Abstract
Automatic and reliable segmentation of the prostate is an important but difficult task for various clinical applications such as prostate cancer radiotherapy. The main challenges for accurate MR prostate localization lie in two aspects: (1) inhomogeneous and inconsistent appearance around prostate boundary, and (2) the large shape variation across different patients. To tackle these two problems, we propose a new deformable MR prostate segmentation method by unifying deep feature learning with the sparse patch matching. First, instead of directly using handcrafted features, we propose to learn the latent feature representation from prostate MR images by the stacked sparse auto-encoder (SSAE). Since the deep learning algorithm learns the feature hierarchy from the data, the learned features are often more concise and effective than the handcrafted features in describing the underlying data. To improve the discriminability of learned features, we further refine the feature representation in a supervised fashion. Second, based on the learned features, a sparse patch matching method is proposed to infer a prostate likelihood map by transferring the prostate labels from multiple atlases to the new prostate MR image. Finally, a deformable segmentation is used to integrate a sparse shape model with the prostate likelihood map for achieving the final segmentation. The proposed method has been extensively evaluated on the dataset that contains 66 T2-wighted prostate MR images. Experimental results show that the deep-learned features are more effective than the handcrafted features in guiding MR prostate segmentation. Moreover, our method shows superior performance than other state-of-the-art segmentation methods.
Index Terms: MR prostate segmentation, stacked sparse auto-encoder (SSAE), sparse patch matching, deformable model
I. Introduction
Prostate cancer is the second leading cause of cancer death in American men, behind only lung cancer [1]. As a main imaging modality for clinical inspection of prostate, Magnetic Resonance (MR) imaging provides better soft tissue contrast than ultrasound in a non-invasive way, and has the emerging role in prostate cancer diagnosis and treatment [2, 3]. The accurate localization of the prostate is an important step for assisting the diagnosis and treatment, such as guiding biopsy procedure [2] and radiation therapy [3]. However, the manual segmentation of the prostate is tedious and time-consuming, and also suffers from intra- and inter-observer variability. Therefore, developing automatic and reliable segmentation methods for MR prostate is clinically desirable and an important task.
However, accurate prostate localization in MR images is difficult due to the following two main challenges. First, the appearance patterns vary a lot around the prostate boundary across patients. As we can see from Fig. 1 (a), the image contrasts at different prostate regions, i.e., the anterior, central and posterior regions, change both across different subjects and within each subject. Fig. 1 (b) gives the intensity distributions of prostate and background voxels around the prostate boundary, respectively. As shown in the figure, the intensity distributions highly vary across different patients and do not often follow the Gaussian distribution.
Fig. 1.
(a) Typical T2-weighted prostate MR images. Red contours indicate the prostate glands delineated manually by an expert. (b) Intensity distributions of prostate and background voxels around the prostate boundary of (a). (c) The 3D illustrations of prostate surfaces corresponding to each image in (a).
To evaluate the shape difference in our dataset, we adopt the PCA analysis by mapping each high-dimensional shape vector onto a space spanned by the first three principal components. Note that the shape vector is formed by the concatenation of all vertex coordinates, and then linearly aligned to the mean shape before PCA analysis. Fig 2 shows the distribution of 66 prostate shapes, which also indicates the inter-patient shape variation among the shape repository.
Fig. 2.
The prostate shape distribution obtained from the PCA analysis.
A. Related Work
Recently, most studies in T2-weighted MR prostate segmentation focus on two types of methods: multi-atlas-based [4–7] and deformable-model-based [8, 9] segmentation methods. Multi-atlas-based methods are widely used in medical imaging [10–12]. Most research focuses on the design of sophisticated atlas selection or label fusion method. Yan et al [5] proposed a label image constrained atlas selection and label fusion method for prostate MR segmentation. During the atlas selection, label images are used to constrain the manifold projection of intensity images, which can relieve the misleading projection due to other anatomical structures. Ou et al [7] proposed an iterative multi-atlas label fusion method by gradually improving the registration based on the prostate vicinity between the target and atlas images. For deformable-model-based methods, Toth [8] proposed to incorporate different features in the context of AAMs (Active Appearance Models). Besides, with the adoption of the level set, the issue of landmark correspondence can be avoided.
But both types of these methods require careful feature engineering to achieve good performance. The multi-atlas based methods require good features for identifying correspondences between a new testing image and each atlas image [13], while the deformable model relies on discriminative features for separating the target object (e.g., the prostate) from the background [14]. Traditionally, intensity patch is often used as features for the above two methods [15, 16]. However, due to the inhomogeneity of MR images, the simple intensity features often fail in segmentation of MR images with different contrasts and illuminations. To overcome this problem, recent MR prostate segmentation methods started to use features that are specifically designed for vision tasks, such as gradient [17], Haar-like wavelets [18], Histogram of Oriented Gradients (HOG) [19], SIFT [20], Local Binary Patterns (LBP) [21], and variance adaptive SIFT [14]. Compared to simple intensity features, these vision-based features show better invariance to illumination, and also provide some invariance to small rotation. In [22], authors showed that better prostate segmentations could be obtained by using the combination of these features.
One major limitation of the aforementioned handcrafted features is incapable of adapting to data at hand. That means the representation power and effectiveness of these features could vary across different kinds of image data. To deal with this limitation, the learning based feature representation methods [23, 24] are developed to extract latent information, which can be adapted to the data at hand. As one important type of feature learning methods, deep learning recently becomes a hot topic in machine learning [23], computer vision [25], and many other research fields including medical image analysis [26]. Compared with handcrafted features, which need expert knowledge for careful design and also lack sufficient generalization power to different domains, deep learning is able to automatically learn effective feature hierarchies from the data. Therefore, it draws an increasing interest in the research communities. For example, Vincent et al. [27] showed that the features learned by deep belief network and the stacked denoising auto-encoder beat the state-of-the-art handcrafted features for the digit classification problem in the MINST dataset. Farabet et al. [28] proposed to use convolutional network to produce feature representation, which is more powerful in the application of scene labeling than the engineered features, and also achieved the state-of-the-art performance. In the field of medical image analysis, Shin et al. [29] applied the stacked auto-encoders to organ identification in MR images, which shows the potential of deep learning method for application to medical images. In summary, compared with handcrafted features, deep learning has the following advantages: (1) Instead of designing effective features for a new task by trial and error, deep learning largely saves researchers’ time by automating this process. Also, it is capable to exploit the complex feature patterns, which the manual feature engineering is not good at. (2) Unlike the handcrafted features, which are usually shallow in representation due to the difficulty of designing abstract high-level features, deep learning is able to learn the feature hierarchy in a layer-by-layer manner, by first learning the low-level features and then recursively building more comprehensive high-level features based on the previously learned low-level features. (3) When unsupervised pre-training is combined with supervised fine-tuning, the deep-learned features can be optimized for a certain task, such as segmentation, thus boosting the final performance.
B. Our Contribution
Motivated by the above factors, we propose to learn the hierarchical feature representation from MR prostate images by deep feature learning. These learned features are further integrated in a sparse patch matching framework to find the corresponding patches in the atlas images for label propagation. Finally, a deformable model is adopted to segment the prostate by combining the shape prior with the prostate likelihood map derived from sparse patch matching. The main contribution of our method lies in threefold:
Instead of using handcrafted features, we propose to learn the latent feature representation from prostate MR images by the stacked sparse auto-encoder (SSAE) [30, 31], which includes an unsupervised pre-training step and also a task-related fine-tuning step.
By using deep-learned features for measuring inter-patch similarity, a sparse patch matching method is proposed for finding the corresponding patches in the atlas images and then transferring their prostate labels from atlases to the new prostate image.
A deformable model is adopted to further enforce a sparse shape constraint during segmentation, which aims to cope with the large variation existing in prostate shape space.
