Abstract
Transfer RNAs (tRNAs) are the macromolecules that transfer activated amino acids from aminoacyl-tRNA synthetases to the ribosome, where they are used for the mRNA guided synthesis of proteins. Transfer RNAs are ancient molecules, perhaps even predating the existence of the translation machinery. Albeit old, these molecules are tremendously conserved, a characteristic that is well illustrated by the fact that some bacterial tRNAs are efficient and specific substrates of eukaryotic aminoacyl-tRNA synthetases and ribosomes. Considering their ancient origin and high structural conservation, it is not surprising that tRNAs have been hijacked during evolution for functions outside of translation. These roles beyond translation include synthetic, regulatory and information functions within the cell. Here we provide an overview of the non-canonical roles of tRNAs and their mimics in bacteria, and discuss some of the common themes that arise when comparing these different functions.
Graphical Abstract
Introduction
All currently known genetic information is coded in nucleic acids (RNA or DNA). In order to be functional, much of this genetic information must first be decoded into proteins that are composed of amino acids that do not have any specific affinity to the message coded in nucleic acids (Crick, 1958; Crick, 1970; Francklyn and Minajigi, 2010). In order to translate this genetic information cells need adaptors, molecules that can translate nucleic acid into amino acid sequences (Crick, 1958; Hoagland et al., 1958). This adaptor function is performed by aminoacyl-transfer RNAs (aa-tRNA), which have a nucleic acid portion composed of a highly structured transfer RNA (tRNA) of about 80 nucleotides. The second part of an aa-tRNA molecule is composed of a single amino acid bound through an ester bond to the 2' or 3' OH of the 3'-end of the tRNA. The secondary structure of the tRNA is composed of an acceptor arm that carries the amino acid, the anticodon arm where there is an anticodon triplet which recognizes codons on mRNA during translation, and the deoxyuridine, TΨC and variable arms (D, T, and V arms respectively) (Fig. 1) (Ladner et al., 1975; Giegé and Frugier, 2000; Marck and Grosjean, 2002). Transfer RNAs are functionally grouped into families of isoacceptors, which may differ in sequence, but all of which carry a unique amino acid for protein synthesis. This amino acid is added by aminoacyl-tRNA synthetases (aaRS), a group of enzymes that specifically recognize and activate a particular amino acid and transfer it to the correct tRNA. This reaction is central to the process of translation as it ensures that the correct amino acid is transferred to the nascent peptide (Cavarelli and Moras, 1993; Schimmel et al., 1993; Ibba et al., 2005). In order to select the correct tRNAs, aaRS recognize specific nucleotides called “identity elements”. These nucleotides are scattered throughout the tRNA structure, but are usually concentrated in the anticodon loop and the acceptor stem (Beuning and Musier-Forsyth, 1999). Ribosomes also contact tRNAs throughout their surface and require them to be flexible molecules. Nevertheless, with the exception of discrimination between initiator and elongator tRNAs, interactions with anticodon nucleotides have a predominant role in selection of the correct tRNA. Additionally, other regions of the tRNA have a role in compensating for the different affinity of the diverse anticodons and amino acids that are used in translation either directly interacting with the ribosome or allowing for optimal flexibility (Dale and Uhlenbeck, 2005; Khade and Joseph, 2010; Shepotinovskaya and Uhlenbeck, 2013).
Figure 1. Structure of tRNA.
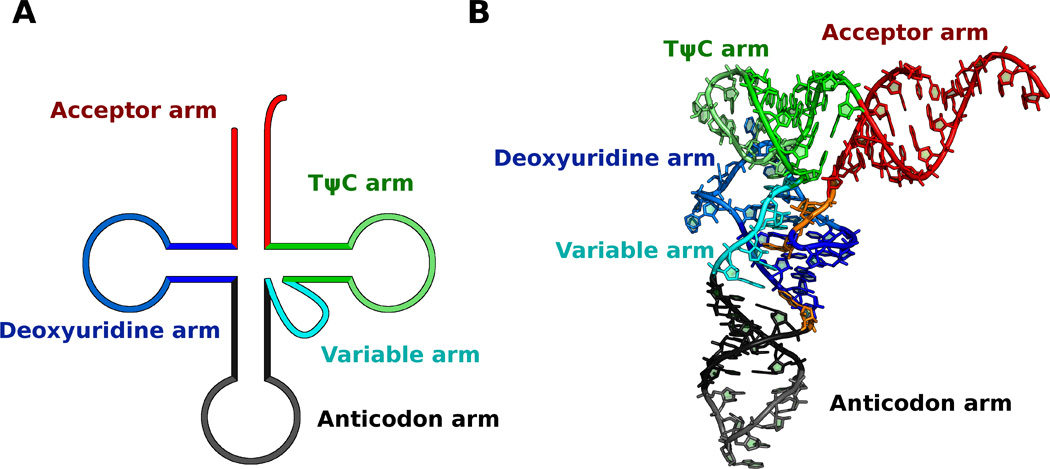
Schematic representation of A) secondary and B) tertiary structure of tRNA. Tertiary structure is based on the backbone of yeast tRNAAsp as in pdb model 1VTQ (Moras et al., 1980). The main parts of tRNA are highlighted in different colors, using darker colors for stems and lighter colors for loops. Used colors are red: acceptor stem, blue: deoxyuridine arm, gray: anticodon arm, cyan: variable arm, green: TΨC arm and orange: segments linking tRNA arms.
Phylogenetic studies and the observation that tRNA structure and the genetic code are essentially maintained in all known organisms, suggest that tRNAs (or their direct and structurally similar predecessor) originated at the time of the last universal common ancestor or earlier (Schimmel et al., 1993; Di Giulio, 1995; Di Giulio, 2006; Sun and Caetano-Anollés, 2008; Moura et al., 2010). Precursors of tRNA might even predate the existence of proteins and could have had a catalytic or metabolic role outside of translation in the “RNA world” (Rodin et al., 2011; de Vladar, 2012; Morgens, 2013). Regardless of their exact origin, the high degree of similarity between extant tRNAs probably derives from their role in a very complex machinery where changes in one component could necessitate modification of other parts of the genetic code and/or translation apparatus to maintain function. An example of this can be observed in the mitochondria of several metazoans where many tRNAs have lost either their T or D arms. In the cases that have been studied in more detail, it was observed that EF-Tu, rRNA, ribosomal proteins and aaRS coevolved to allow the recognition of these non-canonical tRNAs (Watanabe et al., 2014). Changes in the “sense” of codons (where they are reassigned to encode a different amino acid) also require major modifications as the appearance of this new sense for a codon would otherwise force the introduction of amino acids in an erroneous context. For instance, comparative genomic studies have suggested that Leu CUG codon reassignment to Ser in several Candida and Debaryomyces fungi led to a drastic reduction in the usage of these codons (Moura et al., 2010). Replicating this codon reassignment in the model organism Saccharomyces cerevisiae resulted in decreased fitness in several growth media and decreased thermal stability for at least one protein. Nevertheless, these experiments also showed a great growth advantage in some culture conditions (e.g. a 500% growth advantage in the presence of copper sulfate). This change in niche preference and a probable low level of misincorporation in the original conditions may have allowed the fixation of these changes to the genetic code (Mateus et al., 2013). Other codon reassignments may have required the prior elimination of the original codon from the genetic code, for instance due to abnormally high or low CG usage in the genome (Moura et al., 2010). Although such tRNA-dependent modifications to the translation apparatus are rare, they probably had an important role in the original establishment and/or expansion of the genetic code by facilitating the coding of new amino acids (Moura et al., 2010; Rodin et al., 2011; de Vladar, 2012). Beyond its integral role in defining and translating the genetic code, tRNA has also evolved a surprisingly wide range of alternative functions (Fig. 2). The aim of this review is to analyze the pathways that have evolved to utilize tRNAs, sometimes by hijacking them to function beyond the translation machinery. Emphasis will be placed on bacterial systems, and examples from other organisms will be mentioned when relevant.
