Fu et al. show that Drosophila period (dper) codon usage is important for circadian clock function. Codon optimization of dper resulted in conformational changes of dPER protein, altered dPER phosphorylation profile and stability, and impaired dPER function in the circadian negative feedback loop, which manifests into changes in molecular rhythmicity and abnormal circadian behavioral output.
Keywords: circadian clock, codon usage, Drosophila, period, protein structure
Abstract
Codon usage bias is a universal feature of all genomes, but its in vivo biological functions in animal systems are not clear. To investigate the in vivo role of codon usage in animals, we took advantage of the sensitivity and robustness of the Drosophila circadian system. By codon-optimizing parts of Drosophila period (dper), a core clock gene that encodes a critical component of the circadian oscillator, we showed that dper codon usage is important for circadian clock function. Codon optimization of dper resulted in conformational changes of the dPER protein, altered dPER phosphorylation profile and stability, and impaired dPER function in the circadian negative feedback loop, which manifests into changes in molecular rhythmicity and abnormal circadian behavioral output. This study provides an in vivo example that demonstrates the role of codon usage in determining protein structure and function in an animal system. These results suggest a universal mechanism in eukaryotes that uses a codon usage “code” within genetic codons to regulate cotranslational protein folding.
Most amino acids are encoded by two to six synonymous codons. Preferential use of certain synonymous codons, a phenomenon called codon usage bias, was found in all genomes (Ikemura 1985; Sharp et al. 1986; Comeron 2004; Plotkin and Kudla 2011). Selection for efficient and accurate translation was proposed to be a major cause for codon usage bias (Akashi 1994; Drummond and Wilke 2008; Hershberg and Petrov 2008; Gingold and Pilpel 2011; Plotkin and Kudla 2011). However, direct experimental evidence supporting such a hypotheses is still not available. By studying the heterologous protein expression in Escherichia coli, it was suggested that translation rate and synonymous codon usage can affect protein folding and functions (Komar et al. 1999; Zhang et al. 2009; Siller et al. 2010; Spencer et al. 2012). Strongly supporting this hypothesis, we previously showed that the codon usage bias in the Neurospora circadian clock gene frequency (frq) is critical for the structure and function of FRQ in vivo (Zhou et al. 2013). More recently, we demonstrated that codon usage regulates the speed of mRNA translation elongation and, by doing so, affects cotranslational protein folding in Neurospora (Yu et al. 2015). Bioinformatics analyses have uncovered correlations between codon usage and the potential to form certain protein structural motifs (Zhou et al. 2009; Pechmann and Frydman 2013; Pechmann et al. 2014; Zhou et al. 2015). Together, these studies led to the hypothesis that there is a codon usage “code” within genetic codons to allow proper folding of proteins.
Optimization of part of frq codon usage in Neurospora resulted in altered FRQ structure and the abolishment of clock function (Zhou et al. 2013, 2015). Surprisingly, however, codon optimization of the cyanobacterial circadian clock genes kaiC and kaiB resulted in more robust clock function, albeit with impaired growth under certain conditions (Xu et al. 2013). To reconcile the different results of these two studies, we compared the predicted FRQ and Kai protein structures and found that, while most of the FRQ is predicted to be unstructured, Kai proteins are highly structured with known high-resolution crystal structures (Johnson et al. 2011). Such differences raise the possibility that codon usage has distinct effects on different types of protein structures. Consistent with this hypothesis, our genome-wide bioinformatic analyses showed that regions likely to be well structured are encoded more by preferred codons, while predicted unstructured protein regions are preferentially encoded by nonoptimal codons (Zhou et al. 2015). However, there were no in vivo genetic and biochemical studies further supporting this hypothesis in a nonfungal system.
Previous studies on codon usage were based mostly on bioinformatics analyses and experimental studies in microorganisms. Biological functions of codon usage in animal systems are not clear. It is well known that many human diseases were found to be associated with silenced single-nucleotide proteins (SNPs). Previously, a single synonymous SNP in the human multidrug resistance 1 (MDR1) gene was found to result in altered protein conformation and function (Kimchi-Sarfaty et al. 2007), suggesting a role for codon usage in affecting protein structure.
In this study, we used the Drosophila melanogaster system to determine whether codon usage is a universal mechanism for protein structure and function determination in eukaryotes. Similar to Neurospora, the D. melanogaster genome has a strong codon bias for G/C at the wobble positions, with an average gene codon bias index (CBI) of 0.23 (Kanaya et al. 2001; Hambuch and Parsch 2005; Heger and Ponting 2007; Zhou et al. 2013, 2015). A positive correlation between codon usage and gene expression levels was observed previously in flies (Duret and Mouchiroud 1999). In addition, introduction of unpreferred codons in the alcohol dehydrogenase (Adh) gene in vivo led to reduced protein levels (Carlini and Stephan 2003). As in our studies in Neurospora, we reasoned that the sensitivity and robustness of the Drosophila circadian system would allow us to uncover the role of codon usage in vivo. In the Drosophila circadian system, the CLOCK/CYCLE (CLK/CYC) complex is the positive element in the core circadian negative feedback loop that activates the transcription of Drosophila period (dper) and other clock-controlled genes (Supplemental Fig. S1; Young and Kay 2001; Allada and Chung 2010; Hardin and Panda 2013; Tataroglu and Emery 2015). On the other hand, dPER functions as a negative element with TIMELSS (TIM) in the circadian negative feedback loop by interacting with and repressing the activity of the CLK/CYC complex to inhibit the transcription of dper and other CLK/CYC target genes. After its synthesis, dPER is progressively phosphorylated by DOUBLETIME (DBT) and other kinases (Kloss et al. 1998; Price et al. 1998; Preuss et al. 2004; Bae and Edery 2006; Gallego and Virshup 2007; Ko et al. 2010; Chiu et al. 2011). A major function of DBT-mediated dPER phosphorylation is to trigger binding to SLIMB (β-TrCP in mammals), which promotes dPER ubiquitination and subsequent degradation by the proteasome pathway (Grima et al. 2002; Ko et al. 2002; Ko and Edery 2005). After dPER degradation, CLK/CYC is then released from inhibition to start the next round of transcriptional activation of dper and other target genes, resulting in rhythmic transcription.
By codon-optimizing parts of the dper ORF, we showed that dper codon usage is critical for its circadian clock function. Optimization of dper codon usage resulted in conformational changes of the dPER protein, altered dPER phosphorylation profile and stability, and impaired dPER repressor function in the circadian negative feedback loop. Our study establishes an in vivo example that demonstrates the role of codon usage in determining protein structure and function in an animal system and suggests a universal mechanism in eukaryotes that uses codon usage to regulate cotranslational protein folding.