The proposed method has been extensively evaluated on the T2-weighted MR prostate image dataset, which contains 66 3D images. The manual prostate segmentations are provided by a radiation oncologist for the evaluation purpose. Experimental results show that the sparse patch matching with deep-learned features achieve better segmentation accuracy than using the handcrafted features, as well as the simple intensity features. Besides, compared to other state-of-the-art prostate segmentation methods, our method obtains competitive segmentation accuracy.
C. Brief Outline of Our method
The proposed MR prostate segmentation framework is composed of two levels (Fig. 3). The first level (two upper panels of Fig. 3) learns the deep feature representation and then applies sparse patch matching with the deep-learned features for deriving the prostate likelihood map. Based on the produced likelihood map, the second level (lower panel of Fig. 3) consists of a deformable model by enforcing the shape prior during the evolution of prostate segmentation.
Fig. 3.
The schematic description of our proposed segmentation framework.
The rest of the paper is organized as follows. In Section II, we present the stacked sparse auto-encoder for feature learning and the sparse patch matching framework for deriving the prostate likelihood map in the first level. Section III elaborates both the deformable model and the sparse shape model in the second level. Section IV evaluates the proposed segmentation method on the T2-weighted prostate MR dataset. Finally, conclusive remarks are presented in Section V.
II. First Level: Learning Deep Feature Representation and Sparse Patch Matching
The goal of this level is to learn a latent feature representation for MR prostate images, and then use them to infer a likelihood map of prostate gland for a new image. To achieve this goal, two main stages (i.e., learning stage and testing stage) are conducted as illustrated in the two upper panels of Fig. 3. First, in the learning stage, the intrinsic feature hierarchy from MR prostate image patches is learned by using a deep learning framework, namely the stacked sparse auto-encoder (SSAE). Then, in the testing stage, each image patch from both atlas and target images is first represented by the features learned from the SSAE network. Then, these features are integrated into a sparse patch matching method for estimating the prostate likelihood map by transferring the label information from atlas images to the target image.
The organization of this section is as follows. In Section II.A, we first investigate the limitation of handcrafted features in MR prostate segmentation, and give our motivation of adopting deep learning features. Afterwards, we introduce the feature learning method in Section II.B, and the sparse patch matching in Section II.C, respectively.
A. The Limitation of Handcrafted Features in MR Prostate Segmentation
Since our sparse patch matching method belongs to multi-atlas based segmentation methods, in the following, we will illustrate the importance of features in such context. As briefly mentioned in the Introduction, good features in multi-atlas based segmentation should identify the correct correspondences between the target image and the atlas images. In computer vision, various handcrafted features, such as Haar features [18], HOG features [20] and Local Binary Patterns [21], have been proposed in different applications, with promising results such as in object detection of natural images. However, these features are not suitable for MR prostate images, as they are not invariant to both the inhomogeneity of MR images and the appearance variations of prostate gland.
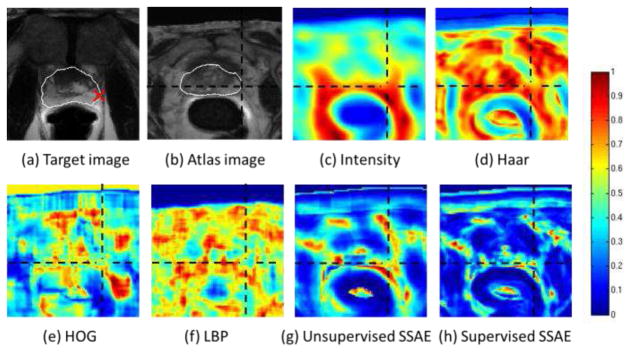
To describe and compare the effectiveness of different features for identifying correspondences in two images, Fig. 4 shows a typical example by computing the similarity maps between one point (shown as red cross in Fig. 4(a)) in the target image (Fig. 4(a)) and all points in an aligned atlas image (Fig. 4(b)). The white contours in (a) and (b) show the prostate boundaries, and the black dashed cross in Fig. 4 (b) indicates the correct correspondence of the red-cross target point in the atlas image. The effectiveness of features can be reflected by the similarity map. If features are distinctive for correspondence detection, the similarity computed by using these features would be high for correct correspondences and low for incorrect correspondences. Fig. 4(c–f) shows the similarity maps computed using different handcrafted features, such as intensity patch features, Haar features, HOG features and LBP features, respectively. It is clear that none of these features could capture correct correspondence, as the similarity between the corresponding voxels indicated by the red crosses is low, compared to that of nearby voxels. This shows that the existing handcrafted features are insufficient in multi-atlas based segmentation for the MR prostate.
Fig. 4.
The similarity maps computed between a reference voxel (red cross) in the target image (a) and all voxels in the atlas image (b) by the four handcrafted feature representations, i.e., intensity (c), Haar (d), HOG (e) and LBP (f), as well as the two deep learning feature representations, namely unsupervised SSAE (g) and the supervised SSAE (h). White contours indicate the prostate boundaries, and the black dashed crosses indicate the ground-truth point in (b), which is corresponding to the red cross in (a).
To relieve the limitation of handcrafted features, it is necessary to learn discriminant features adaptive to MR prostate images. To demonstrate the effectiveness of deep learning features, Fig. 4 (g) and (h) provide the similarity maps computed using the two kinds of deep learning features obtained by our proposed unsupervised and supervised stacked sparse auto-encoder (SSAE), respectively. Compared to similarity maps of handcrafted features, it is clear that the correct correspondence can be better identified with the deep learning features, especially for the supervised SSAE. In the following section, we will elaborate how these features could be adaptively learned from MR prostate images by SSAE.
B. Stacked Sparse Auto-Encoder (SSAE) for Learning the Latent Feature Representation
As illustrated in the previous section, it is necessary to learn the feature representation adaptive to the data, thus alleviating the need of labor-intensive feature engineering. To achieve this purpose, we introduce stacked sparse auto-encoder (SSAE) as a way to learn the latent feature representation from a collection of training prostate image patches. Stacked sparse auto-encoder is a deep learning architecture, which consists of basic feature learning layers, i.e., sparse auto-encoders (SAE). It is built by layer-wise stacking of sparse auto-encoders (Fig. 7). In the following paragraphs, we first introduce the auto-encoder as a basic feature learning algorithm. Then, we explain sparse auto-encoder, which imposes sparsity constraint for learning the robust shallow feature representations. Finally, we elaborate how to learn deep feature hierarchy by stacking multiple sparse auto-encoders layer-wisely.
Fig. 7.
Construction of the unsupervised SSAE with R stacked SAEs.
1) Basic Auto-Encoder
Serving as the fundamental component for SSAE, the basic auto-encoder (AE) trains a feed-forward non-linear neural network, which contains three layers, i.e., input layer, hidden layer, and output layer, as illustrated in Fig. 5. Each layer is represented by a number of nodes. Blue nodes on the left and right sides of Fig. 5 indicate the input and output layers, respectively, and green nodes indicate the hidden layer. Nodes in the two neighboring layers are fully connected, which means that each node in the previous layer can contribute to any node in the next layer. Basically, AE consists of two steps, namely encoding and decoding. In the encoding step, AE encodes the input vector into a concise representation through connections between input and hidden layers. In the decoding step, AE tries to reconstruct the input vector from the encoded feature representation in the hidden layer. The goal of AE is to find a concise representation of input data, which could be used for the purpose of best reconstruction. Since we are interested in the representation of image patches, in this application the input to AE is an image patch, which is concatenated as a vector. In the training stage, given a set of training patches X = {xi ∈ ℝL, i = 1, …, N}, where N and L are the number and the dimension of training patches, respectively, AE automatically learns the weights of all connections in the network by minimizing the reconstruction error in Eq. (1).
Fig. 5.