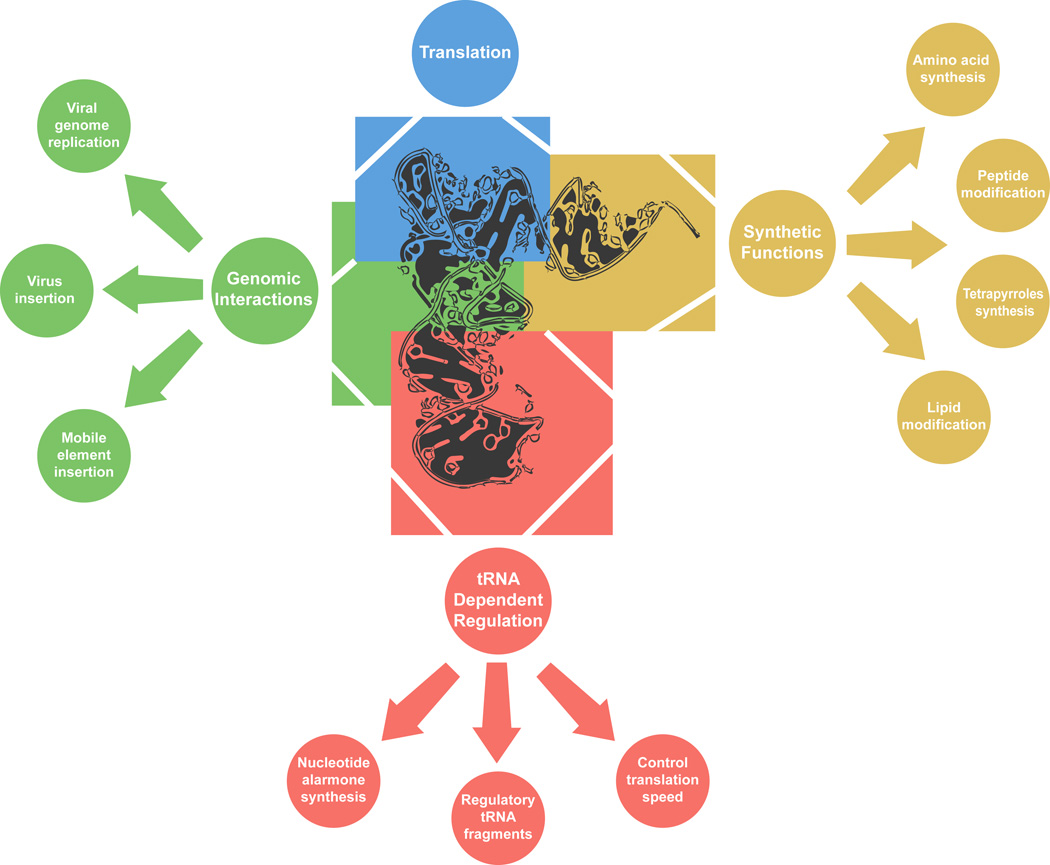
Figure 2. Biological roles of tRNA.
Classification of the diverse roles that tRNAs play in cell biology. Functions of tRNAs were classified in 4 main groups, translation (blue), synthetic functions beyond translation (yellow), tRNA dependent regulation (red) and genomic interactions (green). Positioning of each group in the figure is not related to the parts of tRNA involved in specific functions.
aa-tRNA based synthesis beyond translation
Modification of macromolecules
Many different aa-tRNAs are used outside of translation to provide activated amino acids for a wide variety of purposes. These include the synthesis of several metabolites as well as the modification of macromolecules (RajBhandary and Söll, 2008; Banerjee et al., 2010; Francklyn and Minajigi, 2010; Dare and Ibba, 2012; Katz and Orellana, 2012a; Belin et al., 2012; Raina and Ibba, 2014). Some of the earliest described examples of such roles outside of translation are related to modification of cellular envelopes (RajBhandary and Söll, 2008; Francklyn and Minajigi, 2010). For example, the addition of amino acids to phosphatidylglycerol through ester bonds to the glycerol moiety and synthesis of certain peptide cross bridges for peptidoglycan both require aa-tRNA substrates (RajBhandary and Söll, 2008). Modification of lipids is used to change the membrane charge distribution, thereby protecting the cell against some antibiotics or bactericidal peptides produced by competing species (Dare and Ibba, 2012; Raina and Ibba, 2014; Goldfine, 2014). Lipid aminoacylation also participates in resistance to osmotic or pH stresses (Dare and Ibba, 2012; Goldfine, 2014) and might even have a role in bacterial replication by modulating the ability of DnaA to exchange ADP with ATP (Ichihashi et al., 2003). In addition to lipid modification, aa-tRNAs can also be used as substrates for the addition of amino acids to peptides in bacteria. This includes the synthesis of the pentapeptide bridge from peptidoglycan peptide by MurM or Fem proteins, modification of the amino terminus in proteins starting with basic amino acids by leucyl/phenylalanyl transferases (L/F-transferases; in eukaryotes Arg-tRNA-protein transferase modifies proteins with Arg) and the synthesis or modification of some small peptides using enzymes like AlbC, PacB or VlmA (Berger-Bächi and Tschierske, 1998; Garg et al., 2008; Zhang et al., 2011; Dougan et al., 2012; Dare and Ibba, 2012; Belin et al., 2012; Shepherd and Ibba, 2013b; Raina and Ibba, 2014). These amino acid additions to peptides have diverse functions. The peptidoglycan bridge changes the structural properties of the cell wall, enhancing antibiotic resistance and binding of some extracellular proteins (Berger-Bächi and Tschierske, 1998; Guan et al., 2004; Scheffers and Pinho, 2005; Dare and Ibba, 2012; Shepherd and Ibba, 2013b). By contrast, modification of the amino terminus of cytoplasmic proteins modulates the half-life of specific proteins by targeting them for recognition by ClpS and subsequent degradation by ClpAP protease in bacteria or by UBR proteins and subsequent degradation by the 26S proteasome in eukaryotes (Dougan et al., 2012; Raina and Ibba, 2014). Finally, small peptides synthesized using the activated amino acid on aa-tRNA as a precursor correspond to secondary metabolites with diverse physiological functions ranging from antibiotics and inter-cellular signaling molecules to metal chelators (Garg et al., 2008; Zhang et al., 2011; Belin et al., 2012).