Results
N-terminal codon optimization of per impairs circadian behavioral rhythms
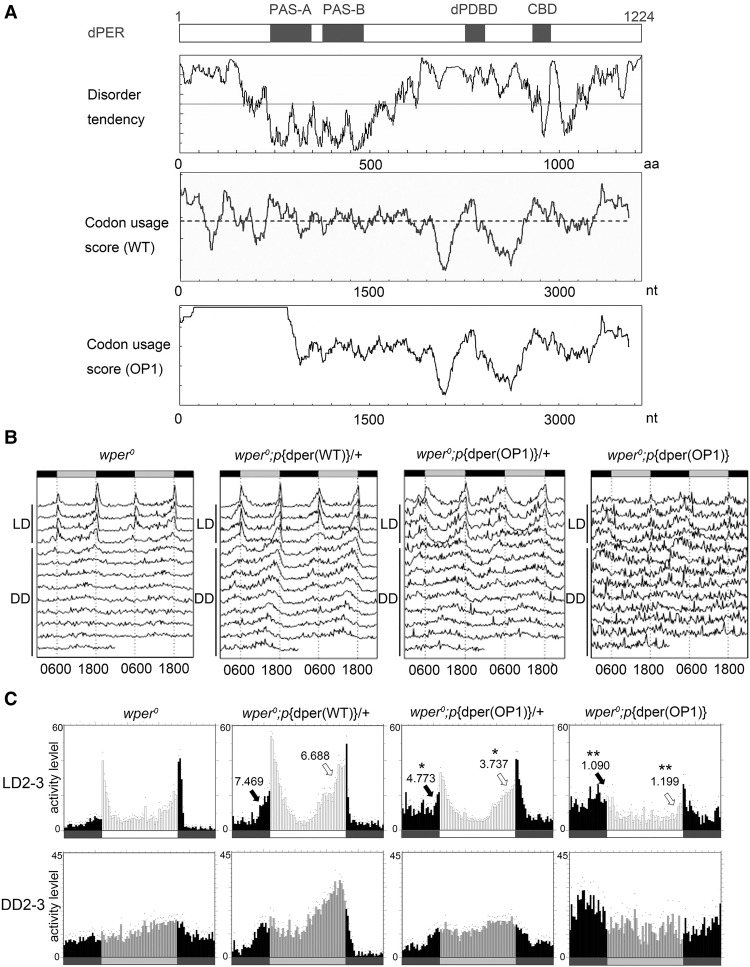
To analyze the codon usage of the dper gene, we calculated codon usage scores based on the codon adaptation index (CAI) through its ORF (Fig. 1A; Supplemental Fig. S2; Sharp and Li 1987). In addition, we used the protein secondary structure prediction program IUPred (http://iupred.enzim.hu) to predict the locations of intrinsically unstructured/disordered residues of the dPER protein. Similar to the Neurospora FRQ protein (Zhou et al. 2013, 2015), most of dPER protein, with the exception of the two known PAS (PERIOD–ARNT–SIM) domains (Huang et al. 1993), was predicted to be intrinsically disordered. This prediction is consistent with the fact that the PAS domains are the only regions of dPER with a known crystal structure (Hennig et al. 2009; Merbitz-Zahradnik and Wolf 2015). Interestingly, the predicted intrinsically disordered regions in the N-terminal and middle part of dPER have domains with relatively low codon usage scores. This is consistent with our previous observation that predicted intrinsically disordered regions preferentially use nonoptimal codons (Zhou et al. 2015), raising the possibility of a role for codon usage in cotranslational protein folding in these regions.
Figure 1.
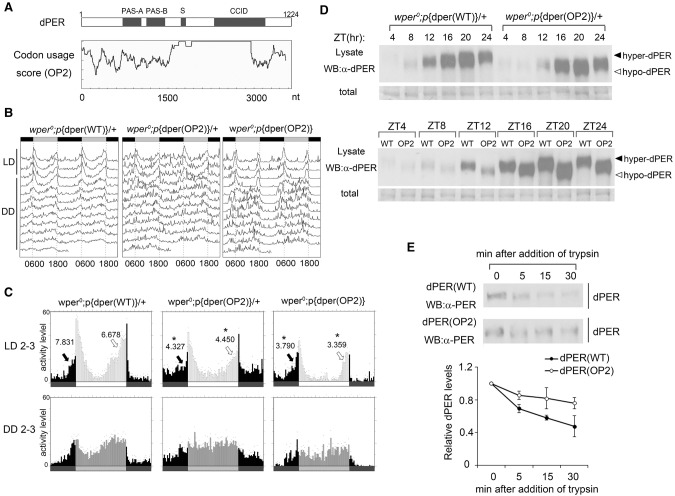
Codon optimization of the N-terminal part of dper led to impaired circadian locomotor activity rhythms. (A, from top to bottom) A diagram depicting the previously identified domains of dPER. PAS-A and PAS-B (PAS domains); (dPDBD) DBT-binding domain; (CBD) dCLK-binding domain. Disorder tendency plot of the dPER protein using IUPred. Codon usage score plot (CAI value, window 35) of wild-type dper. Codon usage score plot (CAI value, window 35) of dper(OP1). The dashed line in the codon usage for the wild-type gene indicates the average CAI of wild-type dper. (B) Double plot actograms showing locomotor activity rhythms of the wper0; p{dper(WT)} and wper0;p{dper(OP1)} fly strains in 4 d of light/dark cycles (LD) and 7 d of constant darkness (DD). (C) Eduction graphs generated from locomotor activity analysis showing the rhythms of the indicated strains. The Y-axis represents activity levels. (Top) The activity data generated by averaging the second and third days in light/dark cycles (LD 2–3). (Bottom) The activity data generated by averaging the second and third days in DD (DD 2–3). Arrows indicate morning anticipation (black) and evening anticipation (white) behaviors with their respective anticipation index (AI) values. The statistical analysis was performed using a two-tailed t- test to compare the AIs between the OP1 mutants and the wild-type per gene rescue strain. (*) P-value < 0.05; (**) P-value < 0.01.
To test this hypothesis, we used a commonly used dper rescue vector that contains a 13.2-kb dper genomic fragment tagged with the HA epitope at the C terminus (pCasPeR-per-13.2) that is capable of rescuing the per0 mutant (Lee et al. 1998) and codon-optimized the gene region that encodes for the N-terminal end of the dPER protein, including the N-terminal disordered region and part of the PAS A domain (Fig. 1A). To avoid the potential effects on translation initiation and mRNA splicing, the first 10 codons of dper and codons near an intron in this region were not changed. The codon-optimized vector was termed p{dper(OP1)}, which has the same 5′ and 3′ regulatory sequences and encodes the identical amino acid sequence as the wild-type gene. Independent lines of transgenic flies harboring the p{dper(OP1)} construct were obtained and were evaluated in a per-null (per0) mutant background, in which the circadian clock function could be rescued only when functional dPER is expressed from the transgene. Transgenic flies with the wild-type dper construct [p{dper(WT)}] were used as control.
To determine whether p{dper(OP1)} can rescue the circadian clock of per0 flies, we analyzed the locomotor activity rhythm of the transgenic flies at 25°C. Flies were entrained for 4 d in 12 h:12 h light:dark (LD) cycles before being shifted into constant darkness (DD) to detect free-running rhythms (Fig. 1B,C; Supplemental Fig. S3; Supplemental Table S1). As shown previously, p{dper(WT)} flies exhibited normal circadian phenotypes, as indicated by the typical bimodal morning and evening locomotor activity peaks in LD and the continued rhythmicity under DD (Rosato and Kyriacou 2006; Lear et al. 2009), indicating the full rescue of clock function by the wild-type dper transgene in the per0 background. In the heterozygous p{dper(OP1)} flies, however, although we observed free-running rhythms in DD, their amplitudes were not as robust as those of the p{dper(WT)} flies. The eduction graphs in Figure 1C show that the heterozygous p{dper(OP1)}flies exhibited significantly reduced anticipation of the morning and evening peaks, as indicated by lower anticipation indexes (AIs) (Cusumano et al. 2009). This indicates that one copy of the p{dper(OP1)} transgene is not able to rescue the per0 mutant. Two copies of p{dper(OP1)} transgenes also failed to rescue the circadian phenotype of the per0 flies. In fact, p{dper(OP1)} homozygous flies exhibited behavioral rhythms that were mostly abolished or severely dampened (Fig. 1B). In addition, morning and evening peaks in LD were dramatically reduced in the mutant homozygous flies. It should be noted that a much lower than expected number of homozygous p{dper(OP1)} flies could be obtained, suggesting that the high dosage of optimized PER protein might have a negative impact on survival. Because of this, we used heterozygous p{dper(OP1)} flies for the rest of the study to dissect the molecular basis of the observed phenotype. Together, these results indicate that dper codon usage is important for its clock function.