Construction of the basic AE.
| (1) |
where W, b, Ŵ, b̂ are the parameters in the AE network, and σ(a) = (1 + exp(−a))−1. Given an input vector xi, AE first encodes it into the concise representation hi = σ(Wxi + b), where hi is the responses of xi at the hidden nodes, and the dimension of h equals to the number of nodes in the hidden layer. In the next step, AE tries to decode the original input from the encoded representation, i.e., with Ŵhi + b̂. To learn effective features for the input training patches, AE requires that the dimension of the hidden layer is less than that of the input layer. Otherwise, the minimization of Eq. (1) would lead to trivial solutions, e.g., identity transformation. Studies [32] have also shown that the basic AE learns very similar features as PCA.
Once the weights {W,b,Ŵ,b̂} have been learned through the training patches, in the testing stage AE could efficiently obtain a concise feature representation for a new image patch xnew by a forward passing step, i.e., hnew = σ(Wxnew + b).
2) Sparse Auto-Encoder
Rather than limiting the dimension of hidden layer (i.e., feature representation), an alternative could be imposing regularization on the hidden layer. Sparse auto-encoder (SAE) falls into this category. Instead of requiring the dimension of hidden layer less than that of the input layer, SAE imposes sparsity regularization on the responses of hidden nodes (i.e., h) to avoid the problem of trivial solutions suffered by the basic AE. Specifically, SAE enforces the average response of each hidden node over the training set to be infinitesimal, i.e., , where is the response of the i-th training input at hidden node j, and ρ is a very small constant. In this way, to balance both the reconstruction power and the sparsity of the hidden layer, only a few useful hidden nodes could have responses for each input, thus forcing the SAE network to learn sparse feature representation of the training data. Mathematically, we can extend Eq. (1) to derive the objective function of SAE by adding a sparsity constraint term shown below:
| (2) |
where δ is a parameter to balance between reconstruction and sparsity terms, and M is the number of hidden nodes. KL(ρ|ρj) is the Kullback-Leibler divergence between two Bernoulli distributions with probability ρ and ρj. As we can see, the sparsity term is minimized only when ρj is close to ρ for every hidden node j. Since ρ is set to be a small constant, minimizing Eq. (2) could lead to the sparse responses of hidden nodes, hence the sparsity of learned feature representation.
3) Stacked Sparse Auto-Encoder
By using SAE, we can learn the low-level features (such as Gabor-like features as shown in Fig. 6) from the original data (MR image patches). However, low-level features are not enough due to large appearance variations of the MR prostate. It is necessary to learn abstract high-level features, which could also be invariant to the inhomogeneity of MR images. Motivated by the human perception, which constitutes a deep network to describe concepts in a hierarchical way using multiple levels of abstraction, we recursively apply SAE to learn more abstract/high-level features based on the features learned from the low-level. This multi-layer SAE model is referred to as a stacked sparse auto-encoder (SSAE), which stacks multiple SAEs on top of each other for building deep hierarchies.
Fig. 6.
The low-level feature representation learned from the SAE. Here, we reshape each row in W into the size of image patch, and only visualize its first slice as an image filter.
Fig. 7 shows a typical SSAE with R stacked SAEs. Let W(r), b(r), Ŵ(r) and b̂(r) denote the connection weights and intercepts between the input layer and hidden layer, and between the hidden layer and output layer in the r-th SAE, respectively. In the encoding part of the SSAE, the input vector xi is first encoded by the first SAE for obtaining the low-level representation , i.e., . Then, the low-level representation of the first SAE is considered as the input vector to the next SAE, which encodes it into higher level representation , i.e., . Generally, the r-th level representation can be obtained by a recursive encoding procedure with . Similarly, the decoding step of SSAE recursively reconstructs the input of each SAE. In this example, SSAE first reconstructs the low-level representation from the high-level representation , i.e., with for r =R,…,2. Then, using the reconstructed low-level representation , the original input vector could be estimated, i.e., .
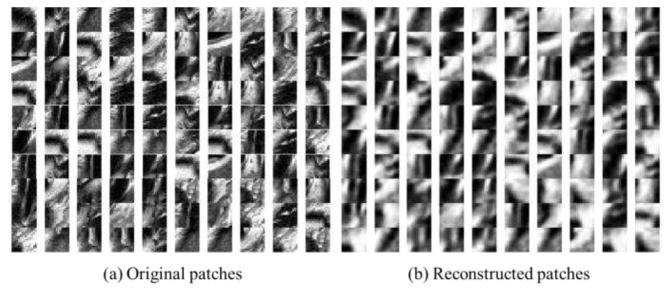
After stacking multiple SAEs together by feeding the output layer from the low-level SAE as the input layer of a high-level SAE, SSAE is able to extract more useful and general high-level features. In the optimization of SSAE, this deep architecture is first pre-trained in an unsupervised layer-wise manner and then fine-tuned by back propagation. Since the aforementioned SSAE network is trained based only on the original image patches, without using the supervised label information, it is denoted as the unsupervised SSAE. Fig. 8 shows some typical prostate image patches and their reconstructions by the unsupervised SSAE with R=4.
Fig. 8.
Typical prostate image patches (a) and their reconstructions (b) by using the unsupervised SSAE with four stacked SAEs.
However, since the unsupervised SSAE trains the whole network on the unlabeled data, the high-level features learned from unsupervised SSAE are only data-adaptive, that is, not necessarily discriminative to separate prostate and background voxels. To make the learned feature representation discriminative [33, 34], the supervised fine-tuning is often adopted by stacking another classification output layer on the top of the encoding part of the SSAE, as shown in red dashed box of Fig. 9. This top layer is used to predict the label likelihood of the input data xi by using the features learned from the most high-level representation . The number of nodes in the classification output layer equals to the number of labels (i.e., “1” denotes prostate, and “0” denotes background). Using the optimized parameters from the pre-training of SSAE as initialization, the entire neural network (Fig. 9) can be further fine-tuned by back-propagation to maximize the classification performance. This tuning step is referred to as the supervised fine-tuning, in contrast with the unsupervised fine-tuning mentioned before. Accordingly, the entire deep network is referred to as the supervised SSAE. Fig. 10 gives a visual illustration of typical feature representations of the first and second hidden layers learned by a four-layer supervised SSAE based on the visualization method in [35]. Here, Figs. 10 (a) and (b) show the visualization of 60 units obtained from the first and second hidden layers under unsupervised pre-training (with unlabeled image patches) and supervised fine-tuning (with labeled image patches), respectively. It can be seen that higher hidden layer tends to be more affected by the classification layer we introduced.
Fig. 9.
Construction of the supervised SSAE with a classification layer, which fine-tunes the SSAE with respect to the task of voxel-wise classification between prostate (label = 1) and background (label = 0).
Fig. 10.
Visualization of typical feature representations of the first hidden layer (first row) and second hidden layer (second row) for the unsupervised pre-training (a) and supervised fine-tuning (b), respectively.
After learning all the parameters {W(r), Ŵ(r), b(r), b̂(r)} of SSAE (r =1,…,R), where R denotes the number of stacked SAEs, the high-level representations of a new image patch xnew can be efficiently obtained by a recursive forwarding pass, i.e., with for r=1,…, R. The final high-level representation will be used as features to guide the sparse patch matching (Section II.C), and propagate labels from atlas images to the target image for estimating the prostate likelihood map.
C. Sparse Patch Matching with the Deep Learning Features
Before sparse patch matching, all atlas images are registered to the target image. This registration includes two steps. First, linear registration is applied for initial alignment, with the guidance from the landmarks automatically-detected around the prostate region [36]. Then, the free-form deformation (FFD) [37] is further adopted to the linearly aligned images for deformable registration.