In all cases that have so far been studied, the specificity for aa-tRNA in the modification of macromolecules depends primarily on recognition of the amino acid with a minor but relevant role for the tRNA, particularly the acceptor stem. For instance, FemX from Weissella viridescens uses Ala-tRNAAla as substrate, excluding from its active site any amino acid that is bigger than Ala. The only aa-tRNA carrying a smaller amino acid, Gly-tRNAGly, is excluded by recognition of tRNA “anti-determinants”, that is, the exclusion of any tRNA that presents a specific sequence (in this case base pair C2-G71) that is absent from the natural substrate of the enzyme (Fonvielle et al., 2009). Base pairs that are outside of the acceptor stem of tRNA, or even 3 to 4 base pairs away from the amino acid, have a negligible effect on the aa-tRNA specificity of FemX (Villet et al., 2007; Fonvielle et al., 2009; Fonvielle et al., 2013). Studies on aminoacyl-phosphatidylglycerol synthases (MprF or aa-PGS) that catalyze lipid modification (Gould et al., 1968; Roy and Ibba, 2008; Hebecker et al., 2011; Dare and Ibba, 2012), L/F-transferases that modify the amino terminus of proteins tagging them for degradation (Suto et al., 2006; Watanabe et al., 2007; Wagner et al., 2011; Fung et al., 2014), and AlbC that catalyzes the synthesis of a cyclic dipeptide (Bonnefond et al., 2011; Sauguet et al., 2011; Moutiez et al., 2014) all showed similar dependence on acceptor stem sequences. From this perspective, AlbC is particularly interesting as the enzyme uses two aa-tRNAs that bind sequentially to different regions of the enzyme. The enzyme is more specific for the recognition of the first aa-tRNA (where it uses only one amino acid) than for the second aa-tRNA (where it uses one amino acid preferentially, but accepts others). In the case of AlbC, recognition of the second aa-tRNA depends strongly on the identity of the first base pair of the tRNA acceptor stem (G1-C72 in AlbC substrates), while the identity of these nucleotides is not relevant for the recognition of the first aa-tRNA substrate (Moutiez et al., 2014).
Synthesis of small metabolites
In addition to their roles in the synthesis or modification of macromolecules, tRNAs participate in the production of several much smaller metabolites. Beside the synthesis of di-peptides mentioned above, tRNAs have central roles in the synthesis of tetrapyrroles (such as heme or chlorophyll) and several amino acids. Many organisms lack the complete set of aaRSs required to aminoacylate every tRNA isoacceptor with the corresponding amino acid. In these organisms some tRNAs are aminoacylated in an indirect pathway where the cognate tRNA is first “mis-acylated” with a precursor of the correct amino acid that is subsequently modified while on the tRNA. These indirect pathways have been observed for the synthesis of glutamine on tRNAGln (in all known archaea, as well as in most bacteria and eukaryal organelles), asparagine on tRNAAsn (in several bacteria and archaea), formylmethionine on tRNAfMet (in bacteria and eukaryotic organelles), cysteine on tRNACys (in some archaea) and selenocysteine on tRNASec (in some bacteria, archaea and eukarya) (Ibba et al., 2000; O’Donoghue et al., 2005; Sheppard et al., 2008; Yuan et al., 2008; Katz and Orellana, 2012a). In several organisms the main function of these pathways is to provide aa-tRNA for the synthesis of proteins. Furthermore, some organisms lack alternative pathways to synthesize these amino acids and for them, tRNA dependent synthesis is the sole means to produce these metabolites (Sauerwald et al., 2005; O’Donoghue et al., 2005; Katz and Orellana, 2012a; Mladenova et al., 2014). The indirect aminoacylations of “amide” (Gln and Asn) tRNAs were among the first of these pathways to be described. In these examples, tRNAGln or tRNAAsn are first aminoacylated with the “acid” amino acid (Glu or Asp) by a non-discriminating aaRS (ND-GluRS or ND-AspRS). In some organisms, these non-discriminating enzymes aminoacylate the tRNAs for both acid (tRNAGlu or tRNAAsp) and amide (tRNAGln or tRNAAsn) amino acids, but in other organisms there are specialized enzymes for each kind of tRNA. In a second step, a tRNA dependent amidotransferase (AdT) catalyzes the formation of the amide (Gln or Asn) on the tRNA (Katz and Orellana, 2012a). The other indirect pathways follow similar steps, first adding a precursor (phosphoserine (Sep) on tRNACys, Met on tRNAfMet or Ser on tRNASec) that is converted into the final product in one or two additional reactions (catalyzed by Sep-tRNA:Cys-tRNA synthase (SepCysS) for Cys-tRNACys, methionyl-tRNA formyltransferase (FMT) for fMet-tRNAfMet and selenocysteine synthase (SelA) in bacteria or phosphoseryl-tRNA kinase (PSTK) plus Sep-tRNA:Sec-tRNA synthase (SepSecS) in archaea and eukarya for Sec-tRNASec). In all these pathways, the production of a “mis-acylated” tRNA as an intermediary could potentially allow the mis-incorporation of amino acids during translation. This is prevented mainly by two processes: 1) the ability of translation elongation factors and, possibly the ribosomal A site, to partially discriminate tRNAs, binding preferentially the correctly acylated ones (LaRiviere et al., 2001; Dale and Uhlenbeck, 2005; Yuan et al., 2008; Katz and Orellana, 2012a) and 2) the formation of complexes between the non-discriminatory aaRS and the modifying enzyme from the second step of the pathway that sequester the mis-acylated tRNAs (Bailly et al., 2007; Zhang et al., 2008; Rampias et al., 2010; Katz and Orellana, 2012a). Additionally, in some cases channeling is not required as kinetic competition between the amino acid modifying enzyme and elongation factor prevents accumulation of mis-acylated intermediates (Bhaskaran and Perona, 2011).
While the enzymes used for aa-tRNA-dependent modification of macromolecules recognize their substrates mainly through direct recognition of the amino acid and the nearby nucleotides of the acceptor stem on the tRNA, enzymes involved in the modification of amino acids on tRNAs may recognize a bigger surface of the tRNA including the D and/or T arms. For example, bacterial AdT (GatCAB) (Katz and Orellana, 2012a), eukaryotic and archaeal PSTK (Sherrer et al., 2008; Yuan et al., 2008) and FMT from bacteria and mitochondria of mammals (Lee et al., 1991; Varshney et al., 1991; Guillon et al., 1992; Li et al., 1996; Newton et al., 1999; Takeuchi et al., 2001) have all been shown to recognize their substrates through interactions with the acceptor and D arms. In contrast, FMT from yeast mitochondria (Vial et al., 2003) and SepCys (Fukunaga and Yokoyama, 2007; Helgadóttir et al., 2012) have been suggested to recognize tRNA through its acceptor stem and SepSecS through its acceptor and T stems (Palioura et al., 2009).