Impaired molecular rhythms in OP1 flies
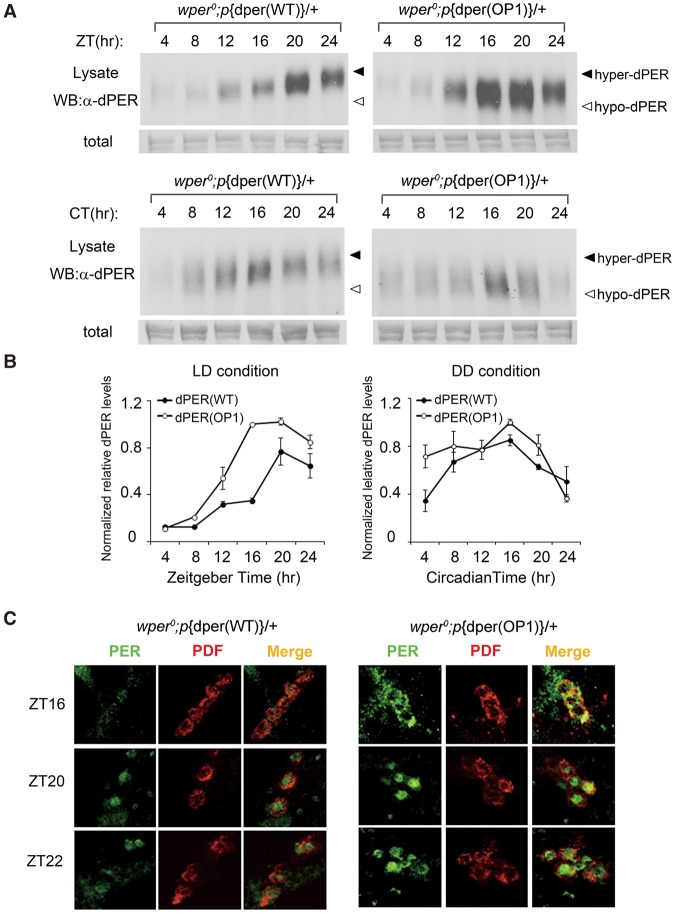
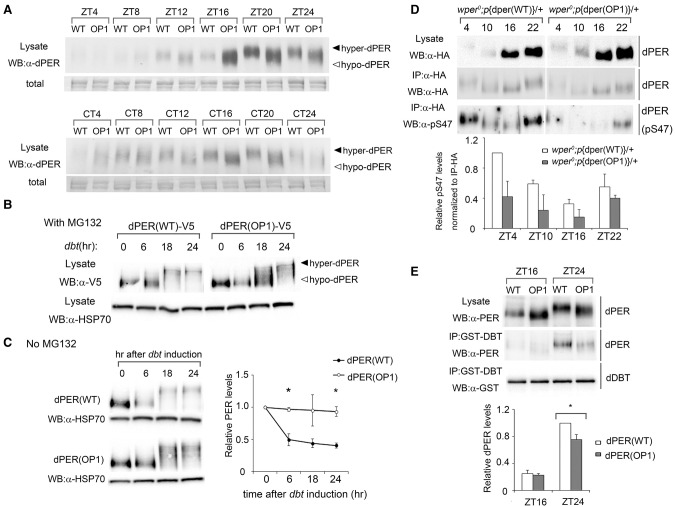
To examine oscillator function at the molecular level, fly head extracts from the heterozygous p{dper(WT)} and p{dper(OP1)} flies were prepared every 4 h in LD after three full days of entrainment and on the second day in DD. Although p{dper(OP1)} showed a cycling of PER protein abundance in LD, we observed a phase advance of ∼4 h for the dPER rhythm, which peaked at Zeitgeber time 16 (ZT16) instead of ZT20 for the p{dper(WT)} flies (Fig. 2A). In addition, dPER protein levels were also elevated in the mutant flies, suggesting a role for codon optimization in enhancing dPER expression (Fig. 2B). Consistent with previous studies, there was a robust rhythm of dPER phosphorylation profiles in the p{dper(WT)} flies in LD, as indicated by the dPER mobility changes at different time points (dPER mostly hyperphosphorylated at DD24). In contrast, such dPER mobility changes in LD were largely absent in the p{dper(OP1)} flies. The impaired dPER phosphorylation rhythm was also obvious in DD in the mutant flies (Fig. 2A, bottom panels). These results indicate that dPER molecular rhythms are also impaired in OP1 flies.
Figure 2.
Impaired dPER rhythms in OP1 flies. (A) Western blot results showing the dPER molecular rhythm in LD (top) and DD (bottom) for wild-type and OP1 flies. The filled and open arrowheads indicate the hyperphosphorylated and hypophosphorylated dPER proteins, respectively. Membrane staining was used as a loading control. (B) Densitometric analyses of the results from three independent experiments. The levels of dPER were normalized to the loading control. Error bars indicate ±SD. (C) Immunohistochemistry assay of dPER expression in pigment dispersing factor (PDF)-positive (PDF+) circadian neurons in fly brains. Adult flies were entrained to LD cycle, and brains were dissected for immunohistochemistry analysis at the indicated time points.
Because most dPER protein signals of fly head extracts came from the eyes, we examined levels of dPER and its cellular localization in the circadian behavior-related pigment dispersing factor (PDF)-positive neurons in the brain by immunohistochemistry (Li et al. 2014). As shown in Figure 2C, PDF mostly resided in the cytoplasm. For the p{dper(WT)} files, the dPER level was low, was mostly cytoplasmic at ZT16, and became mostly nuclear localized at ZT20 and ZT22. Even though a similar temporal change in dPER nuclear localization was also observed in the p{dper(OP1)} flies, the levels of p{dper(OP1)} were higher at all three time points, reflecting what we observed in the Western blots (Fig. 2A). The greatest difference in dPER signal levels were observed at ZT16, consistent with an advanced phase of dPER rhythm at the protein levels in the p{dper(OP1)} flies (Fig. 2A). In contrast, no significant differences in PDF levels were apparent between the two fly strains, suggesting that the impaired circadian behaviors in the p{dper(OP1)} flies are not due to the changes of PDF (Stoleru et al. 2004).
Impaired dPER function in the circadian negative feedback loop in OP1 flies
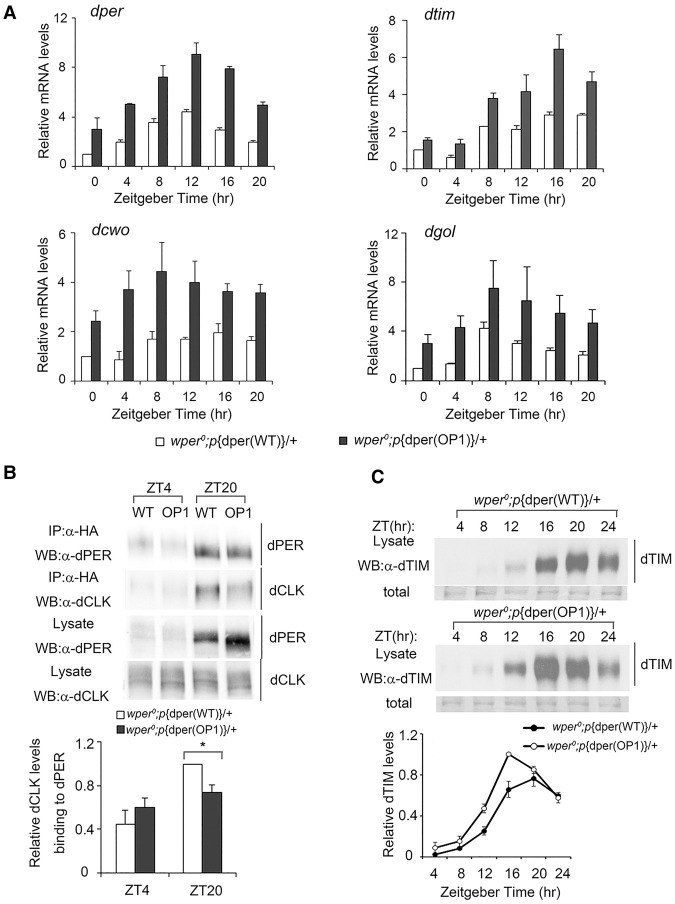
dPER functions as the core negative element in the fly circadian negative feedback loop by repressing the activity of the CLK/CYC complex, which results in the transcriptional repression of dper and other CLK–CYC target genes. Thus, the elevation of dPER expression in the p{dper(OP1)} flies should result in decreased transcription of CLK–CYC target genes if dPER function in the negative feedback loop is normal. However, the mRNA levels of dper and three other direct CLK–CYC target genes (dtim, dcwo, and dgol) (Abruzzi et al. 2011) were all significantly elevated in the p{dper(OP1)} flies at all time points in LD as compared with the p{dper(WT)} flies (Fig. 3A). These results suggest that dPER function in the circadian negative feedback loop was impaired in the p{dper(OP1)} flies despite the increase in dPER levels.
Figure 3.