After learning the SSAE networks (either in unsupervised or supervised manner), each new image patch in the testing stage can be encoded as a high-level feature vector (i.e., the last hidden layer of the SSAE). These features can be fed into a segmentation framework for labeling voxels as either prostate or background. As one of the popular segmentation frameworks, multi-atlas based segmentation demonstrates its effectiveness on dealing with image variations in different applications [38, 39]. However, traditionally the multi-atlas based segmentation adopts only the intensity or handcrafted features for measuring the similarity between different local patches, or computing the weights of different patches during label propagation. Since MR prostate images exhibit large structural and appearance variations, we propose to incorporate the deep learning features, instead of the conventional handcrafted features, into the multi-atlas segmentation framework. As the features extracted by the deep learning methods are usually more robust and discriminative, the performance of multi-atlas segmentation can be improved at the same time. Fig. 11 gives the general description of our multi-atlas based method, called sparse patch matching. In this method, instead of computing pair-wise intensity similarity as a matching weight, we propose to select only a small number of similar atlas patches by sparse representation, which is more robust to outliers. In the following, we give the detailed description for our sparse patch matching method.
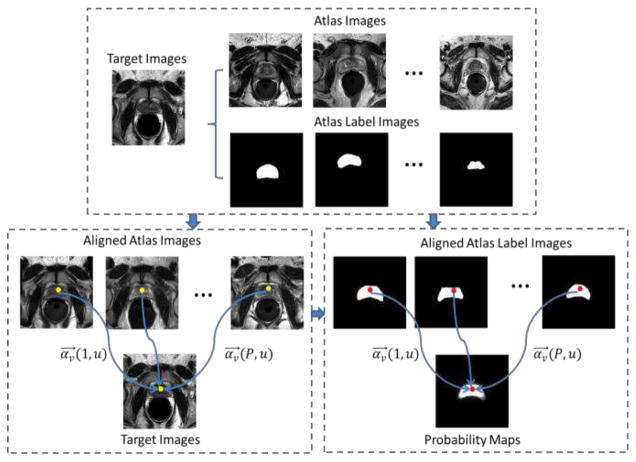
Fig. 11.
The schematic description of sparse patch matching
In order to estimate the prostate likelihood map of a target image Is, we first align all the atlas images {Ip, p=1,…,P} and their label maps {Gp, p=1,…,P} onto the target image Is. Then, to determine the prostate likelihood of a particular voxel v in the target image Is, we first extract the image patch centered at voxel v from the target image, and then all image patches within a certain searching neighborhood 𝒩(v) across all the aligned atlas images.
Next, the deep learned feature representations for those extracted intensity patches are obtained through the encoding procedure of the learned SSAE as introduced in Section II.B. Denote fs(v) as the deep learning features for the target image patch at point v, and denote Av as the feature matrix resulted from column-wise combination of deep learning features of atlas patches, i.e., Av =[fp(u)|p=1,…,P;u ∈ 𝒩(v)]. To estimate the prostate likelihood Qs(v) for voxel v in the target image Is, we linearly combine the label of each voxel u ∈ 𝒩(v) from each atlas image Ip with a weighting vector αv= [αv(p,u)]u=1,…,|𝒩(v)|;p=1,…,P as follows
| (3) |
According to Eq. (3), it is easy to see that the robustness and accuracy of prostate likelihood estimation depends on how well the weighting vector αv is determined. In the literature, different weight estimation methods have been proposed [40, 41]. Most multi-atlas based segmentation methods directly compute αv as the pair-wise similarity between intensity patches, such as using the Euclidean distance. In our method, we compute the weighting vector αv different from the previous methods in respect to the following two aspects. First, instead of using the intensity or handcrafted features, the high-level features are learned from the deep learning architecture. Second, with the help of recently proposed sparse representation method [4], we enforce sparsity constraint upon the weighting vector αv. In this way, we seek for the best representation of the target patch using a limited set of similar atlas patches. Mathematically, the optimization of αv can be formulated as the sparse representation problem below:
| (4) |
The first term is the data fitting term, which measures the difference between the target feature vector fs(v) and the linearly combined feature representation Av αv from all atlas image patches. The second term is the sparsity term, which attributes to the sparsity property of the weighting vector αv. η controls the strength of sparsity constraint on the weighting vector αv. If η is larger, the number of non-zero elements in αv will be smaller. In this way, only a few patches in patch dictionary Av will be selected to reconstruct the target features fs(v) in a non-parameter fashion, thus reducing the risk of including those misleading atlas patches in the likelihood estimation.
Based on the derived weighting vector αv, the prostate likelihood Qs(v) for a target point v can be estimated by Eq. (4). Since the weighting vector αv is sparse, the prostate likelihood Qs(v) is finally determined by the linear combination of labels corresponding to atlas patches with non-zero elements in vector αv. After estimating the prostate likelihood for all voxels in the target image Is, a likelihood map Qs is generated, which can be used to robustly locate the prostate region (as shown in Fig. 11). Usually, a simple thresholding or level set method [42, 43] can be applied to binarize the likelihood map for segmentation. However, since each voxel in the target image is independently estimated in the multi-atlas segmentation method, the final segmentation could be weird as no shape prior is considered. In order to robustly and accurately estimate the final prostate region from the prostate likelihood map, it is necessary to take into account the prostate shape prior during the segmentation.
III. Second Level: Data and Shape Driven Deformable Model
The main purpose of this section is to segment the prostate region based on the prostate likelihood map estimated in the previous section. The likelihood map can be used in two aspects for deformable model construction. First, the initialization of deformable model can be easily built by thresholding the likelihood map. In this way, the limitation of model initialization problem in the traditional deformable segmentation can be naturally relieved. Second, the likelihood map can be used as the appearance force to drive the evolution of deformable model. Besides, in order to deal with the large inter-patient shape variation, we propose to use sparse shape prior for deformable model regularization. In the following paragraphs, we first introduce the sparse shape composition as a non-parametric shape modeling method. Then, we present the optimization of our deformable model by jointly considering both shape and appearance information. Finally, the proposed deformable segmentation method is summarized.
A. Shape Prior by Sparse Shape Composition
Here, our deformable model is represented by a 3D surface, which is composed of K vertices {dk|k=1,…,K}. After concatenating these K vertices {dk|k=1,…,K} into a vector d, each deformable model can be represented as a shape vector with length of 3·K. Let D denotes a large shape dictionary that includes prostate shape vectors of all training subjects. Each column of shape dictionary D corresponds to the shape vector of one subject. The shape dictionary can be used as a shape prior to constrain the deformable model in a learned shape space. Instead of assuming the Gaussian distribution of shapes and then simply using PCA for shape modeling as in the Active Shape Model [44], we adopt a recently proposed method, named sparse shape composition [45], for shape modeling. In the sparse shape composition, the shapes are sparsely represented by shape instances in the shape dictionary without the need of Gaussian assumption. Specifically, given a new shape vector d and shape dictionary D, sparse shape composition method reconstructs shape vector d as the sparse representation of shape dictionary D by minimizing the following objective function:
| (5) |
where ψ(d) denotes the target shape d that is affine aligned onto the mean shape space of shape dictionary D. ε indicates the sparse coefficient for the linear shape combination. Once (ε, ψ) are estimated by Eq. (5), the regularized shape can be computed by ψ−1(Dε), where ψ−1 is the inverse affine transform of ψ.
B. Optimization of Deformable Model Method
For each target image, the segmentation task is formulated as the deformable model optimization problem. During the optimization procedure, each vertex of deformable model dk is driven iteratively by the information from both prostate likelihood map and shape model until converged at the prostate boundaries. Mathematically, the evolution of the deformable model can be formulated as the minimization of an energy function, which contains a data energy Edata and a shape energy Eshape as in Eq. (6):
| (6) |
The data term Edata is used to attract the 3D surface towards the object boundary based on the likelihood map. Specifically, each vertex dk is driven by the force related to the gradient vector of prostate likelihood map. Denote as the gradient vector at vertex dk in the prostate likelihood map, and n⃗s(dk) as the normal vector on the vertex dk of surface. When vertex dk deforms exactly to the prostate boundary and also its normal direction aligns with the gradient direction of prostate boundary, the local matching term 〈 , n⃗s(dk)〉 is maximized. In this case, we formulate to minimize the data energy Edata as:
| (7) |
Since all the vertices on the deformable model are jointly evolved during the deformation, the matching of the deformable model with prostate boundary will be robust to possible incorrect likelihood on some vertices, as well as inconsistency between neighboring vertices.
