrate slower differentiation kinetics compared with their female wild-type counterparts, but no other phenotypes have been noted (Schulz et al. 2014). Here, our goal was to examine the effects of total-body Xist loss by decoupling the known effects on imprinted XCI in extraembryonic tissues (Marahrens et al. 1997) from those on random XCI in the soma. Surprisingly, deleting both copies of Xist during early embryogenesis did not preclude female embryonic development and organogenesis. A quarter of the Xist-null mice survived to term. Since the conditional deletion was driven by a Sox2-Cre recombinase, the loss most likely occurred prior to the initiation of XCI in pregastrulation embryos. Indeed, Xist ablation is mostly complete by E8.5, and the recovery of XistΔ/fl; Sox2-Cre embryos already displaying developmental defects at this stage points to loss of Xist at early time points, either before or around the time of XCI initiation (Fig. 2). This is further supported by extremely skewed XCI patterns in heterozygous Xist mutants (Fig. 1C), indicative of a primary choice for the wild-type X to become the inactive X chromosome. Nevertheless, we cannot rule out deletion events occurring after the establishment of XCI in rare cells where Cre recombinase might persist briefly. Although the proportion of cells with pre-XCI versus post-XCI deletion events could vary between embryos, we suspect that a large number of cells in any embryo never initiated XCI through an Xist-dependent mechanism.
Another surprising finding to emerge from this study is that, while hundreds of X-linked genes were consistently overexpressed across multiple organs and tissues, the magnitude of X-linked overexpression fell far short of the expected doubling that would be commensurate with two active X chromosomes. In plants and fruit flies, it has long been observed in aneuploids that changes in chromosomal dosage do not scale linearly with changes in transcriptional output (Devlin et al. 1988; Birchler et al. 1990; Guo and Birchler 1994; Stenberg et al. 2009). This intrinsic dosage-balancing system is proposed to stem from inherent buffering capabilities that evolved to reduce genetic imbalances in the absence of a specific dosage-correcting mechanism (Birchler 2013). The phenomenon extends to X-linked genes (Birchler et al. 1989; Sun et al. 2013). In both plants and flies, it is observed that creating trisomies across large regions of many chromosomes results in reduced expression of genes within those regions—leading to coinage of the term “inverse effect” for the paradoxical decreased per-allele output when there is increased copy number. We suggest that a similar phenomenon occurs in the Xist-null mice by which failure to neutralize the extra copy of the X chromosome in females invokes an inherent compensatory system to reduce the imbalance between X and autosomal gene dosages.
We do not know what the backup system is, although previous studies already hinted at various possible Xist-independent mechanisms (Kalantry et al. 2009; Namekawa et al. 2010). However, the backup system does not achieve full dosage parity. All mice eventually succumb. The pathological effects on splenic and perinatal cardiac development are especially intriguing, and an inability to meet increased circulatory and immunological demands of the growing neonate could potentially account for sudden death. In conclusion, our study demonstrates that the loss of Xist during random XCI triggers a dosage imbalance of X-linked versus autosomal genes. Although the magnitude of overexpression ranged only between 10% and 30% overall, this small deviation from dosage equivalence resulted in significantly decreased overall organismal fitness.
Materials and methods
Animal studies
Xistfl/fl mice (in a 129Sv/Jae background) were a gift from R. Jaenisch (Csankovszki et al. 1999). Tg(Sox2-Cre)1Amc/J animals (Jax 004783) were procured from the Jackson Laboratory. All progeny are maintained on a 129Sv/Jae and C57BL/6 mixed background. Genotypings of the XistWT and Xistfl alleles were carried out using the 370/389 primer pair. Sox2-Cre genotyping was performed using the Cre-3F/3R primer pair. For harvesting of embryos, pregnant females were humanely sacrificed at 8.5 d post-coitum (dpc) (for XistΔ/WT; Sox2-Cre) or 14.5 dpc (for Xistfl/fl). The morning of copulation plug detection was considered 0.5 dpc. Sexing of embryos was performed by PCR using the NS18/19 (Tsix) and Zfy1.8b/YNLS.5 (Zfy1) primer pairs and was confirmed by DNA-FISH using a Y-chromosome BAC probe (see below). Primers are listed in Supplemental Table S4. Embryonic fibroblasts were isolated using standard procedures. Mouse husbandry and experiments were carried out as stipulated by the Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC).
Isolation of TTFs
One centimeter of tail tissue was snipped from neonates between P1 and P3. Tissue was rinsed sequentially in betadine solution, 70% ethanol, and 2× penicillin/streptomycin and minced finely. The slurry was incubated with TrypLE Select (Thermo Fisher Scientific) for 30 min at 37°C, plated, and cultured at physiological O2 (4.0% O2, 5.0% CO2).