Impaired circadian negative feedback loop in OP1 flies. (A) Quantitative RT–PCR assays showing the mRNA levels of dper, dtim, dcwo, and dgol. Error bars indicate SD. (B) Immunoprecipitation assay showing the reduced interaction between dPER and CLK in the wper0;p{dper(OP1)} flies. Head extracts were prepared from wper0; p{dper(WT)} and wper0;p{dper(OP1)} flies collected at the indicated times (ZT). (Top) Representative Western blot results are shown. (Bottom) Densitometric analyses from four independent biological experiments. The amount of dCLK was normalized to the HA (dPER) signal in the immunoprecipitation. (C) Western blot analysis showing the protein levels of TIM in the indicated fly strains in LD. Membrane staining was used as a loading control. (Bottom) Densitometric analyses of the Western blot results. Error bars indicate ±SD.
dPER exerts it role in the negative feedback loop by directly interacting with the CLK/CYC complex and sequestering CLK/CYC off the E-box element from the target gene promoters (Lee et al. 1999; Taylor and Hardin 2008; Menet et al. 2010). Therefore, we compared the interaction between PER and CLK in the p{dper(WT)} and p{dper(OP1)} flies by immunoprecipitation. The interaction between dPER and CLK is low during mid-day and high near the end of night (Menet et al. 2010). As shown in Figure 3B, despite a higher level of dPER in the p{dper(OP1)} flies, the amount of CLK associated with dPER was significantly decreased after PER immunoprecipitation. CLK is the limiting factor in the CLK–PER interaction (Bae et al. 2000). CLK immunoprecipitation also showed that there was less PER associated with CLK in the p{dper(OP1)} flies (Supplemental Fig. S4). Together, these results suggest that codon optimization resulted in reduced dPER–CLK interaction, providing an explanation for the impaired circadian negative feedback loop in the p{dper(OP1)} flies.
We also examined expression profiles of TIM in p{dper(OP1)} flies in LD (Fig. 3C). Consistent with the dPER rhythm, TIM rhythm was also phase-advanced in the p{dper(OP1)} flies with elevated TIM levels.
High levels of wild-type dPER protein production does not cause abnormal circadian phenotypes
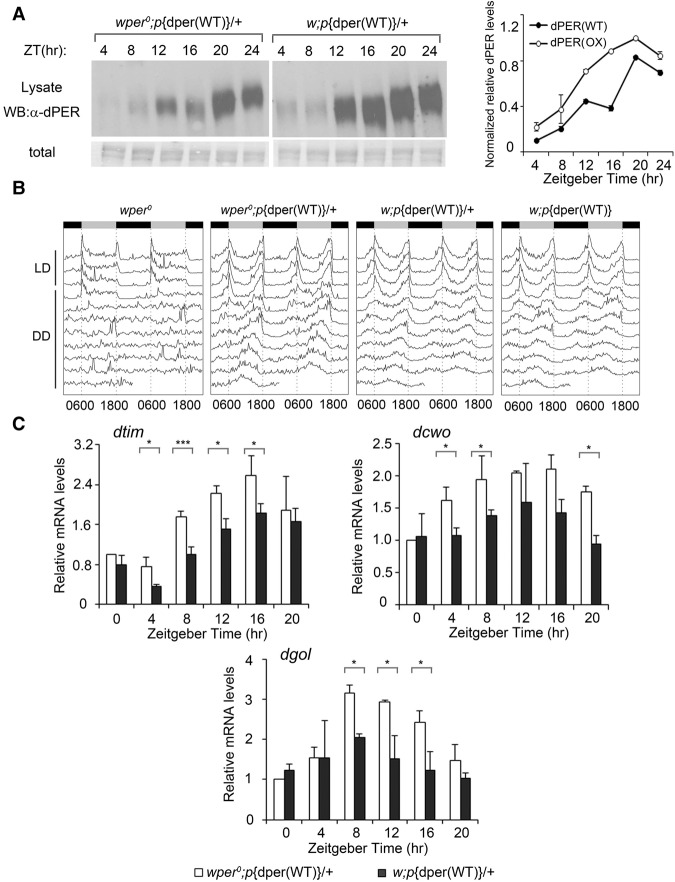
The elevated dPER levels in the p{dper(OP1)} flies raise the possibility that the impaired clock functions might be due to high dPER levels. To rule out this possibility, we created the p{dper(OX)} fly strain, which carries extra copies of the p{dper(WT)} transgenes in addition to the endogenous dper gene in the w1118 background. As shown in Figure 4A, the increase of dper copy number in the p{dper(OX)} flies resulted in high levels of dPER that were comparable with those of the p{dper(OP1)} strain (Fig. 2A,B). Locomotor activity rhythm assays showed that the p{dper(OX)} flies had no apparent defect in circadian behavior (Fig. 4B). Furthermore, in contrast to the p{dper(OP1)} flies, mRNA levels of dtim, dcwo, and dgol were all significantly decreased in the p{dper(OX)} flies (Fig. 4C), which is consistent with the increased repressor function of dPER due to high expression levels. This strongly suggests that the impaired clock function in the p{dper(OP1)} flies was caused by impaired dPER activity as a result of codon optimization rather than high dPER expression.
Figure 4.
Overexpression of wild-type dPER does not result in phenotypes that resemble OP1 flies. (A, left panels) Western blot analysis shows that the levels of dPER were elevated to levels comparable with those of OP1 strains in the w;p{dper(WT)} (OX) fly strains due to the extra copy number of wild-type dPER. Note that endogenous per is located on the X chromosome. Membrane staining was used as a loading control. (Right panels) Densitometric analyses of the Western blot results. Error bars indicate ±SD. (B) Double plot actogram showing circadian locomotor activity rhythms of the indicated strains in 4 d of LD and 7 d of DD. (C) Quantitative RT–PCR assays showing the mRNA levels of dtim, dcwo, and dgol in the indicated strains. Error bars indicate ±SD. (*) P < 0.05.
Codon optimization results in altered PER structure
The impaired PER function in the circadian clock of the p{dper(OP1)} flies—despite having an amino acid sequence identical to that of the wild-type protein—suggests that protein structure of dPER is altered. To test this possibility, we performed a limited trypsin digestion assay in which differential sensitivities can indicate protein structural changes. Fly head extracts from p{dper(WT)}, p{dper(OP1)}, and p{dper(OX)} flies were obtained and subjected to treatment with the same concentration of trypsin. As shown in Figure 5A and Supplemental Figure S5A, dPER was significantly more resistant to trypsin in the p{dper(OP1)} strain than in the p{dper(WT)} strain. Importantly, dPER trypsin sensitivities in the p{dper(WT)} and p{dper(OX)} samples were almost identical, indicating that dPER structural changes in the p{dper(OP1)} strain were not due to overexpression.
Figure 5.
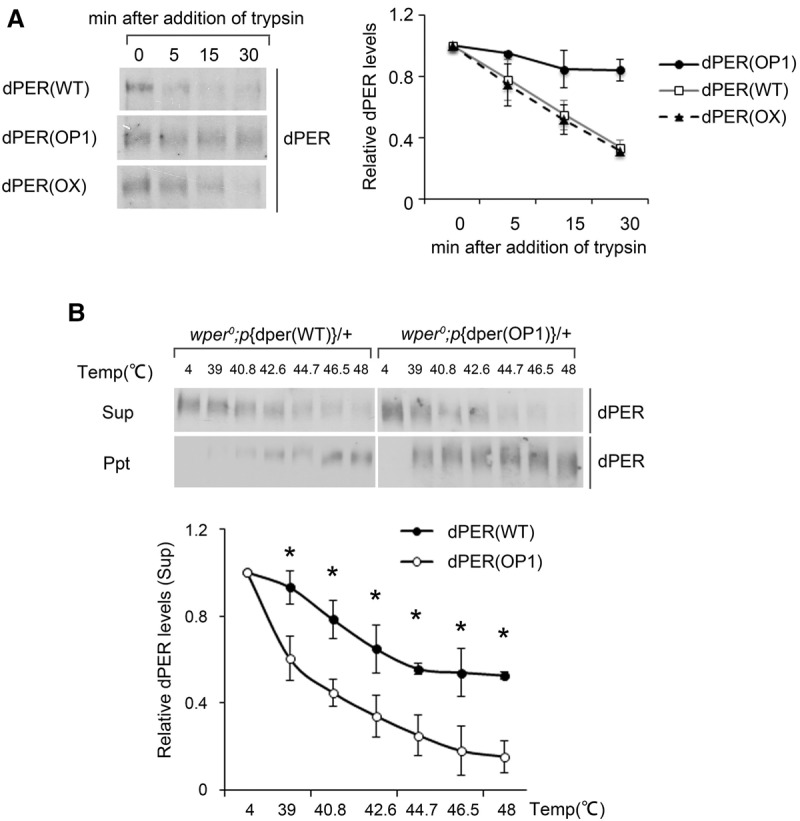
Codon optimization of dper results in altered dPER sensitivity to trypsin digestion and heat treatment. (A, left panels) Western blots showing the levels of dPER from the indicated strains after partial trypsin (0.5 µg/mL) digestion at the indicated time points. (Right panels) Densitometric analyses of the Western blot results from three independent experiments. The levels of dPER at time point 0 were set as 1. (B) Thermal shift assays comparing the sensitivity of dPER from the indicated strains to heat treatment. (Top panels) Western blots showing the levels of dPER in the supernatant (top blot) or precipitate (bottom blot) from wild-type and OP1 strains. (Bottom panels) Densitometric analyses of the results from three independent experiments. The levels of dPER at 4°C were set as 1. Error bars indicate ±SD. (*) P < 0.05.