The shape term Eshape is used to encode the geometric property of prostate shape based on the estimated coefficient ε and the transformation φ in Eq. (5). Specifically, the shape term is formulated as below:
| (8) |
where the first term constrains the deformed shape d to be close to the regularized shape ψ−1(Dε) by the sparse shape composition, and the second term imposes the smoothness constraint on shape, which prevents large deviations between each vertex dk and the center of its neighboring vertices dj ∈𝒩(dk).
By combining Eq. (7) and Eq. (8) into Eq. (6), the vertices on the deformable model are iteratively driven towards the prostate boundary while constraining the shape in a non-parametric shape space.
C. Summary of the Proposed Deformable Segmentation Method
Generally, the overall optimization of deformable model can be summarized as an expectation-maximization (EM) algorithm, which minimizes the data energy and shape energy alternatively. Given the target image Is, the initial deformable surface d0 is estimated by solving the sparse learning problem in Eq. (5). Then the “M” step and “E” step are alternatively executed as follows. Based on the likelihood map generated from stacked sparse auto-encoder and sparse patch matching, in the “M” step, the deformable model dt is first evolved to minimize the data energy function Edata (Eq. (7)). Then, in the “E” step, the parameters (εt, ψt) is estimated for the shape refinement by solving Eq. (5), and the deformable model dt is further refined by minimizing the shape energy (Eq. (8)) with the computed parameters (εt, ψt). After T iterations of the above EM step, the output shape dT is converted to a binary label map Gs, which gives the final segmentation result of the target image Is.
IV. Experiments and Analysis
A. Materials and Parameter Settings
We evaluate our method on the dataset, which includes 66 T2-weighted MR images from the University of Chicago Hospital. The images are acquired with 1.5T magnetic field strength from different patients under different MR image scanners (34 images from Philips Medical Systems and 32 images from GE Medical Systems). Under this situation, the difficulty for the segmentation task increases since both shape and appearance differences are large. In Fig. 12, images (b) and (e) were acquired from a GE MRI scanner, while the other three were acquired from a Philips MRI scanner. As shown in Fig. 12, image (c) was obtained without the endorectal coil. It has different prostate shape with other four images acquired with the endorectal coil. Besides, the prostate appearance suffers from the inhomogeneity (as in (b) and (d)) and noises (as in (d) and (e)), which further produce large variations. The image dimension and spacing are different from image to image. For example, the image dimension varies from 256×256×28 to 512×512×30. The image spacing varies from 0.49×0.49×3 mm to 0.56×0.56×5 mm. The manual delineation of the prostate in each image is provided by a clinical expert as the ground truth for quantitative evaluation. As the preprocessing of the dataset, the bias field correction [46] and histogram matching [47] are applied to each image successively. We adopted the two-fold cross-validation. Specifically, in each case, the images of one fold are used for training the models, while the images of other fold are used for testing the performance.
Fig. 12.
Five typical T2-weighted MR prostate images acquired from different scanners, showing large variations of both prostate appearance and shape, especially for the cases with or without using the endorectal coils.
The parameters for deep feature learning are listed below. The patch size is 15×15×9. The number of layers in SSAE framework is 4. The number of nodes in each layer of SSAE is 800, 400, 200, 100, respectively. Thus, the deep learning features have the dimensionality of 100. The target activation ρ for the hidden units is 0.15. The sparsity penalty β is 0.1. The Deep Learning Toolbox [48] is used for training our SSAE framework.
The searching neighborhood 𝒩(v) is defined as the 7×7×7 neighborhood centered at voxel v. For sparse patch matching, the parameter η in Eq. (4), which controls the sparsity constraint, is 0.001. For the final deformable model segmentation, the parameter λ in Eq. (6), which weights the shape energy during deformation, is 0.5, and the parameter μ in Eq. (5) is 0.5.
B. Evaluation Criteria
Given the ground-truth segmentation S and the automatic segmentation F, the segmentation performance is evaluated by four metrics: Dice ratio, precision, Hausdorff distance, and average surface distance. Dice ratio and precision measure the overlap between two segmentations. Hausdorff distance and average surface distance measures the boundary distance between two surfaces of segmentation. Detailed definitions are given as Table I, where V indicates the volume size, eS and eF are the surfaces for the ground-truth and automatic segmentations, respectively, and dist(di,dj) denotes the Euclidean distance between vertices di and dj.
Table I.
Definition of Evaluation Measurement
| Dice ratio |
|
|
| Precision |
|
|
| Hausdorff distance | max(H(eS, eF), H(eS, eF)), H(eS, eF) = maxdi∈eS{mindj∈eFdist(di, dj)} | |
| Average surface distance |
|
S: ground truth segmentation; F: automatic segmentation; V: volume size;
eS: surfaces of ground-truth segmentation;
eF: surface of automatic segmentation;
di: vertices on the surfaces eS; dj: vertices on the surfaces eF;
dist(di,dj) : Euclidean distance between vertices di and dj
C. Experiment Results
1) Evaluation of the Performance of First Level
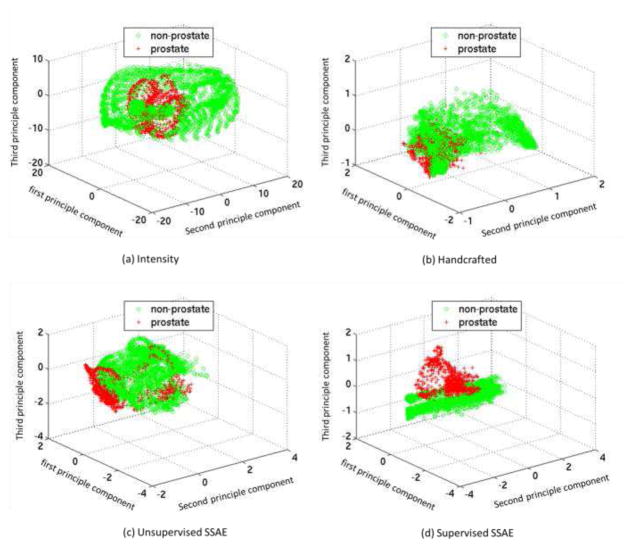
Inspired by paper [49], we plot the PCA-projected features to show the effectiveness of different features in separating voxels from different classes (e.g., the prostate class and the non-prostate class). After mapping each feature vector to the subspace spanned by the first three principal components, the effective features would 1) cluster the voxels with the same class label as close as possible and 2) separate the voxels with different class labels as far as possible. First, we demonstrate the discrimination power of our deep learning features in Fig. 13, by visualizing the PCA-projected feature distributions of different feature representations, i.e., intensity patch (Fig. 13 (a)), handcrafted (Fig. 13 (b)), features learned by unsupervised SSAE (Fig. 13 (c)), and features learned by supervised SSAE (Fig. 13 (d)). Specifically, for the case of handcrafted features, we include three commonly used features, i.e., Haar [50], HoG [47] and LBP [21]. The same patch size is used for computing all features under comparison. It can be seen that the deep learning features from supervised SSAE have better clustering results in the projected feature space, and thus better discriminative power than other two predefined features (i.e., intensity, and handcrafted), as well as deep learning features by unsupervised SSAE. The superior performance of supervised SSAE over the unsupervised SSAE indicates the necessity of utilizing label information to improve the discrimination power of learned features.
Fig. 13.
Distributions of voxel samples by using four types of features: (a) intensity, (b) handcrafted, (c) unsupervised SSAE, and (d) supervised SSAE. Red crosses and green circles denote prostate and non-prostate voxel samples, respectively.