Unlike the amino acid synthesis reactions described above, the first step of tRNA-dependent tetrapyrrole synthesis in archaea, chloroplast and most bacteria involves removal of the amino acid attached to Glu-tRNAGlu. In this reaction Glu is detached from tRNAGlu and transferred to the enzyme glutamyl-tRNA reductase (GluTR) where it is reduced to form glutamate 1-semialdehyde (GSA). Subsequently, GSA is channeled to the second enzyme of the pathway GSA aminotransferase where it is isomerized to δ-aminolevulinic acid, which is the universal precursor for tetrapyrrole synthesis. GluTR can use Gln-tRNAGlu as a substrate indicating that the enzyme is not very stringent in amino acid recognition. Additionally, experiments performed with several mutants indicated that GluTR accepts several variations in the tRNA sequence. In contrast to all the other enzymes discussed previously, GluTR does not tightly recognize the amino acid or any specific sequence features. Instead, the enzyme seems to recognize the global folding of tRNAGlu mainly due to the nucleotide in position 47, which is absent in tRNAGln (O’Brian and Thöny-Meyer, 2002; Randau et al., 2004; Heinemann et al., 2008; Katz and Orellana, 2012a).
In some organisms that use these alternative pathways, specialized tRNAs have been found. For instance, in Staphylococcus epidermidis and apparently Staphylococcus aureus there are specialized tRNAGly isoacceptors that are used for peptidoglycan synthesis, but not for mRNA translation (Bumsted et al., 1968; Giannouli et al., 2009). Conversely, in some strains of Acidithiobacillus ferrooxidans a specialized tRNAGlu used in translation, but unable to participate in tetrapyrrole synthesis, has been found (Levicán et al., 2005). In both cases such specialized tRNAs might ensure aa-tRNA availability for both pathways or alternatively prevent sudden changes in tRNA aminoacylation levels when usage by one of the alternative pathways changes. Nevertheless, neither of these hypotheses has been tested. When a tRNA is released from the constraints imposed by its participation in translation, its sequence can deviate from the canonical. As an example, all tRNAGly from S. aureus that apparently are specialized in peptidoglycan synthesis have an altered sequence that prevents binding to EF-Tu, including one that has deviated in structure (mainly at its D and T arms) to the degree that tRNA prediction software categorizes the sequence as belonging to a pseudogene (Giannouli et al., 2009). Another potential example of this is tRNAOther in Bacillus cereus, which presents several deviations from canonical tRNA structure including a G2:A71 bulge and the fact that it presents a Trp anticodon whereas it is aminoacylated with Lys (Ataide et al., 2005). Although originally shown to be a substrate for aminoacylation in vitro (Ataide et al., 2005), this tRNA was later shown to be absent from ribosome fractions in vivo. Instead of participating in translation, it was shown to be part of a larger regulatory RNA, where it was proposed to represent an important structural element (Rogers et al., 2012). The role of the smaller tRNA like fragment has yet to be determined.
Hijacking of tRNA for non-synthetic processes
The information in tRNAs can be used for several “non-synthetic” roles which do not require donating an activated amino acid. Some of these roles, such as the usage of tRNAs as primers for viral genome replication (Mak and Kleiman, 1997) or the use of their genes as insertion sites in the genome for invading viruses (Hacker and Kaper, 2000; Williams, 2002) depend on the mere existence of these molecules, while others such as the use for regulatory functions take advantage of tRNA’s ability to act as a messenger for the metabolic status of the cell.
tRNA in regulatory processes
The ratio of aa-tRNA to deacylated tRNA depends on the availability of amino acids for aminoacylation and the speed of translation. Thus, the aminoacylation status of tRNA is a good indicator of the balance between the pathways that use amino acids (including translation and central metabolism) and amino acid synthesis or acquisition from the environment (Henkin and Yanofsky, 2002; Elf and Ehrenberg, 2005). In this context, it is not surprising that tRNAs have been hijacked out of their biosynthetic roles for use in regulatory processes. These regulatory systems usually require very small amounts of molecules and sometimes use only RNA. Thus, it has been proposed that such tRNA-based regulatory systems are cheap to produce and easy to evolve, and that for these reasons they might have already been in use in a probable ancestral RNA world (Henkin and Yanofsky, 2002). Furthermore, it has recently been proposed that the high specificity of aaRSs additionally allows tRNA-based regulatory systems to be highly sensitive and substrate-specific (Bullwinkle and Ibba, 2016).
Some of the best-described regulatory mechanisms involving tRNA are the stringent response and transcription attenuation. Both take advantage of different “side effects” of changing the aa-tRNA/tRNA ratio. In the stringent response, a sudden decrease in amino acid availability allows accumulation of deacylated tRNAs that bind the ribosome where they stimulate the transfer of PPi from ATP to either GDP or GTP by RelA/SpoT enzymes. These reactions form ppGpp and pppGpp, alarmones with pleiotropic effects on bacterial physiology including changes in metabolism of amino acids and nucleotides and regulation of synthesis of stable RNAs (rRNAs and tRNAs) (Liu et al., 2015). Under transcription attenuation, instead the state of tRNA aminoacylation can be directly sensed by riboswitches that control the formation of Rho independent transcription terminators and thus, modulate expression of genes coded by the transcript downstream of the terminator site. The riboswitch may sense tRNA aminoacylation levels by two different strategies. In one strategy, represented by the classic example of the trp operon in E. coli, a small open reading frame at the 5' leader region of the operon contains several Trp codons. Changes in the abundance of the specific aa-tRNATrp will change the speed of translation of this leader peptide modulating the time of residence of ribosomes in this area of the mRNA and controlling the formation of the terminator structure. In a second strategy, most frequently found in Gram-positive bacteria, the mRNA may present a riboswitch structure termed the T-box. Binding of the non-aminoacylated tRNA to the T-box prevents the formation of the transcription terminator, allowing transcription of the genes located downstream (Henkin and Yanofsky, 2002).
Similar to the effect of aa-tRNA abundance on the speed of translation of the leader peptide for transcription attenuation, changes in the availability of a specific aa-tRNA can also modify the speed of translation of genes that are rich in codons translated by this aa-tRNA. These genes are usually less sensitive than leader peptides and require large changes in amino acid availability that can potentially have strong effects on the abundance of aa-tRNA. Additionally, genes involved in signal transduction pathways can be extremely sensitive to small changes in protein levels (Elf and Ehrenberg, 2005; Sørensen et al., 2005; Subramaniam, Pan, et al., 2013; Subramaniam et al., 2014). As an example of this extreme sensitivity, Bacillus subtilis can sense decreases in environmental serine abundance through changes in the speed of translation of the sinR biofilm repressor and use this signal to trigger the formation of biofilms. Changes in codon usage of sinR produce only minor changes in the protein's cellular concentration. Nevertheless, as SinR has a cooperative behavior and is part of a negative feedback mechanism, this small change in protein concentration induces a strong change in colony morphology (Subramaniam, Deloughery, et al., 2013).