To further confirm our conclusion, we carried out a thermal shift assay (Molina et al. 2013; Jafari et al. 2014) for the p{dper(WT)} and p{dper(OP1)} extracts. This assay quantifies changes in thermal denaturation and aggregation temperature of a protein as a result of treatment by increasing temperatures, and such changes indicate structural changes of a protein. As shown in Figure 5B and Supplemental Figure S5C, although increasing temperatures resulted in gradual precipitation of dPER in both extracts, the precipitation rates were much higher in the p{dper(OP1)} extracts than those of the p{dper(WT)}at every temperature >39°C (Fig. 5B, bottom panel). Together, these results demonstrate that, despite having the same amino acid sequence, dPER in the p{dper(WT)} and p{dper(OP1)} extracts are structurally different. Thus, nonoptimal codons of dper play an important role in affecting dPER protein structures, most likely through their effects on cotranslational protein folding.
Impaired dPER phosphorylation in the p{dper(OP1)} flies
A side-by-side comparison of dPER phosphorylation profiles at different time points in LD and DD indicates that the dPER is hypophoshorylated in the p{dper(OP1)} strain compared with that in the p{dper(WT)} strain (Fig. 6A; Supplemental Fig. S6A). The difference was more prominent for the time points during subjective night (circadian time [CT] 16–24), when dPER is mostly hyperphoshorylated in the p{dper(WT)} strain. These results further highlight the structural differences of dPER proteins in these two strains.
Figure 6.
Impaired dPER phosphorylation profiles and degradation in OP1 flies. (A) Western blots showing a side-by-side comparison of dPER phosphorylation profiles at different time points in LD and DD between the wper0;p{dper(WT)} and wper0;p{dper(OP1)} flies. Membrane staining was used as a loading control. (B,C) Drosophila Schneider (S2) cells were cotransfected with dbt and dper (pAC-dper-V5) variants and collected at the indicated times (hours) after dbt induction. For the experiments in B, the culture medium contained MG132 to inhibit dPER degradation. (C, bottom) Densitometric analyses of the Western blot results for experiments without MG132 from three independent experiments. HSP70 signal was used as a loading control. (D) Western blot analysis using anti-pS47 antibody showing the reduction of S47 phosphorylation of dPER in the wper0;p{dper(OP1)} flies. Head extracts were prepared at the indicated times (ZT). dPER-HA-containing immune complexes were recovered using anti-HA beads, and dPER(S47) were detected by Western blots using an anti-pS47 antibody. (Bottom) Densitometric analyses of the results from three independent experiments. Error bars indicate ±SD. (E) GST pull-down assay showing the reduced interaction between dPER and DBT in the wper0;p{dper(OP1)} flies. Head extracts were prepared from wper0;p{dper(WT)} and wper0;p{dper(OP1)} flies collected at the indicated times (ZT). (Top) Representative Western blot results are shown. (Bottom) Densitometric analyses from four independent biological experiments. (*) P < 0.05.
dPER phosphorylation has several important functional impacts, affecting its nuclear translocation, transcriptional repressor activity, and protein stability (Cyran et al. 2005; Blau 2008; Chiu et al. 2008; Kivimae et al. 2008; Ko et al. 2010; Garbe et al. 2013). The dper region that is optimized in the p{dper(OP1)} strain encodes the N-terminal part of dPER that contains major DBT phosphorylation sites that are necessary and sufficient to mediate its ubiquitination and degradation (Chiu et al. 2008). The localized codon usage effect on translation and folding raised the possibility that codon optimization in this region should result in impaired dPER phosphorylation by DBT. To test this, we compared the ability of dPER (wild type or OP1) to be phosphorylated by DBT in cultured Drosophila Schneider (S2) cells. In this assay, the expression of recombinant dPER and DBT can recapitulate the DBT-dependent progressive phosphorylation and subsequent SLIMB-mediated degradation of dPER (Ko et al. 2002; Chiu et al. 2008). We first performed the assay in the presence of the proteasome inhibitor MG132 to block dPER degradation so that the phosphorylation and degradation processes could be separated. Similar to previous results, DBT induction resulted in progressive phosphorylation of wild-type dPER, which became mostly hyperphosphorylated after 24 h (Fig. 6B). As predicted, the progressive phosphorylation process of OP1 dPER induced by DBT is markedly delayed, as indicated by the near absence of dPER phosphorylation at 6 h after DBT induction and the presence of hypophosphorylated species at later time points (Fig. 6B; Supplemental Fig. S6B,C).
When such assays were performed in the absence of MG132 (Fig. 6C), the DBT-triggered dPER degradation was significantly impaired for dPER(OP1), indicating that dPER(OP1) is more stable than wild-type dPER due to impaired DBT phosphorylation (Supplemental Figs. S6D, S7A,B). This result also provides an explanation for the elevated dPER protein levels in the p{dper(OP1)} flies.
Phosphorylation of Ser47 (S47) is a critical DBT phosphorylation event in the N-terminal end of dPER that generates an atypical SLIMB-binding site (Chiu et al. 2008). To confirm the impact of codon optimization on DBT-mediated phosphorylation in vivo, we compared pS47 levels at different LD time points in the head extracts of p{dper(WT)} and p{dper(OP1)} flies by using a S47 phospho-specific antibody. As shown in Figure 6D, the levels of S47 were markedly lower in p{dper(OP1)} despite having higher dPER levels than p{dper(WT)}. Together, these results demonstrate that codon optimization of the N-terminal part of the dper gene lead to altered dPER protein structure, resulting in impaired DBT phosphorylation at the N-terminal end of dPER and less efficient protein degradation.
We further tested whether this ineffective phosphorylation of p{dper(OP1)} is linked to altered DBT binding (Kim et al. 2007) using GST pull-down assays (Preuss et al. 2004). In these assays, DBT was purified using a GST tag from a stable Drosophila cell line. The amount of dPER bound to DBT was then detected by Western blot analysis. The enhanced binding of p{dper(WT)} to DBT at ZT24 compared with at ZT16 (Fig. 6E) was consistent with the progressive phosphorylation and accelerated degradation of dPER during the late night/early day (Muskus et al. 2007). Binding of PER in p{dper(OP1)}to DBT was less efficient than the wild-type PER at ZT24 even though it was relatively more abundant.
Codon optimization in the central part of dper also results in impaired circadian rhythms and PER structural changes
To further establish the role of codon usage in determining dPER structure, we created transgenic flies harboring p{dper(OP2)} in which the dper region that encodes for the central part of dPER (downstream from the PAS domains) was codon-optimized (Fig. 7A). This part of dper also has several regions with relatively low codon usage scores and encodes for protein domains that were mostly predicted to be intrinsically disordered (Fig. 1A). This region includes the per short domain and part of the CLK/CYC inhibition domain (CCID) (Yu et al. 1987; Chang and Reppert 2003). It also contains a number of phosphorylation sites that are critical for controlling clock speed, such as T583, S585, S589, and S596 (Chiu et al. 2011). Locomotor activity assays showed that the p{dper(OP2)} flies also exhibited impaired behavioral rhythms as observed in p{dper(OP1)} flies, including reduced morning anticipation in LD and dampened rhythms in DD (Fig. 7B,C).