Next, we evaluate the segmentation accuracy of different feature representations in the context of sparse patch matching. Table II lists the quantitative results (Dice ratio, precision, Hausdorff distance, and average surface distance) for all feature representations. The p-values (computed with paired t-test at 5% significance level), comparing the supervised SSAE with all other methods, are provided below each quantitative result. It can be observed that our supervised SSAE method significantly outperforms all the intensity and handcrafted feature methods. According to the paired t-test at 5% significance level, both our proposed method (unsupervised and supervised SSAE) outperformed the rest of competing method, but the supervised SSAE is not statistically superior to the unsupervised SSAE.
Table II.
Mean and Standard Deviation of Quantitative Results for MR Prostate Segmentation with Different Feature Representations.
| Method | Intensity | Haar | HOG | LBP | Handcraft | Unsupervised SSAE | Supervised SSAE |
|---|---|---|---|---|---|---|---|
| Dice (%) | 85.3±6.2 | 85.6±4.9 | 85.7±4.9 | 85.5±4.3 | 85.9±4.5 | 86.7±4.4 | 87.1±4.2 |
| (1.1e-04) | (2.2e-05) | (6.1e-05) | (5.5e-05) | (7.0e-06) | (2.1e-01) | (NA) | |
| Precision (%) | 85.1±8.3 | 85.9±8.5 | 85.3±8.7 | 83.7±7.7 | 87.3±7.4 | 87.3±7.3 | 87.1±7.3 |
| (1.9e-03) | (3.63e-02) | (4.5e-03) | (1.3e-06) | (7.0e-01) | (7.3e-01) | (NA) | |
| Hausdorff | 8.68±4.24 | 8.50±2.86 | 8.51±2.69 | 8.59±2.38 | 8.55±2.91 | 8.65±2.69 | 8.12±2.89 |
| (1.8e-01) | (1.9e-01) | (1.6e-01) | (1.1e-01) | (1.5e-01) | (6.3e-02) | (NA) | |
| ASD | 1.87±0.93 | 1.76±0.52 | 1.74±0.50 | 1.75±0.44 | 1.77±0.54 | 1.68±0.49 | 1.66±0.49 |
| (6.0e-03) | (8.9e-03) | (3.1e-02) | (2.5e-02) | (5.0e-04) | (5.8e-01) | (NA) |
Best performance is indicated by bold font.
Fig. 14 further shows the typical likelihood maps estimated by four different feature representations for three different patients. It can be observed that the features learned from supervised SAE can better capture the prostate boundary, especially on the anterior and right posterior parts of the prostate. Fig. 15 shows some typical segmentation results obtained by the sparse label matching method with four different feature representations, respectively. Similarly, the proposed method (i.e., supervised SSAE) achieves the best segmentation, especially on the anterior parts of the prostate, which demonstrates the effectiveness of our proposed method. Moreover, Fig. 16 gives the typical prostate segmentation results of different patients produced by four different feature representations, respectively. 3D visualization of the segmentation result has been added below each segmentation result shown in 2D. For each 3D visualization, the red surface indicates automatic segmentation results with different features, such as intensity, handcrafted, unsupervised SSAE and supervised SSAE, respectively. The transparent grey surfaces indicate the ground-truth segmentations. Our proposed supervised SSAE method improves the segmentation accuracy on both the anterior and posterior parts of the prostates.
Fig. 14.
(a) Typical slices of T2 MR images with manual segmentations. The likelihood maps produced by sparse patch matching with four feature representations: (b) intensity patch, (c) handcrafted, (d) unsupervised SSAE, and (e) supervised SSAE. Red contours indicate the manual ground-truth segmentations.
Fig. 15.
Typical prostate segmentation results of the same patients produced by four different feature representations: (a) intensity, (b) handcrafted, (c) unsupervised SSAE, and (d) supervised SSAE. Three rows show the results for three different slices of the same patient, respectively. Red contours indicate the manual ground-truth segmentations, and yellow contours indicate the automatic segmentations.
Fig. 16.
Typical prostate segmentation results of three different patients produced by four different feature representations: (a) intensity, (b) handcrafted, (c) unsupervised SSAE, and (d) supervised SSAE. Three odd rows show the results for three different patients, respectively. Red contours indicate the manual ground-truth segmentations, and yellow contours indicate the automatic segmentations. Three even rows show the 3D visualization of the segmentation results corresponding to the images above. For each 3D visualization, the red surfaces indicate the automatic segmentation results using different features, such as intensity, handcrafted, unsupervised SSAE and supervised SSAE, respectively. The transparent grey surfaces indicate the ground-truth segmentations.
2) Evaluation of the Performance of Second Level
In this section, we further evaluate our deformable model to show its effectiveness. The comparison methods contain three different deformable model based methods. The first one is the conventional Active Shape Model (ASM). The second one uses intensity features for multi-atlas label fusion, and then finalizes the segmentation by adopting a deformable model on the produced likelihood map, similar to our proposed method. The second method follows the same procedure as the first one except using the handcrafted features, such as Haar, HOG, and LBP, instead of intensity patch for multi-atlas label fusion. Table III shows the segmentation results of intensity, handcrafted and supervised SSAE with/without deformable model and the p-value (with paired t-test at 5% significance level) between the supervised SSAE with deformable model and all other methods. According to the paired t-test at 5% significance level on Dice ratio, our proposed deformable model is statistically the best among all the competing methods. Specifically, our proposed supervised SAE outperforms the ASM, the intensity based deformable model, and the handcrafted based deformable model by 10.7%, 2.1% and 1.6%, respectively. Besides, it can be seen that, after adopting the second level of deformable segmentation, the segmentation accuracy can be further improved for all the comparing methods.
Table III.
Mean and standard deviation of quantitative results for the segmentations obtained by supervised SSAE with/without using deformable model.
| Intensity | Handcrafted | Supervised SSAE | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Method | ASM | w/o Deformable model | w/ Deformable model | w/o Deformable model | w/ Deformable model | w/o Deformable model | w/ Deformable model* |
| Dice (%) | 78.4±9.7 | 85.3±6.2 | 86.0±4.3 | 85.9±4.5 | 86.4±4.4 | 87.1±4.2 | 87.8±4.0* |
| (2.7e-11) | (3.0e-06) | (7.3e-10) | (7.7e-08) | (5.1e-06) | (2.6e-03) | (NA) | |
| Precision (%) | 71.9±13.8 | 85.1±8.3 | 89.3±7.4 | 87.3±7.4 | 92.3±7.3 | 87.1±7.3 | 91.6±6.5 |
| (1.3e-21) | (8.0e-17) | (4.5e-07) | (1.4e-11) | (7.7e-02) | (5.7e-20) | (NA) | |
| Hausdorff | 11.50±5.48 | 8.68±4.24 | 7.72±2.90 | 8.55±2.91 | 7.97±2.92 | 8.12±2.89 | 7.43±2.82* |
| (8.5e-07) | (3.6e-04) | (2.5e-02) | (3.8e-05) | (1.2e-04) | (7.4e-03) | (NA) | |
| ASD | 3.12±1.71 | 1.87±0.93 | 1.78±0.55 | 1.77±0.54 | 1.71±0.50 | 1.66±0.49 | 1.59±0.51* |
| (9.4e-10) | (6.4e-04) | (2.0e-07) | (2.0e-05) | (1.3e-03) | (1.6e-02) | (NA) | |
Best performance is indicated by bold font.
denotes the statistically best performance among all the methods with/without deformable model (according to the paired t-test at 5% significant level).
D. Discussion
1) The Issues of Deep Learning Method
The key advantage of deep learning methods is learning good features automatically from data and avoiding using the manually-designed feature extractors, which often require high engineering skills. SAE differs from AE and PCA in the aspect that it imposes sparsity on the mapped features (i.e., responses of hidden nodes), thus avoiding the problem of trivial solutions when the dimensionality of hidden features is more than that of the input features. By stacking SAE together, SSAE is able to learn the hierarchical feature representation, similar to other deep learning models. Besides, we use unsupervised initialization in the pre-training stage, which prevents the later supervised training from falling into the bad local minimum. This also contributes to good performance of our method.