DNA-FISH, RNA-FISH, and immunostaining
For TTFs, cells were grown on glass coverslips and fixed in 4% paraformaldehyde before permeabilization with 0.2% Triton-X in PBS at room temperature. For E8.5 embryos, cells were dispersed using TrypLE Select (Thermo Fisher Scientific) and immobilized onto glass slides by centrifugation (Shandon Cytospin). Cells were pre-extracted in ice-cold cytoskeletal buffer with 0.5% Triton-X and then fixed with 4% paraformaldehyde. DNA-FISH and RNA-FISH were performed according to established protocols (Zhang et al. 2007; Sunwoo et al. 2015). The probes used for RNA-FISH were Xist-, Med14-, and Msn-labeled oligo probes; the probe for Atrx was nick translation of BAC RP23-450B21. The probes used for DNA-FISH were RP24-386O11 for chromosome 8, RP24-148H21 for chromosome X, and RP24-332J21 for chromosome Y (all BAC probes labeled by nick translation) and nick translation of PCR product from Xist1.3F/Xist1.3R primers for the Xist locus between loxP sites (Supplemental Table S4). Mouse monoclonal H3K27me3 antibody (1:300 in 1% BSA/PBS; Active Motif, 39535) was used for immunostaining. Images were acquired with a Nikon Eclipse 90i microscope and a Hamamatsu CCD camera. Image analysis was performed using Volocity (Perkin-Elmer).
RNA isolation from mouse tissues, reverse transcription, and qPCR
Mouse tissues (∼100 mg) were collected into RNALater (Qiagen) at the time of dissection and mechanically homogenized in Trizol using a Tissue Lyzer II (Qiagen) at 25 Hz twice for 2 min. RNA is isolated by phenol–chloroform extraction. For reverse transcription, cDNA was prepared using SuperScript III (Thermo Fisher Scientific) with random hexamers (Promega). qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad), and relative gene expression was calculated by the standard curve method. The primers used are listed in Supplemental Table S4.
Strand-specific RNA-seq
Four micrograms of total RNA (RIN value ≥8) from fibroblasts or mouse tissues was depleted of ribosomal RNA (RiboMinus eukaryote kit version 2, Thermo Fisher Scientific) as per the manufacturer's recommendations, DNase-treated for 30 min at room temperature, and purified using an RNeasy minelute cleanup kit (Qiagen). Eluted RNA was then quantified (Quant-iT RiboGreen RNA assay, Thermo Fisher Scientific), and 30 ng of RNA was fragmented in 6 mM MgCl2 buffer for 10 min at 94°C before first strand (SuperScript III, Thermo Fisher Scientific) and second strand (NEBNext Ultra Directional RNA second strand synthesis module, New England Biolabs) cDNA synthesis. Adaptor oligos were ligated to the cDNA library (NEBNext Chip-seq library master mix, New England Biolabs). Libraries were amplified with multiplexing barcodes (NEBNext Multiplex Oligos for Illumina, New England Biolabs) and quantified (KAPA library quantification kit, Kapa Biosystems) before 50-nucleotide paired-end sequencing was performed on an Illumina HiSeq 2000 or HiSeq 2500. All sequencing data can be accessed via Gene Expression Omnibus under accession number GSE84665.
RNA-seq analysis
Sequencing reads were first checked for quality by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) before alignment with TopHat2 (Kim et al. 2013) to the mouse reference genome (GRCm38/mm10). Read counting was performed with HTseq-count (Anders et al. 2015) against all ENSEMBL transcripts (ENSEMBL release 84). Normalization and differential expression analysis were performed using the EdgeR package (Robinson et al. 2010). For analyzing distributions of gene expression fold changes on chromosome X and autosomes, only genes with CPM ≥ 1 were considered. For undifferentiated ES cell analysis, a published RNA-seq data set (Gendrel et al. 2014) (SRA124266) was downloaded from the European Nucleotide Archive and processed similarly for analysis.
Histopathology
Tissues were fixed in 10% neutral buffered formalin (Fisher Scientific) overnight and stored in 70% ethanol for further processing. Sections were stained with H&E. Images were acquired using a Nikon Eclipse 90i microscope and a Q-imaging MicroPublisher RTV color camera. Image analysis was performed with Volocity (Improvision, Perkin-Elmer).
Supplementary Material
Acknowledgments
We thank R. Jaenisch for Xistfl/fl mice; R. Mostoslavsky, K. Hochedlinger, W. Bender, B. Payer, and the Lee laboratory for valuable advice and feedback; D. Lessing, T. Dial, and B. del Rosario for critical reading of the manuscript, and W. Press for mouse colony maintenance. This work was supported by grant funding from the Agency for Science, Technology, and Research of Singapore (to L.Y.), the National Institutes of Health (DA-R01-36895 to J.T.L.), and the Rett Syndrome Research Trust (to J.T.L.). J.T.L. is also an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.281162.116.