Figure 7.
Codon optimization of the central part of dper resulted in impaired circadian rhythms and altered dPER structure. (A) Diagrams showing the dPER protein domains and the codon usage score plot of dper (CAI value, window 35) after codon optimization. (B) Double plot actogram showing the circadian rhythms of wper0;p{dper(WT)} and wper0;p{dper(OP2)} strains after 4 d of LD and 7 d of DD. (C) Eduction graphs generated from locomotor activity analysis showing the circadian rhythms of the indicated strains in LD 2–3 (top) and in DD 2–3 (bottom). Arrows indicate morning anticipation (black) and evening anticipation (white) behaviors with their respective AI values. (*) P < 0.05. (D, top panels) Western blot results using dPER antibody showing the dPER rhythm in LD. (Bottom panels) Side-by-side Western blot analysis results showing the dPER mobility differences between two fly strains. Membrane staining was used as a loading control. (E, top panels) Western blots comparing the sensitivity of dPER from the indicated strains with partial trypsin (0.5 µg/mL) digestion. (Bottom panels) Densitometric analyses of the Western blot results from three independent experiments. The levels of dPER at time point 0 were set as 1. Error bars indicate ±SD.
Interestingly, unlike the p{dper(OP1)} flies, dPER protein levels were comparable between the p{dper(WT)} and p{dper(OP2)} flies in LD and DD with a similar phase (Fig. 7D). However, dPER phosphorylation rhythms were severely impaired in the p{dper(OP2)} flies, and dPER accumulates in hypophosphorylated forms. Furthermore, trypsin sensitivity assays showed that dPER in p{dper(OP2)} head extracts was significantly more resistant to trypsin digestion compared with that in the p{dper(WT)} head extracts (Fig. 7E). Together, these results further demonstrate a role for codon usage in affecting dPER protein structure and function.
Discussion
The in vivo role of codon usage was previously unclear in animal systems. In this study, we demonstrate that dper codon usage affects dPER structure and is critical for its function in the Drosophila circadian clock. Together with our previous studies in Neurospora, our results here suggest that codon usage is a universal mechanism that regulates protein structure and function from fungi to animals.
The role of codon usage in regulating dPER protein structure and function is supported by several lines of evidence. First, codon optimization of part of the dper ORF resulted in severe impairment of circadian locomotor activity rhythms and dPER function in the circadian negative feedback loop. Second, dper codon optimization affects the ability of dPER to interact with CLK/CYC, providing a mechanism for the impaired circadian negative feedback loop in the optimized flies. Third, despite having the same amino acid sequence, dPER protein in the codon-optimized flies exhibited significantly differential sensitivities to partial trypsin digestion and in thermal shift assays compared with that extracted from flies with the wild-type gene, indicating that dPER structure changes are due to codon manipulation. Furthermore, dper codon optimization impaired dPER phosphorylation at the site of the codon changes, which led to altered dPER stability. Finally, we showed that the observed effects of dper codon optimization is not due to dPER protein overexpression. Together, these results, to our knowledge, establish the first in vivo example in an animal system that demonstrates the role of codon usage in determining protein structure and function.
How does codon usage influence dPER structure and function? By comparing mRNA translation elongation speed in Neurospora, we demonstrated that codon usage regulates speed of elongation: Preferred codons speed up translation elongation, while unpreferred codons slow it down (Yu et al. 2015). Although the effects of codon usage on elongation rate have not been demonstrated in insects, a similar mechanism should exist due to the conservation of the translation process. Thus, codon usage affects the amount of time available for cotranslational folding. This is consistent with previous studies showing that translation rate and synonymous codon usage can affect protein folding and functions (Komar et al. 1999; Zhang et al. 2009; Siller et al. 2010; Spencer et al. 2012; Kim et al. 2015; Presnyak et al. 2015). We previously observed genome-wide correlations showing that optimal codons are preferentially used in regions that are predicted to be well folded, while relatively more unpreferred codons are used in protein regions predicted to be unstructured. Our studies and previously uncovered correlations between codon usage and certain protein structural motifs suggest that codon usage and protein structures coevolve and are adapted to each other (Zhou et al. 2009, 2015; Pechmann and Frydman 2013; Pechmann et al. 2014). Therefore, there is a codon usage code within genetic codons that generates elongation speed rhythms to optimize the cotranslational folding process.
The predicted unstructured or intrinsically disordered protein (IDP) domains widely exist in all analyzed proteomes. Despite the fact that they are not predicted to form stable three-dimensional structures, IDPs have been shown to play important roles in many biological processes (Dyson and Wright 2005; Dunker et al. 2008; Tompa 2011). Similar to Neurospora FRQ, most of the dPER protein is predicted to be unstructured. Here we showed that codon optimization of parts of dper that encode for the N-terminal and middle parts of the unstructured regions resulted in altered dPER structure and function. Therefore, both of our previous studies in Neurospora and this present study highlight the importance of codon usage in the unstructured protein regions. These putative IDP regions may require relatively longer cotranslational folding time to either fold into certain structures or serve as platforms for intermolecular or intramolecular protein–protein interactions.
Synonymous codon mutations have been associated with many human diseases with unknown mechanisms. These diseases include cystic fibrosis, amyotrophic lateral sclerosis (ALS), and Crohn's disease (Bartoszewski et al. 2010; Lazrak et al. 2013; Bali and Bebok 2015; Liu et al. 2016). A single synonymous SNP in the form of a rare codon in the human multidrug resistance 1 (MDR1) gene was found to result in altered drug and inhibitor interactions (Kimchi-Sarfaty et al. 2007). For circadian clock-related diseases, a T2434C synonymous polymorphism in exon 18 of hPER1 was found to be associated with extreme diurnal performance (Carpen et al. 2006), and G2114A in hPER2 was linked to diurnal preference (Matsuo et al. 2007). Our results here suggest that the change of codon usage in these genes may be a mechanism that contributes to impaired function of the encoded proteins.
Materials and methods
Calculation of the CAI, CBI, and protein structural disorder tendency
The CAI was calculated as described previously (Sharp and Li 1987). The codon usage frequency table for D. melanogaster was obtained from the Codon Usage Database (http://www.kazusa.or.jp/codon). The CBI was calculated by CodonW (http://codonw.sourceforge.net) (Bennetzen and Hall 1982). IUPred (http://iupred.enzim.hu) was used to predict protein disorder tendency.
Constructs and transgenic flies
A previously described CaSpeR-4-based transformation vector, 13.2(per+-HA10His), was used as the parent vector for codon optimization (Lee et al. 1998). This vector contains a 13.2-kb genomic dper region in which a dPER protein was tagged by a HA epitope tag and a stretch of histidine residues at the C terminus. Codon-optimized sequences encoding amino acids 15–21, 43–316 (OP1), and 539–982 (OP2) were synthesized by GenScript and cloned into the above-mentioned vector to yield 13.2[per(op1)-HA10His] and 13.2[per(op2)-HA10His]. Transgenic flies were generated by BestGene, Inc., using standard P-element-mediated transformation techniques using w1118embryos. At least three independent germline transformants bearing the dper-containing plasmids were obtained and then crossed into a wper0 genetic background to yield wper0;p{dper(OP1)} and wper0;p{dper(OP2)} [referred to here as p{dper(OP1)} and p{dper(OP2)}]. Transgenic flies carrying 13.2(per+-HA10His) were used as a control [wper0;p{dper(WT)}]. The p{dper(OX)}flies were flies with a w1118 genetic background carrying extra copies of the wild-type version of the dper transgene.