However, the issue of small dataset vs. large number of variables arises during the training of the deep network. To prevent the potential issue raised from the limited number of training samples, two strategies are adopted in our method. First, we pre-trained the deep network in a layer-by-layer manner [31, 51], which can learn a hierarchy of feature representation one layer a time. Specifically, in the training of each layer, the features learned from previous layer are feed into the next layer. The first three layers consist of 320,000, 80,000, 20,000 parameters, respectively. In our experiment, totally 396,000 training samples were used, which should be sufficient for this lay-wise pre-training step. Second, in the fine-tuning stage, the entire deep network is refined only by several iterations, thus better preventing the overfitting issue.
To further relieve the possible overfitting issues, we can also use the idea of transfer learning [52–55]. Specifically, in the unsupervised pre-training step, we can borrow MR images from other body parts (e.g., heart) to initialize our deep network, thus capturing more general MR image appearance. We believe that this initialization could benefit the fine-tuning step and thus overcome the small sample problem. Note that similar strategies have been widely used in the field of computer vision and machine learning [31, 51–56].
2) The Issue of Deformable Segmentation Method
According to Eq. (7), the data term of deformable model is driven by the gradient of the prostate likelihood map. One potential issue may happen if evolving the deformable model according to this data term. That is, the gradient will be zero if the initial shape is a bit away from the boundary. We proposed two strategies to address this potential issue. First, we obtained the initial prostate shape by thresholding the probability map, which makes the initialization not far away from the boundary. Second, the deformation is regularized by the shape model. Thus, even some model vertices cannot find the boundary in the capture range, the shape model can still pull them towards the boundary as long as other vertices have been deformed to the boundary. That is, shape regularization makes all vertices deform as a whole, thus addressing the capture range issue.
3) The Computational Time
For our algorithm, the computational time mainly contains three parts: 1) registration part; 2) multi-atlas label fusion part; 3) deformable segmentation part. For registration, the run-time for each affince and non-linear registration is about 20 seconds and four minutes, respectively. For multi-atlas label fusion, the computational time is about 45 minutes, which is the major computational cost of our method. This mainly is due to the individual labeling for a large amount of voxels in each subject image. Currently, we implement multi-atlas label fusion in MATLAB, which is time-consuming for the loop job of sequentially labeling each voxel. For improving the efficiency of our algorithm, one possible solution is to implement the whole label fusion step by using the C++ language. In this way, we expect the computational time to be reduced to 10 minutes for the entire label fusion part. As for the deformable model segmentation part, the computational time is less than one minute.
V. Conclusion
In this paper, we present an automatic segmentation algorithm for T2 MR prostate images. To address the challenges of making the feature representation robust to large appearance variations of the prostate, we propose to extract the deep learning features by the SSAE framework. Then, the learned features are used under sparse patch matching framework to estimate the prostate likelihood map of the target image. To further relieve the impact of large shape variation in the prostate shape repository, a deformable model is driven toward the prostate boundary under the guidance from the estimated prostate likelihood map and sparse shape prior. The proposed method is extensively evaluated on the data set containing 66 prostate MR images. By comparing with several state-of-the-art MR prostate segmentation methods, our method demonstrates the superior performance regarding to the segmentation accuracy.
Acknowledgments
This work was supported in part by NIH grant CA140413.
Contributor Information
Yanrong Guo, Email: yrguo@email.unc.edu, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC 27599 USA.
Yaozong Gao, Email: yzgao@cs.unc.edu, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC 27599 USA.
Dinggang Shen, Email: dgshen@med.unc.edu, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC 27599 USA; and also with Department of Brain and Cognitive Engineering, Korea University, Seoul 02841, Republic of Korea.
References
- 1.Prostate Cancer. Available: http://www.cancer.org/acs/groups/cid/documents/webcontent/003134-pdf.pdf.
- 2.Pondman KM, Fütterer JJ, ten Haken B, Schultze Kool LJ, Witjes JA, Hambrock T, et al. MR-guided biopsy of the prostate: an overview of techniques and a systematic review. European Urology. 2008;54:517–527. doi: 10.1016/j.eururo.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Hricak H, Wang L, Wei DC, Coakley FV, Akin O, Reuter VE, et al. The role of preoperative endorectal magnetic resonance imaging in the decision regarding whether to preserve or resect neurovascular bundles during radical retropubic prostatectomy. Cancer. 2004;100:2655–2663. doi: 10.1002/cncr.20319. [DOI] [PubMed] [Google Scholar]
- 4.Liao S, Gao Y, Shi Y, Yousuf A, Karademir I, Oto A, et al. Automatic prostate MR image segmentation with sparse label propagation and domain-specific manifold regularization. In: Gee J, Joshi S, Pohl K, Wells W, Zöllei L, editors. Information Processing in Medical Imaging. Vol. 7917. Springer; Berlin Heidelberg: 2013. pp. 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan P, Cao Y, Yuan Y, Turkbey B, Choyke PL. Label Image Constrained Multiatlas Selection. Cybernetics, IEEE Transactions on. 2015;45:1158–1168. doi: 10.1109/TCYB.2014.2346394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Li X, Yan P. Multi-atlas Based Image Selection with Label Image Constraint. Machine Learning and Applications (ICMLA), 2012 11th International Conference on; 2012. pp. 311–316. [Google Scholar]
- 7.Ou Y, Doshi J, Erus G, Davatzikos C. Multi-Atlas Segmentation of the Prostate: A Zooming Process with Robust Registration and Atlas Selection. PROMISE12. 2012 [Google Scholar]
- 8.Toth R, Madabhushi A. Multifeature landmark-free active appearance models: application to prostate MRI segmentation. Medical Imaging, IEEE Transactions on. 2012;31:1638–1650. doi: 10.1109/TMI.2012.2201498. [DOI] [PubMed] [Google Scholar]
- 9.Toth R, Bloch BN, Genega EM, Rofsky NM, Lenkinski RE, Rosen MA, et al. Accurate prostate volume estimation using multifeature active shape models on T2-weighted MRI. Academic radiology. 2011;18:745–754. doi: 10.1016/j.acra.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Sabuncu MR, Yeo BTT, Van Leemput K, Fischl B, Golland P. A generative model for image segmentation based on label fusion. Medical Imaging, IEEE Transactions on. 2010;29:1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y, Shi Y, Zhan L, Gutman BA, de Zubicaray GI, McMahon KL, et al. Automatic clustering of white matter fibers in brain diffusion MRI with an application to genetics. NeuroImage. 2014 Oct 15;100:75–90. doi: 10.1016/j.neuroimage.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Shi Y, Zhan L, TPM Automated multi-atlas labeling of the fornix and its integrity in alzheimer’s disease. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on; 2015. pp. 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Wee CY, Shi F, Thung KH, Ni D, Yap PT, et al. Identification of infants at high-risk for autism spectrum disorder using multiparameter multiscale white matter connectivity networks. Human Brain Mapping. 2015;36:4880–4896. doi: 10.1002/hbm.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Li X, Turkbey B, Choyke PL, Yan P. Prostate Segmentation in MR Images Using Discriminant Boundary Features. Biomedical Engineering, IEEE Transactions on. 2013;60:479–488. doi: 10.1109/TBME.2012.2228644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langerak TR, van der Heide UA, Kotte ANTJ, Viergever MA, van Vulpen M, Pluim JPW. Label fusion in atlas-based segmentation using a selective and iterative method for performance level estimation (SIMPLE) Medical Imaging, IEEE Transactions on. 2010;29:2000–2008. doi: 10.1109/TMI.2010.2057442. [DOI] [PubMed] [Google Scholar]