References
- Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, Bergmann O. 2015. No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163: 1026–1036. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. 2013. Aneuploidy in plants and flies: the origin of studies of genomic imbalance. Semin Cell Dev Biol 24: 315–319. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Hiebert JC, Krietzman M. 1989. Gene expression in adult metafemales of Drosophila melanogaster. Genetics 122: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Hiebert JC, Paigen K. 1990. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics 124: 679–686. [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. 1996. Vive la difference: males vs females in flies vs worms. Annu Rev Genet 30: 637–702. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22: 323–324. [DOI] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Holm DG, Grigliatti TA. 1988. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 118: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. 2012. Dosage compensation of the sex chromosomes. Annu Rev Genet 46: 537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RL, Lyon MF. 1971. Biological sciences: X chromosome inactivation studied by injection of a single cell into the mouse blastocyst. Nature 231: 385–386. [DOI] [PubMed] [Google Scholar]
- Gendrel A-V, Attia M, Chen C-J, Diabangouaya P, Servant N, Barillot E, Heard E. 2014. Developmental dynamics and disease potential of random monoallelic gene expression. Developmental Cell 28: 366–380. [DOI] [PubMed] [Google Scholar]
- Guo M, Birchler JA. 1994. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266: 1999–2002. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 119: S97–S101. [DOI] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. 2009. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136: 139–146. [DOI] [PubMed] [Google Scholar]
- Hoki Y, Ikeda R, Mise N, Sakata Y, Ohhata T, Sasaki H, Abe K, Sado T. 2011. Incomplete X-inactivation initiated by a hypomorphic Xist allele in the mouse. Development 138: 2649–2659. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. 2009. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi R, Park PJ. 2011. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet 43: 1167–1169. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103: 17–27. [DOI] [PubMed] [Google Scholar]
- Lee JT. 2002. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat Genet 32: 195–200. [DOI] [PubMed] [Google Scholar]
- Lee JT. 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12: 815–826. [DOI] [PubMed] [Google Scholar]
- Lin H, Gupta V, VerMilyea MD, Falciani F, Lee JT, O'Neill LP, Turner BM. 2007. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol 5: e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, et al. 2007. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns 7: 680–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Lyon MF, Searle AG, Ford CE, Ohno S. 1964. A mouse translocation suppressing sex-linked variegation. Cytogenet Genome Res 3: 306–323. [DOI] [PubMed] [Google Scholar]
- Maduro C, de Hoon B, Gribnau J. 2016. Fitting the puzzle pieces: the bigger picture of XCI. Trends Biochem Sci 41: 138–147. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Martin GR, Epstein CJ, Travis B, Tucker G, Yatziv S, Martin DW, Clift S, Cohen S. 1978. X-chromosome inactivation during differentiation of female teratocarcinoma stem cells in vitro. Nature 271: 329–333. [DOI] [PubMed] [Google Scholar]
- Monk M, Harper MI. 1979. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281: 311–313.551278 [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. 2010. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol 30: 3187–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, et al. 2014. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 157: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Montoro DT, Contreras-Trujillo H, Harari-Steinberg O, Newman AM, Tsai JM, Lim X, Van-Amerongen R, Bowman A, Januszyk M, et al. 2014. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep 7: 1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani P, Rinkevich Y, Dekel B. 2015. The use of lineage tracing to study kidney injury and regeneration. Nat Rev Nephrol 11: 420–431. [DOI] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. 2006. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol 26: 7167–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Blüthgen N, Heard E. 2014. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14: 203–216. [DOI] [PubMed] [Google Scholar]
- Senner CE, Nesterova TB, Norton S, Dewchand H, Godwin J, Mak W, Brockdorff N. 2011. Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation. Development 138: 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, et al. 2010. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature 467: 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504: 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, Zebrowski DC, Platt C, Rosenzweig A, Engel FB, Field LJ. 2015. Cardiomyocyte cell-cycle activity during preadolescence. Cell 163: 781–782. [DOI] [PubMed] [Google Scholar]
- Starmer J, Magnuson T. 2009. A new model for random X chromosome inactivation. Development 136: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P, Lundberg LE, Johansson AM, Ryden P, Svensson MJ, Larsson J. 2009. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Johnson AF, Li J, Lambdin AS, Cheng J, Birchler JA. 2013. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc Natl Acad Sci 110: 16514–16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Wu JY, Lee JT. 2015. The Xist RNA–PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc Natl Acad Sci 112: E4216–E4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N. 1980. Primary and secondary nonrandom X chromosome inactivation in early female mouse embryos carrying Searle's translocation T(X; 16)16H. Chromosoma 81: 439–459. [DOI] [PubMed] [Google Scholar]
- Takagi N, Abe K. 1990. Detrimental effects of two active X chromosomes on early mouse development. Development 109: 189–201. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Takagi N, Wake N, Sasaki M. 1978. Cytologic evidence for preferential inactivation of the paternally derived X chromosome in XX mouse blastocysts. Cytogenet Genome Res 20: 240–248. [DOI] [PubMed] [Google Scholar]
- West JD, Frels WI, Chapman VM, Papaioannou VE. 1977. Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell 12: 873–882. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Sadreyev RI, Pinter SF, Lee JT. 2012. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol 19: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. 2013. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.