The modification of several tRNA nucleotides is also known to be involved in regulatory pathways. Usually these modified nucleotides are located in the anticodon loop, although there are some exceptions. One of the first to be identified is located at position 37 (next to the 3' end of the anticodon) of tRNAs reading codons with a 5' U (tRNATyr, tRNATrp, tRNAPhe, tRNALeu, tRNASer, tRNACys) from E. coli. Cells cultured in a medium lacking iron were found to have lost a methylthio modification (presenting an i6A nucleotide instead of the normal hypermodified ms2i6A nucleotide), an alteration that induced the synthesis of enterobactin (a high affinity siderophore) and its aromatic amino acid precursors (Wettstein and Stent, 1968; Rosenberg and Gefter, 1969; Buck and Ames, 1984). Since then, several other modifications have been found to be involved in modulation of aerobic/respiratory metabolism (Buck and Ames, 1984; Persson et al., 1998; Björk et al., 1999; Nakayashiki et al., 2013), expression of virulence factors (Björk et al., 1999; Durand et al., 2000; Shippy and Fadl, 2014), the response to changes in amino acid availability (Laxman et al., 2013) or other stressful conditions (Golovina et al., 2009; Murata et al., 2011; Chan et al., 2012; Caballero et al., 2012; Dedon and Begley, 2014; Gu et al., 2014) in both bacteria and eukaryotes including mammals (Wei and Tomizawa, 2011; Gu et al., 2014; Endres et al., 2015). Additionally, high-throughput experiments have found changes of tRNA nucleotide modifications or the enzymes involved in the modification pathways related to stress conditions or diseases (Wang and He, 2014) suggesting that modification of tRNA might play a central role in regulation of gene expression.
In addition to nucleotide modifications, a series of bacterial toxins can cleave initiator or elongation tRNAs, thus inhibiting translation (Ogawa et al., 1999; Kaufmann, 2000; Winther and Gerdes, 2011; Ruhe et al., 2013; Cruz et al., 2015). This usually forms part of stress defense mechanisms, as in the case of PrrC, that inhibit protein synthesis under phage infection (Kaufmann, 2000) or some toxins from the VapC family that induce a dormancy state under other stressful conditions (Winther and Gerdes, 2011; Cruz et al., 2015). Nevertheless, other toxins have a very different ecological role and are used to attack nearby bacteria. This is the case of colicins D and E5 (Ogawa et al., 1999; Tomita et al., 2000) as well as some toxins from contact dependent growth inhibition systems such as WapA in Bacillus subtilis or some CDI toxins from Burkholderia pseudomallei (Koskiniemi et al., 2013; Ruhe et al., 2013). The reduction of functional tRNAs could have other consequences, as it has been shown that it can trigger the formation of dinucleotide second messengers (Kramer et al., 1988; Katz and Orellana, 2012b) or an increased usage of elongator tRNAs at initiation (Winther and Gerdes, 2011; Samhita et al., 2013; Shetty et al., 2015). Nevertheless, a physiological role for these phenomena in the context of tRNA fragmentation has not been studied yet. In eukaryotes, tRNA fragments are also produced. In addition to the effects derived from a decrease in tRNA concentration, in these organisms the fragments per se have a physiological role, binding polysomes or proteins involved in the siRNA and miRNA pathways and consequently affecting gene expression by mechanisms that are still not well understood (Raina and Ibba, 2014; Keam and Hutvagner, 2015).
tRNA mimics
The structural and functional adaptability of tRNA have allowed not only their use for alternative roles beyond protein synthesis, but also the appearance of processes that take advantage of their existence albeit not using them directly. For instance, several virus and mobile genetic elements use genes coding for tRNAs as an insertion site in genomes. These mobile elements have to include segments that mimic part of the tRNA gene sequence in order to enable recombination and to allow for reconstitution of the tRNA gene in cases where it is essential (Hacker and Kaper, 2000; Williams, 2002). Along the same lines, several molecules mimic the structure of tRNAs in order to control translation or use its machinery for other purposes. An impressive example of this are the tRNA-like structures (TLS) in the RNA genomes of several viruses (Dreher, 2009). A prototype of these mimics is the TLS located at the 3`end of the turnip yellow mosaic virus (TYMV) (Dreher, 2009; Colussi et al., 2014). When folded, the TLS adopts the classic L-shaped tRNA structure, although with a different topology based on idiosyncratic intramolecular interactions. Although the elements analogous to the D loop, T loop and V loop are positioned similar to tRNA, they interact in a different manner. This configuration allows the TLS to present two faces. One face of the TLS is the ‘tRNA-like face’, which closely mimics tRNA and achieves tRNA-like valylation efficiencies and eIF1A binding. The opposing side presents tRNA-deviating features, an upstream pseudoknot domain and the genomic RNA interacting to enable additional functionality (Dreher and Goodwin, 1998; Colussi et al., 2014). In contrast to tRNA, the unique topology and intramolecular interactions of the viral RNA also allows the TLS to unfold, possibly to allow the replication of the RNA genome (Colussi et al., 2014).
Beside mimicry of foreign molecules (mobile elements and viruses) there are also several molecules within a cell that mimic its own tRNAs. One remarkable example is established by the participation of 2 molecules: tmRNA (small transfer-messenger RNA also known as SsrA and 10Sa RNA) and a tmRNA-specific binding protein called SmpB (small protein B). Together they mimic the upper (tmRNA) and lower (SmpB) halves of a tRNA, respectively. tmRNA owes its name to its action as both a transfer and a messenger RNA. The tmRNA-SmpB complex targets and rescues stalled ribosomes in a process known as trans-translation (Himeno et al., 2014; Keiler, 2015). This conserved translation surveillance pathway prevents accumulation of non-functional proteins from truncated mRNA (Karzai et al., 1999; Chadani et al., 2010). The tmRNA-SmpB complex mimics tRNA throughout the process. First, it is aminoacylated with alanine by alanyl-tRNA synthetase. Ala-tmRNA then enters the A-site of the stalled ribosome on a truncated mRNA to receive the nascent polypeptide from peptidyl-tRNA in the P-site. Then peptidyl-Ala-tmRNA translocates to the P-site, which exchanges the template switching the stalled ribosome from the translation of the defective mRNA to the translation of the mRNA domain of tmRNA. Translation of the tmRNA reading frame tags the defective protein to be degraded by cellular proteases (Richards et al., 2008; Giudice et al., 2014).
In principle, any macromolecule could mimic tRNAs and in fact, even a tRNA has been found to mimic another tRNA. In this interesting case of structural mimicry between the anticodon arm of tRNAAsp and the acceptor stem of tRNAGlu, a paralog of glutamyl-tRNA synthetases, glutamyl-Q-tRNAAsp synthetase (Glu-Q-RS, previously YadB), activates glutamate and transfers it to the queuosine at the wobble position of tRNAAsp (Blaise et al., 2004; Campanacci et al., 2004; Dubois et al., 2004; Salazar et al., 2004). This is considered an hypermodification as queuosine corresponds already to a modified guanine and Glu-Q-RS was the first aaRS paralog known to catalyze this kind of reaction. Several aspects of Glu-Q-RS are unusual. Firstly, in contrast to GluRS the enzyme can activate glutamate in the absence of tRNA (Campanacci et al., 2004; Salazar et al., 2004) and transfer it to tRNAAsp instead of tRNAGlu (Dubois et al., 2004; Salazar et al., 2004). Secondly, it does not glutamylate tRNAAsp at its 3' terminal adenosine, but instead it transfers the amino acid to a modified queuosine nucleoside, which is the first anticodon nucleoside in tRNAAsp (Blaise et al., 2004; Salazar et al., 2004). Finally, the orientation in which a tRNA interacts with Glu-Q-RS is different from that with GluRS since the tRNA needs to come close to Glu-Q-RS’s active site with its anticodon loop instead of its acceptor helix (Blaise et al., 2004). The role of this hypermodification is poorly understood. It is speculated that it might reverse the effect of a single queuosine on GAU codon binding by tRNAASP Q34 during translation (Blaise et al., 2005). The high KM for glutamate of Glu-Q-RS suggests that this glutamylation might only occur when the glutamate concentration rises in the cell (Salazar et al., 2004). If this is true, the hypermodification may be a sensor, allowing Glu to affect the speed of GAU decoding during translation (Blaise et al., 2005) and, consistent with this, Glu-Q-RS seems to have a role in regulating the response to osmotic stress in Shigella flexneri (Caballero et al., 2012).