Drosophila locomotor activity analysis
The locomotor activities of individual flies were measured as previously described using the monitoring system from Trikinetics (Chiu et al. 2010). Briefly, 1- to 5-d-old adult male flies were used for the analysis and were kept in incubators at 25°C, exposed to 4 d of LD (where ZT0 is defined as the time when the light phase begins), and subsequently kept in DD for 5–8 d to measure free-running rhythm. The locomotor activity data for each individual fly were analyzed using FaasX, which was generously provided by F. Rouyer (Centre National de Ia Recherche Scientifique, Paris, France). Periods were calculated for each individual fly using periodogram analysis and pooled to obtain a group average for each independent transgenic line or genotype. Power was a measure of the relative strength of the rhythm during DD. Individual flies with a power ≥10 and a “width” value of ≥2 (denotes number of peaks in 30-min increments above the periodogram 95% confidence line) were considered rhythmic.
Quantification of morning and evening anticipations in LD cycles were calculated using the method described previously (Cusumano et al. 2009). An AI was obtained as the slope of a linear regression through the last eight 30-min bins before lights off (evening anticipation) or the last five bins before lights on (morning anticipation). It thus represents an average variation in activity counts per 30-min bin from one bin to the next within that time window.
Immunoblotting
Flies were collected by freezing at the indicated times in LD or DD, and total head extracts were prepared using either EB1 (20 mM HEPES at pH 7.5, 100 mM KCl, 5% glycerol, 2.5 mM EDTA, 5 mM DTT, 0.1% Triton X-100, 0.5 mM PMSF, 10 µg/mL aprotinin, 5 µg/mL leupeptin, 1 µg/mL pepstatin A, 25 mM NaF) or modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl,1 mM EDTA, 0.25% sodium deoxycholate, 1% NP-40, 0.5 mM PMSF, 25 mM NaF, Roche Complete protease inhibitor [EDTA-free], Roche PhosStop phosphatase inhibitor) with sonication, depending on which proteins we sought to detect; i.e., EB1 buffer was used for dPER and TIM, whereas modified RIPA with sonication was used for CLK (Fig. 3B). In the case of S2 cell extracts, the cells were harvested at the indicated times after kinase induction (Fig. 6B,C) and lysed using EB2 lysis buffer (20 mM HEPES at pH 7.5, 100 mM KCl, 5% glycerol, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 0.5 mM PMSF, 10 µg/mL aprotinin, 5 µg/mL leupeptin, 1 µg/mL pepstatin A, 25 mM NaF). The following primary antibodies were used: anti-V5 (1:7000; Invitrogen), anti-mouse HRP (1:2000; GE), anti-HA 3F10 (1:1000 for fly extracts; Roche), anti-rat HRP (1:1000; GE), anti-Per (1:3000; GP5620) anti-GP HRP (1:2000; Sigma), anti-Tim (1:1000; R1) anti-GP HRP (1:2000; Sigma), anti-goat HRP (1:5000; Santa Cruz Biotechnology), anti-CLK (1:1000; Santa Cruz Biotechnology), and anti-goat HRP (1:1000; Santa Cruz Biotechnology). SDS-PAGE gels (6%) were used to resolve dPER and TIM, and, in the case of CLK, 5% Tris-HCl criterion gels (Bio-Rad) were used. All Western blots were imaged using the Chemidoc software for the Bio-Rad Chemidoc, which included correction for background signals.
Immunohistochemistry
Confocal imaging of adult brains was performed as described previously (Ko et al. 2007). Briefly, adult files were entrained and dissected from each Zeitgeber time point and incubated briefly in cold isopropyl alcohol. Fly heads were removed in an embryo dish (Electron Microscopy Sciences) filled with PBS and then transferred to a 1% collagenase solution for 10 min with agitation. Collagenase was removed, and fixative (4% formaldehyde, 0.2% Triton X-100 in PBS) was added. Heads were fixed for 45 min at room temperature with agitation. Fixative was then removed, and heads were rinsed twice and then washed twice for 30 min. Heads were transferred to blocking solution (0.2% Triton X-100, 5% normal goat or horse serum in PBS). Brains were dissected using #5 Rubis nano tweezers (Electron Microscopy Sciences) in an embryo dish filled with blocking solution. Brains were blocked between 1 and 2 h and incubated overnight with primary antibodies, anti-HA (clone 3F10, Roche) at 1:750, and anti-PDF (Developmental Studies Hybridoma Bank) at 1:200 for all brains. Brains were incubated in primary antibodies overnight in blocking at 4°C with agitation. After ∼16 h, brains were rinsed twice and then washed twice for 30 min in wash solution (0.2% Triton X-100 in PBS). Next, secondary antibodies, which were conjugated to Alexa dyes (Life Technologies) for visualization on a confocal microscope, were added to brains in blocking solution. Brains were incubated with secondary antibodies Alexa fluor 488-conjugated goat anti-rat (against anti-HA) and Alexa fluor 594-conjugated goat anti-mouse (against anti-PDF) for 4 h at 4°C with agitation. After secondary antibody incubation, brains were once again rinsed twice and washed twice for 30 min in wash solution. Finally, brains were mounted on microscope slides in VectaShield mounting medium (Vector Laboratories) under a #1.5 (17-mm) coverslip. Prepared slides were stored at 4°C in a light-blocking container to prevent bleaching of fluorescent dye.
Quantitative real-time PCR
Total RNA was isolated from frozen heads using TRIzol (Invitrogen). Five-hundred nanograms of total RNA was reverse-transcribed using a high-capacity cDNA reverse transcription kit (ABI) in accordance with the manufacturer's instructions and subjected to real-time PCR analysis. Primer sequences used here for quantitation were as follows: dper forward (5′-CGCAGCATCATGGACTTCTA-3′), dper reverse (5′-CCGTCTGACCCTTCTTCATTAC-3′), tim forward (5′-CCTTTTCGTACACAGATGCCA-3′), tim reverse (5′-GGTCCGTCTGGTGATCCCAG-3′) (Kadener et al. 2007), dcwo forward (5′-GTCTGTGGATCGAGGAGCAG-3′), dcwo reverse (5′-GGCATATTCAGCATCGTCCT-3′) (Kadener et al. 2007), dgol forward (5′-GCCACGGATCTATGCAGTTT-3′), and dgol reverse (5′-CTTGGATAGCGACTGCTGTG-3′) (Abruzzi et al. 2011).
Immunoprecipitation assays
For immunoprecipitation, total head extracts were prepared using a modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.25% sodium deoxycholate, 1% NP-40, 0.5 mM PMSF, 25 mM NaF, Roche Complete protease inhibitor [EDTA-free]) with sonication. Twenty microliters of HA-agarose (Sigma) was added to total fly head extracts. Immune complexes were incubated with gentle rotation for 6 h at 4°C. Beads were collected and mixed with 2× SDS sample buffer. Six percent gels were used to resolve dPER, and 5% Tris-HCl Criterion gels (Bio-Rad) were used to detect CLK.
To detect signals with the phospho-specific antibodies, Roche PhosStop phosphatase inhibitor was added to modified RIPA buffer. To immunoprecipitate dPER, 30 µL of anti-HA-agarose beads (Sigma, A2095) was added to the extracts and incubated with gentle rotation for 4 h at 4°C. Proteins were eluted with SDS sample buffer and resolved by 6% SDS-PAGE. To perform λ phosphatase treatment, 0.6 µL of λ phosphatase (New England Biolabs) was added after dPER immunoprecipitation and incubated for 30 min at 30°C. Proteins were eluted with SDS sample buffer and resolved by 6% SDS-PAGE.
Trypsin sensitivity assay
Protein concentration of the extract was diluted to 2.5 µg/µL. A 100-µL aliquot of extract was treated with trypsin (final concentration was experimentally determined) at room temperature with gentle shaking. A 20-µL sample was taken from the reaction at each time point (0, 5, 15, and 30 min) after addition of trypsin. Each 20-µL sample was mixed with protein loading buffer, and proteins were resolved on an SDS-PAGE gel (7.5%). Western blot analysis was performed to examine dPER protein levels at each time point (Zhou et al. 2013). Assays for different extracts were performed side by side, and protein samples were transferred to the same membrane for Western blot analysis.
Thermal shift assay
Flies were collected by freezing at ZT20, and total head extracts were prepared using modified RIPA buffer. Aliquots of protein lysates (18 µL) were heated for 2 min at different temperatures (C1000 Thermal Cycler PCR machine, Bio-Rad) followed by cooling for 3 min at room temperature. The lysates were then centrifuged at 15,000g for 20 min at 4°C to separate the soluble fractions from precipitates. The amount of dPER protein in the supernatants and precipitates was then analyzed by Western blot analysis.