- 16.Chandra SS, Dowling JA, Kai-Kai S, Raniga P, Pluim JPW, Greer PB, et al. Patient specific prostate segmentation in 3-D magnetic resonance images. Medical Imaging, IEEE Transactions on. 2012;31:1955–1964. doi: 10.1109/TMI.2012.2211377. [DOI] [PubMed] [Google Scholar]
- 17.Somkantha K, Theera-Umpon N, Auephanwiriyakul S. Boundary detection in medical images using edge following algorithm based on intensity gradient and texture gradient features. Biomedical Engineering, IEEE Transactions on. 2011;58:567–573. doi: 10.1109/TBME.2010.2091129. [DOI] [PubMed] [Google Scholar]
- 18.Viola P, Jones M. Rapid object detection using a boosted cascade of simple features. Computer Vision and Pattern Recognition, Proceedings of the 2001 IEEE Computer Society Conference on; 2001. pp. I-511–I-518. [Google Scholar]
- 19.Dalal N, Triggs B. Histograms of oriented gradients for human detection. Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on; 2005. pp. 886–893. [Google Scholar]
- 20.Lowe D. Distinctive image features from scale-invariant keypoints. International Journal of Computer Vision. 2004;60:91–110. [Google Scholar]
- 21.Ojala T, Pietikäinen M, Harwood D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognition. 1996;29:51–59. [Google Scholar]
- 22.Liao S, Gao Y, Lian J, Shen D. Sparse Patch-Based Label Propagation for Accurate Prostate Localization in CT Images. Medical Imaging, IEEE Transactions on. 2013;32:419–434. doi: 10.1109/TMI.2012.2230018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bengio Y. Representation learning: a review and new perspectives. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 24.Mairal J, Bach F, Ponce J, Sapiro G, Zisserman A. Supervised dictionary learning. Advances in neural information processing systems. 2009 [Google Scholar]
- 25.Farabet C, Couprie C, Najman L, LeCun Y. Learning hierarchical features for scene labeling. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2013;35:1915–1929. doi: 10.1109/TPAMI.2012.231. [DOI] [PubMed] [Google Scholar]
- 26.Carneiro G, Nascimento JC, Freitas A. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search Methods. Image Processing, IEEE Transactions on. 2012;21:968–982. doi: 10.1109/TIP.2011.2169273. [DOI] [PubMed] [Google Scholar]
- 27.Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol PA. Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J Mach Learn Res. 2010;11:3371–3408. [Google Scholar]
- 28.Farabet C, Couprie C, Najman L, Lecun Y. Scene parsing with Multiscale Feature Learning, Purity Trees, and Optimal Covers. Proceedings of the 29th International Conference on Machine Learning (ICML-12); 2012. pp. 575–582. [Google Scholar]
- 29.Hoo-Chang S, Orton MR, Collins DJ, Doran SJ, Leach MO. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2013;35:1930–1943. doi: 10.1109/TPAMI.2012.277. [DOI] [PubMed] [Google Scholar]
- 30.Vincent P, Larochelle H, Bengio Y, Manzagol P-A. Extracting and composing robust features with denoising autoencoders. Proceedings of the Twenty-fifth International Conference on Machine Learning (ICML’08); 2008. pp. 1096–1103. [Google Scholar]
- 31.Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. Advances in Neural Information Processing Systems 19 (NIPS’06) 2006:153–160. [Google Scholar]
- 32.Bengio Y. Deep learning of representations for unsupervised and transfer learning. Workshop on Unsupervised and Transfer Learning; 2012. [Google Scholar]
- 33.Bulò SR, Kontschieder P. Neural decision forests for semantic image labelling. IEEE Conference on Computer Vision and Pattern Recognition; 2014. pp. 81–88. [Google Scholar]
- 34.Suk H-I, Shen D. Deep learning-based feature representation for AD/MCI classification. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Vol. 8150. Springer; Berlin Heidelberg: 2013. pp. 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Grosse R, Ranganath R, Ng AY. Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. presented at the Proceedings of the 26th Annual International Conference on Machine Learning; Montreal, Quebec, Canada. 2009. [Google Scholar]
- 36.Zhan Y, Zhou X, Peng Z, Krishnan A. Active scheduling of organ detection and segmentation in whole-body medical images. In: Metaxas D, Axel L, Fichtinger G, Székely G, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008. Vol. 5241. Springer; Berlin Heidelberg: 2008. pp. 313–321. [DOI] [PubMed] [Google Scholar]
- 37.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, et al. Fast free-form deformation using graphics processing units. Computer Methods and Programs in Biomedicine. 2010 Jun;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Jorge Cardoso M, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, et al. STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Medical Image Analysis. 2013;17:671–684. doi: 10.1016/j.media.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. Medical Imaging, IEEE Transactions on. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asman AJ, Landman BA. Non-local statistical label fusion for multi-atlas segmentation. Medical Image Analysis. 2013;17:194–208. doi: 10.1016/j.media.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nouranian S, Mahdavi SS, Spadinger I, Morris W, Salcudean S, Abolmaesumi P. An automatic multi-atlas segmentation of the prostate in transrectal ultrasound images using pairwise atlas shape similarity. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Vol. 8150. Springer; Berlin Heidelberg: 2013. pp. 173–180. [DOI] [PubMed] [Google Scholar]
- 42.Chan TF, Vese LA. Active contours without edges. Image Processing, IEEE Transactions on. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 43.Kim M, Wu G, Li W, Wang L, Son Y-D, Cho Z-H, et al. Automatic hippocampus segmentation of 7.0 Tesla MR images by combining multiple atlases and auto-context models. NeuroImage. 2013;83:335–345. doi: 10.1016/j.neuroimage.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models-their training and application. Computer Vision and Image Understanding. 1995;61:38–59. [Google Scholar]
- 45.Zhang S, Zhan Y, Metaxas DN. Deformable segmentation via sparse representation and dictionary learning. Medical Image Analysis. 2012;16:1385–1396. doi: 10.1016/j.media.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. Medical Imaging, IEEE Transactions on. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 47.Gao Y, Liao S, Shen D. Prostate segmentation by sparse representation based classification. Medical Physics. 2012;39:6372–6387. doi: 10.1118/1.4754304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.https://github.com/rasmusbergpalm/DeepLearnToolbox.
- 49.Hinton GE, Salakhutdinov RR. Reducing the Dimensionality of Data with Neural Networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 50.Mallat SG. A theory for multiresolution signal decomposition: the wavelet representation. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1989;11:674–693. [Google Scholar]
- 51.Hinton GE, Osindero S, Teh YW. A Fast Learning Algorithm for Deep Belief Nets. Neural Computation. 2006;18:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 52.Sinno Jialin P, Qiang Y. A Survey on Transfer Learning. Knowledge and Data Engineering, IEEE Transactions on. 2010;22:1345–1359. [Google Scholar]
- 53.van Opbroek A, Ikram MA, Vernooij MW, de Bruijne M. Transfer Learning Improves Supervised Image Segmentation Across Imaging Protocols. Medical Imaging, IEEE Transactions on. 2015;34:1018–1030. doi: 10.1109/TMI.2014.2366792. [DOI] [PubMed] [Google Scholar]
- 54.Heimann T, Mountney P, John M, Ionasec R. Real-time ultrasound transducer localization in fluoroscopy images by transfer learning from synthetic training data. Medical Image Analysis. 2014;18:1320–1328. doi: 10.1016/j.media.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Sawada Y, Kozuka K. Transfer learning method using multi-prediction deep Boltzmann machines for a small scale dataset. Machine Vision Applications (MVA), 2015 14th IAPR International Conference on; 2015. pp. 110–113. [Google Scholar]
- 56.Wang M, Li W, Liu D, Ni B, Shen J, Yan S. Facilitating Image Search With a Scalable and Compact Semantic Mapping. Cybernetics, IEEE Transactions on. 2015;45:1561–1574. doi: 10.1109/TCYB.2014.2356136. [DOI] [PubMed] [Google Scholar]