Similar to the example of SmpB in the mimicry of tRNA, there are several other proteins that mimic tRNA. Also similar to SmpB, all these proteins bind to the ribosome and have a function in translation. Presumably, primordial ribosomes prior to the last common ancestor catalyzed peptide-bond formation without the addition of translation factors, however these specialized factors greatly enhance the rate of translation. For instance, elongation factor G (EF-G) is a GTPase that binds the ribosome and upon GTP hydrolysis, accelerates translocation 4–5 fold (Shoji et al., 2009). EF-G is organized into 5 domains, of which domains I and II resemble in EF-Tu structure, while III-V resemble that of A-site tRNA. By virtue of structurally mimicking the tRNA-EF-Tu complex, domains III-V occupy the A-site tRNA to occlude tRNA from entering (Gao et al., 2009; Zhou et al., 2013). EF-G is not the only protein to have evolved A-site tRNA mimicry, in addition ribosome release (RRF) and recycling factors, (RF1/2, RF3) mimic tRNA and bind to similar regions. Despite these factors all binding at or near the A-site, each have functions that differ considerably. RF1/2 and RF3 bind ribosomes that have reached a stop codon to form an anticodon-codon interaction and hydrolyze the tRNA-peptidyl linkage with a conserved GQQ motif that mimics the CCA end of a tRNA (Zhou et al., 2012). RRF traverses between the A- and P-site to disassemble the ribosome into separate subunits, mediated by EF-G catalyzed translocation (Weixlbaumer et al., 2007). Therefore access to the A-site of the ribosome provides translation factors the opportunity to control each aspect of protein synthesis, however the cavity of the ribosome’s entrance imposes a structural barrier that requires such proteins to mimic tRNA.
Protein-tRNA mimics are not all confined to interact with the entrance of the ribosome. Elongation factor P (EF-P) is an acidic protein comprised of three β-barrel domains, arranged in a format that notably resembles tRNA, though the domains corresponding to the anticodon arm are somewhat smaller (Hanawa-Suetsugu et al., 2004). According to biochemical studies and a co-crystal structure with the ribosome, EF-P is positioned between the P/E site, making various contacts with the peptidyl-tRNA to entropically provide the ribosome the necessary velocity when translating consecutive proline residues, as proline forms the slowest peptide bond (Blaha et al., 2009; Doerfel et al., 2013; Doerfel et al., 2015). The considerable structural similarity between EF-P and tRNA gives EF-P a distinct opportunity to interact with enzymes that tRNA may also interact with. In particular, EF-P is post-translationally modified in some organisms by a mechanism reminiscent of tRNA-aminoacylation (Navarre et al., 2010; Roy et al., 2011). The addition of (R)-β-lysine onto the conserved lysine residue at the acceptor stem tip of EF-P augments EF-P’s function by increasing the observed KM for paused ribosomes (Doerfel et al., 2013). Interestingly, EF-P is glycosylated in some beta- and gamma-proteobacteria with a rhamnose molecule at an analogous arginine residue, calling into question how this evolutionary partnership arose while still accommodating important contacts with the ribosome (Rajkovic et al., 2015; Lassak et al., 2015).
Recent crystal and cyroEM structures revealed the energy-dependent translational throttle A (Etta) protein enters the E-site of the ribosome to regulate translation elongation in a manner dependent on the ratio of ATP/ADP (Boël et al., 2014). EttA is composed of four domains that bind ADP or ATP and a long C-terminal α-helix that interacts with the E and P-site of the ribosome. The C-terminal extension mimics tRNA to control protein synthesis directly by interacting with the peptidyl-transfer center when initiator tRNA is bound. For example, when ADP levels are high, such as in stationary phase for bacteria, EttA-ADP restricts synthesis of the first peptide bond, while in the presence of ATP, EttA marginally enhances the synthesis of the first peptide bond. This provides the first example of a protein tRNA mimic as having a regulatory function within the context of translation. Furthermore, it will be interesting to see whether EttA and EF-P accelerate peptide bond formation in a similar way.
Mimics from pathogens and parasites (such as TLS) have a strong selective pressure to maintain similitude to their host models. The hosts instead, have a strong selective pressure to change the structure of the model (in this case tRNA) or of the molecules that interact with it (in this case the ribosome or an aaRS), such that they stop recognizing the mimic and become more resistant to the pathogen/parasite (Elde et al., 2009; Elde and Malik, 2009). Nevertheless, most tRNA mimics are from within the same organism. Thus, there is strong selective pressure over both the mimic and the model for maintenance of mimicry (Katz et al., 2014).
Final remarks
Aa-tRNAs are central in translation of the genetic information, connecting cellular memory stored in nucleic acids with structural and catalytic functions performed mainly by proteins. Thus, these molecules are believed to have performed a major role in the evolution from a probable “RNA world” where RNA had both memory and catalytic functions to a world where these functions are performed mainly by specialized molecules: catalysis by proteins and memory by DNA. In this scenario, it is probable that beside tRNA, there are only a few other information molecules in living organisms that are as old and as well conserved in the three domains of life plus viruses. Although there are some differences between organisms, the fact that bacterial and eukaryal tRNAs can easily be interchanged for both aminoacylation and mRNA translation (Shafritz and Anderson, 1970; Berthelot et al., 1973; Weisblum, 1999) is evidence for how well these molecules have retained their structure. Such a long period of time has created multiple chances for nature to evolve new functions for these old molecules that carry an activated amino acid with very high specificity. As discussed in this review, several of these newer functions correspond to pathways unrelated to translation per se. These functions include several synthetic and regulatory pathways, some of which might be very ancient themselves. For instance, indirect pathways for synthesis of aminoacyl-tRNA are thought to be very old, preceding the corresponding direct pathways (Woese et al., 2000; Ibba et al., 2000; Sheppard et al., 2008). The tRNA dependent synthesis of tetrapyrroles has also been proposed to be significantly older than the corresponding tRNA independent pathway (Schulze et al., 2006) maybe even predating LUCA and having arisen in the RNA world (Benner et al., 1989). Surprisingly, although many of these alternative pathways are old and usually use the same tRNAs as in translation, there are very few cases where a “cross-talk” between translation, alternative synthetic pathways and regulatory processes has been proposed. Potential cross-talk has been suggested for pathways that use large amounts of aa-tRNA, such as synthesis of peptidoglycan in gram-positive bacteria (Shepherd and Ibba, 2013a; Shepherd and Ibba, 2013b) or tetrapyrroles in organisms with high demand of them for either photosynthesis (Jahn, 1992) or oxygen consumption (Levicán et al., 2005; Levicán et al., 2007; Katz and Orellana, 2012a; Farah et al., 2014). Nevertheless, for the most part we are unaware of the effect that changes in tRNA have on most of the pathways using tRNA outside of translation. For instance, the effects of tRNA fragmentation or modification on the regulation of protein stability or tRNA dependent synthesis of amino acids are completely unknown. We expect that the mass-spec and high-throughput sequencing tools that have been developed in the last years, as well as the vast amount of genomic sequences that have been made available, will enhance research about the roles of tRNAs in regulating translation and the alternative pathways where these molecules have a central role.