S2 culture and transfection
The pAct-dper-V5 and pMT-dbt-V5 plasmids were described previously (Kim et al. 2007). pAct-dper(op1)-V5 was generated by replacing the wild-type dper sequence in pAct-dper-V5 with the OP1 sequence. All constructs were verified by DNA sequencing. S2 cells were transfected using Effectene (Qiagen) following the manufacturer's protocol (Ko et al. 2002; Chiu et al. 2008). For each transient transfection, 0.8 µg of different dper-containing plasmids and 0.2 µg of pMT-dbt-V5 plasmids were used. Expression of dbt under the pMT promoter was induced by adding 500 µM CuSO4 to the culture medium for 36 h after transfection. For experiments in which the proteasome inhibitor MG132 (Sigma) and cycloheximide (Sigma) were used, 50 µM MG132 and 10 µg/mL cycloheximide were added 4 h prior to cell harvesting. To quantify different phosphorylation isoforms, ImageJ was used to plot for the relative attribution of signals along each loading lane. To quantify the relative ratio of phosphorylation isoforms in the in vitro degradation assay, the percentage of the top half (hyperphosphorylated) or bottom half (hypophosphorylated) was calculated over the total signal.
GST pull-down assay
To generate GST-DBT proteins, expression was induced from stable cell lines for 36 h, and cells were lysed in GST lysis buffer (20 mM Tris-HCl at pH 7.5, 0.05% IGEPAL CA-60, 1 mM EDTA, 5 mM DTT, 150 mM NaCl, 25 mM NaF, Complete EDTA free protease inhibitor cocktail [Roche]). The extracts were then incubated with glutathione beads overnight at 4°C to achieve binding. Roughly 250 µL of heads was used to prepare extracts for each GST pull-down reaction. Heads were homogenized in modified RIPA buffer (see the Materials and Methods for immunoprecipitation) with the addition of complete EDTA-free protease inhibitor cocktail (Roche). Prior to the pull-down, proteins were quantified using a spectrophotometer to ensure that equal amounts of head extracts were used for each reaction. Bound dPER was resolved using 6% SDS-PAGE to determine dPER–DBT interaction.
Supplementary Material
Acknowledgments
We thank Dr. Dong Wang, Dr. Robin Hiesinger, Dr. Huaqi Jiang, and Dr. Jin Jiang for technical assistance. We thank Dr. Adrian Rothenfluh for locomotor activity assays, and members of our laboratories for assistance. This work is supported by grants from the National Institutes of Health (R35GM118118) and the Welch Foundation (I-1560) to Y.L., and National Institutes of Health R01 GM102225 and National Science Foundation Integrative Organismal Systems 1456297 to J.C.C.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.281030.116.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. 2011. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. 2010. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Edery I. 2006. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem 140: 609–617. [DOI] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I. 2000. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK–CYC transcription factor and the PER–TIM complex. J Neurosci 20: 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali V, Bebok Z. 2015. Decoding mechanisms by which silent codon changes influence protein biogenesis and function. Int J Biochem Cell Biol 64: 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, Collawn JF, Bebok Z. 2010. A Synonymous single nucleotide polymorphism in ΔF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem 285: 28741–28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. 1982. Codon selection in yeast. J Biol Chem 257: 3026–3031. [PubMed] [Google Scholar]
- Blau J. 2008. PERspective on PER phosphorylation. Genes Dev 22: 1737–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini DB, Stephan W. 2003. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 163: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. 2006. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet 51: 1122–1125. [DOI] [PubMed] [Google Scholar]
- Chang DC, Reppert SM. 2003. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol 13: 758–762. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. 2008. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22: 1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Low KH, Pike DH, Yildirim E, Edery I. 2010. Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp 10.3791/2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I. 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. 2004. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chelot E, Picot M, Richier B, Rouyer F. 2009. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci 12: 1431–1437. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. 2005. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci 25: 5430–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18: 756–764. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Fang Y, Zheng X, Sowcik M, Anjum R, Gygi SP, Sehgal A. 2013. Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet 9: e1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. 2011. Determinants of translation efficiency and accuracy. Mol Syst Biol 7: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. 2002. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420: 178–182. [DOI] [PubMed] [Google Scholar]
- Hambuch TM, Parsch J. 2005. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics 170: 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Panda S. 2013. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol 23: 724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. 2007. Variable strength of translational selection among 12 Drosophila species. Genetics 177: 1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S, Strauss HM, Vanselow K, Yildiz O, Schulze S, Arens J, Kramer A, Wolf E. 2009. Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol 7: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2008. Selection on codon bias. Annu Rev Genet 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. 1993. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364: 259–262. [DOI] [PubMed] [Google Scholar]
- Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2: 13–34. [DOI] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, Molina DM. 2014. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9: 2100–2122. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Stewart PL, Egli M. 2011. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys 40: 143–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol 53: 290–298. [DOI] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu WJ, Hardin PE, Edery I. 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27: 5014–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR. 2015. Translational tuning optimizes nascent protein folding in cells. Science 348: 444–448. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. 2007. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528. [DOI] [PubMed] [Google Scholar]
- Kivimae S, Saez L, Young MW. 2008. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol 6: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Young MW. 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Ie. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Ko HW, Edery I. 2005. Analyzing the degradation of PERIOD protein by the ubiquitin-proteasome pathway in cultured Drosophila cells. Methods Enzymol 393: 394–408. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678. [DOI] [PubMed] [Google Scholar]
- Ko HW, DiMassa S, Kim EY, Bae K, Edery I. 2007. Cis-combination of the classic per(S) and per(L) mutations results in arrhythmic Drosophila with ectopic accumulation of hyperphosphorylated PERIOD protein. J Biol Rhythms 22: 488–501. [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. 2010. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci 30: 12664–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Reiss C. 1999. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462: 387–391. [DOI] [PubMed] [Google Scholar]
- Lazrak A, Fu LW, Bali V, Bartoszewski R, Rab A, Havasi V, Keiles S, Kappes J, Kumar R, Lefkowitz E, et al. 2013. The silent codon change I507-ATC → ATT contributes to the severity of the ΔF508 CFTR channel dysfunction. FASEB J 27: 4630–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. 2009. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol 7: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. 1998. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER–TIM complex. Neuron 21: 857–867. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. 1999. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK–CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol 19: 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo F, Shen J, Rosbash M. 2014. PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc Natl Acad Sci 111: 8311–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Zang T, Zhang CL. 2016. Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep 14: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Shino Y, Yamada N, Ozeki Y, Okawa M. 2007. A novel SNP in hPer2 associates with diurnal preference in a healthy population. Sleep Biol Rhythms 5: 141–145. [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 24: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbitz-Zahradnik T, Wolf E. 2015. How is the inner circadian clock controlled by interactive clock proteins?: structural analysis of clock proteins elucidates their physiological role. FEBS Lett 589: 1516–1529. [DOI] [PubMed] [Google Scholar]
- Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao YH, Nordlund P. 2013. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341: 84–87. [DOI] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. 2007. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27: 8049–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. 2013. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J. 2014. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL. 2004. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol 24: 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Adodeely M, Kloss B, Young MW. 1998. double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Rosato E, Kyriacou CP. 2006. Analysis of locomotor activity rhythms in Drosophila. Nat Protoc 1: 559–568. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. 1987. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TM, Mosurski KR. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res 14: 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. 2010. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol 396: 1310–1318. [DOI] [PubMed] [Google Scholar]
- Spencer PS, Siller E, Anderson JF, Barral JM. 2012. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol 422: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- Tataroglu O, Emery P. 2015. The molecular ticks of the Drosophila circadian clock. Curr Opin Insect Sci 7: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Hardin PE. 2008. Rhythmic E-box binding by CLK–CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol 28: 4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. 2011. Unstructural biology coming of age. Curr Opin Struct Biol 21: 419–425. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. 2001. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715. [DOI] [PubMed] [Google Scholar]
- Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, Rosbash M. 1987. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci 84: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280. [DOI] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO. 2009. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol 26: 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang T, Fu J, Xiao G, Liu Y. 2015. Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol Microbiol 97: 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.