Acknowledgments
M.I. is supported by National Institutes of Health Grant GM65183. A.K. is supported by grant 11140222 from Fondo Nacional de Desarrollo Científico y Tecnológico and grant 79130044 from Comisión Nacional de Investigación Científica y Tecnológica.
Footnotes
The authors declare no conflicts of interest with respect to the work reported here.
References
- Ataide SF, Jester BC, Devine KM, Ibba M. Stationary-phase expression and aminoacylation of a transfer-RNA-like small RNA. EMBO Rep. 2005;6:742–747. doi: 10.1038/sj.embor.7400474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Chen S, Dare K, Gilreath M, Praetorius-Ibba M, Raina M, et al. tRNAs: cellular barcodes for amino acids. FEBS Lett. 2010;584:387–395. doi: 10.1016/j.febslet.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin P, Moutiez M, Lautru S, Seguin J, Pernodet J-L, Gondry M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat Prod Rep. 2012;29:961–979. doi: 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- Benner SA, Ellington AD, Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci U S A. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B, Tschierske M. Role of fem factors in methicillin resistance. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother. 1998;1:325–335. doi: 10.1016/s1368-7646(98)80048-4. [DOI] [PubMed] [Google Scholar]
- Berthelot F, Bogdanovsky D, Schapira G, Gros F. Interchangeability of factors and tRNA’s in bacterial and eukaryotic translation initiation systems. Mol Cell Biochem. 1973;1:63–72. doi: 10.1007/BF01659939. [DOI] [PubMed] [Google Scholar]
- Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bhaskaran H, Perona JJ. Two-step aminoacylation of tRNA without channeling in Archaea. J Mol Biol. 2011;411:854–869. doi: 10.1016/j.jmb.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Durand JMB, Hagervall TG, Leipuviene R, Lundgren HK, Nilsson K, et al. Transfer RNA modification: influence on translational frameshifting and metabolism. FEBS Lett. 1999;452:47–51. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise M, Becker HD, Keith G, Cambillau C, Lapointe J, Giegé R, Kern D. A minimalist glutamyl-tRNA synthetase dedicated to aminoacylation of the tRNAASP QUC anticodon. Nucleic Acids Res. 2004;32:2768–2775. doi: 10.1093/nar/gkh608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise M, Becker HD, Lapointe J, Cambillau C, Giegé R, Kern D. Glu-Q-tRNA(Asp) synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNA(Asp) anticodon. Biochimie. 2005;87:847–861. doi: 10.1016/j.biochi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Boël G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, et al. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol. 2014;21:143–151. doi: 10.1038/nsmb.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond L, Arai T, Sakaguchi Y, Suzuki T, Ishitani R, Nureki O. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc Natl Acad Sci U S A. 2011;108:3912–3917. doi: 10.1073/pnas.1019480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Ames BN. A modified nucleotide in tRNA as a possible regulator of aerobiosis: synthesis of cis-2-methyl-thioribosylzeatin in the tRNA of Salmonella. Cell. 1984;36:523–531. doi: 10.1016/0092-8674(84)90245-9. [DOI] [PubMed] [Google Scholar]
- Bullwinkle TJ, Ibba M. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci U S A. 2016;113:2252–2257. doi: 10.1073/pnas.1525206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumsted RM, Dahl JL, Söll D, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. X. Further study of the glycyl transfer ribonucleic acids active in peptidoglycan synthesis in Staphylococcus aureus. J Biol Chem. 1968;243:779–782. [PubMed] [Google Scholar]
- Caballero VC, Toledo VP, Maturana C, Fisher CR, Payne SM, Salazar JC. Expression of Shigella flexneri gluQ-rs gene is linked to dksA and controlled by a transcriptional terminator. BMC Microbiol. 2012;12:226. doi: 10.1186/1471-2180-12-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V, Dubois DY, Becker HD, Kern D, Spinelli S, Valencia C, et al. The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity. J Mol Biol. 2004;337:273–283. doi: 10.1016/j.jmb.2004.01.027. [DOI] [PubMed] [Google Scholar]
- Cavarelli J, Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J Off Publ Fed Am Soc Exp Biol. 1993;7:79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S-I, Takahashi Y, Takai K, Nanamiya H, et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol. 2010;78:796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014;511:366–369. doi: 10.1038/nature13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. On Protein Synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- Cruz JW, Sharp JD, Hoffer ED, Maehigashi T, Vvedenskaya IO, Konkimalla A, et al. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat Commun. 2015;6(7480) doi: 10.1038/ncomms8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale T, Uhlenbeck OC. Amino acid specificity in translation. Trends Biochem Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Dare K, Ibba M. Roles of tRNA in cell wall biosynthesis. Wiley Interdiscip Rev RNA. 2012;3:247–264. doi: 10.1002/wrna.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Begley TJ. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem Res Toxicol. 2014;27:330–337. doi: 10.1021/tx400438d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio M. The phylogeny of tRNAs seems to confirm the predictions of the coevolution theory of the origin of the genetic code. Orig Life Evol Biosph. 1995;25:549–564. doi: 10.1007/BF01582024. [DOI] [PubMed] [Google Scholar]
- Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA) J Theor Biol. 2006;240:343–352. doi: 10.1016/j.jtbi.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kubyshkin V, Starosta AL, Wilson DN, Budisa N, Rodnina MV. Entropic Contribution of Elongation Factor P to Proline Positioning at the Catalytic Center of the Ribosome. J Am Chem Soc. 2015;137:12997–13006. doi: 10.1021/jacs.5b07427. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: from recognition by N-recognins, to destruction by AAA+proteases. Biochim Biophys Acta. 2012;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139:217–229. doi: 10.1016/j.virusres.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–4364. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois DY, Blaise M, Becker HD, Campanacci V, Keith G, Giegé R, et al. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAASP. Proc Natl Acad Sci U S A. 2004;101:7530–7535. doi: 10.1073/pnas.0401634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand JM, Dagberg B, Uhlin BE, Björk GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol Microbiol. 2000;35:924–935. doi: 10.1046/j.1365-2958.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- Elf J, Ehrenberg M. Near-critical behavior of aminoacyl-tRNA pools in E. coli at rate-limiting supply of amino acids. Biophys J. 2005;88:132–146. doi: 10.1529/biophysj.104.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12:603–614. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah C, Levicán G, Ibba M, Orellana O. Effect of hydrogen peroxide on the biosynthesis of heme and proteins: potential implications for the partitioning of Glu-tRNA(Glu) between these pathways. Int J Mol Sci. 2014;15:23011–23023. doi: 10.3390/ijms151223011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonvielle M, Chemama M, Villet R, Lecerf M, Bouhss A, Valéry J-M, et al. Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis. Nucleic Acids Res. 2009;37:1589–1601. doi: 10.1093/nar/gkn1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonvielle M, de La Sierra-Gallay I, Sagheer AHEl-, Lecerf M, Patin D, Mellal D, et al. The structure of FemX(Wv) in complex with a peptidyl-RNA conjugate: mechanism of aminoacyl transfer from Ala-tRNA(Ala) to peptidoglycan precursors. Angew Chem Int Ed Engl. 2013;52:7278–7281. doi: 10.1002/anie.201301411. [DOI] [PubMed] [Google Scholar]
- Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584:366–375. doi: 10.1016/j.febslet.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J Mol Biol. 2007;370:128–141. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- Fung AWS, Leung CCY, Fahlman RP. The determination of tRNALEU recognition nucleotides for Escherichia coli L/F transferase. RNA N Y N. 2014;20:1210–1222. doi: 10.1261/rna.044529.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci U S A. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie. 2009;91:344–351. doi: 10.1016/j.biochi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Giegé R, Frugier M. Madame Curie Bioscience Database [Internet] Austin (TX), USA: Landes Bioscience; 2000. Transfer RNA Structure and Identity. http://www.ncbi.nlm.nih.gov/books/NBK6236/ [Google Scholar]
- Giudice E, Macé K, Gillet R. Trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front Microbiol. 2014;5:113. doi: 10.3389/fmicb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfine H. Charge counter charge: bacterial response to antimicrobial cationic peptides and more. Virulence. 2014;5:451–453. doi: 10.4161/viru.28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovina AY, Sergiev PV, Golovin AV, Serebryakova MV, Demina I, Govorun VM, Dontsova OA. The yfiC gene of E. coli encodes an adenine-N6 methyltransferase that specifically modifies A37 of tRNA1Val(cmo5UAC) RNA N Y N. 2009;15:1134–1141. doi: 10.1261/rna.1494409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RM, Thornton MP, Liepkalns V, Lennarz WJ. Participation of aminoacyl transfer ribonucleic acid in aminoacyl phosphatidylglycerol synthesis. II. Specificity of alanyl phosphatidylglycerol synthetase. J Biol Chem. 1968;243:3096–3104. [PubMed] [Google Scholar]
- Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588:4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Roychowdhury A, Ember B, Kumar S, Boons G-J, Mariuzza RA. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc Natl Acad Sci U S A. 2004;101:17168–17173. doi: 10.1073/pnas.0407856101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon JM, Meinnel T, Mechulam Y, Lazennec C, Blanquet S, Fayat G. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J Mol Biol. 1992;224:359–367. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, et al. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci U S A. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebecker S, Arendt W, Heinemann IU, Tiefenau JHJ, Nimtz M, Rohde M, et al. Alanyl-phosphatidylglycerol synthase: mechanism of substrate recognition during tRNA-dependent lipid modification in Pseudomonas aeruginosa. Mol Microbiol. 2011;80:935–950. doi: 10.1111/j.1365-2958.2011.07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Jahn M, Jahn D. The biochemistry of heme biosynthesis. Arch Biochem Biophys. 2008;474:238–251. doi: 10.1016/j.abb.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Helgadóttir S, Sinapah S, Söll D, Ling J. Mutational analysis of Sep-tRNA:Cys-tRNA synthase reveals critical residues for tRNA-dependent cysteine formation. FEBS Lett. 2012;586:60–63. doi: 10.1016/j.febslet.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays News Rev Mol Cell Dev Biol. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Himeno H, Kurita D, Muto A. tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front Genet. 2014;5:66. doi: 10.3389/fgene.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A Soluble Ribonucleic Acid Intermediate in Protein Synthesis. J Biol Chem. 1958;231:241–257. [PubMed] [Google Scholar]
- Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Söll D. The adaptor hypothesis revisited. Trends Biochem Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- Ibba M, Francklyn C, Cusack S, editors. The aminoacyl-tRNA synthetases. Georgetown, Tex., U.S.A: 2005. Landes Bioscience: Eurekah.com. [Google Scholar]
- Ichihashi N, Kurokawa K, Matsuo M, Kaito C, Sekimizu K. Inhibitory effects of basic or neutral phospholipid on acidic phospholipid-mediated dissociation of adenine nucleotide bound to DnaA protein, the initiator of chromosomal DNA replication. J Biol Chem. 2003;278:28778–28786. doi: 10.1074/jbc.M212202200. [DOI] [PubMed] [Google Scholar]
- Jahn D. Complex formation between glutamyl-tRNA synthetase and glutamyl-tRNA reductase during the tRNA-dependent synthesis of 5-aminolevulinic acid in Chlamydomonas reinhardtii. FEBS Lett. 1992;314:77–80. doi: 10.1016/0014-5793(92)81465-x. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Orellana O. Glutamyl-tRNA in Bacteria. Multiple Identities for Multiple Functions. Croat Chem Acta. 2012a;85:159–169. [Google Scholar]
- Katz A, Orellana O. Protein Synthesis and the Stress Response. In: Biyani M, editor. Cell-Free Protein Synthesis. InTech; 2012b. [Google Scholar]
- Katz A, Solden L, Zou SB, Navarre WW, Ibba M. Molecular evolution of protein-RNA mimicry as a mechanism for translational control. Nucleic Acids Res. 2014;42:3261–3271. doi: 10.1093/nar/gkt1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G. Anticodon nucleases. Trends Biochem Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life. 2015;5:1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- Khade P, Joseph S. Functional interactions by transfer RNAs in the ribosome. FEBS Lett. 2010;584:420–426. doi: 10.1016/j.febslet.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, Roodenbeke C t’Kint de, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer GF, Baker JC, Ames BN. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J Bacteriol. 1988;170:2344–2351. doi: 10.1128/jb.170.5.2344-2351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner JE, Jack A, Robertus JD, Brown RS, Rhodes D, Clark BF, Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975;72:4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Lassak J, Keilhauer EC, Fürst M, Wuichet K, Gödeke J, Starosta AL, et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat Chem Biol. 2015;11:266–270. doi: 10.1038/nchembio.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, et al. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Seong BL, RajBhandary UL. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991;266:18012–18017. [PubMed] [Google Scholar]
- Levicán G, Katz A, Armas M de, Núñez H, Orellana O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci U S A. 2007;104:3135–3140. doi: 10.1073/pnas.0611611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levicán G, Katz A, Valenzuela P, Söll D, Orellana O. A tRNA(Glu) that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett. 2005;579:6383–6387. doi: 10.1016/j.febslet.2005.09.100. [DOI] [PubMed] [Google Scholar]
- Li S, Kumar NV, Varshney U, RajBhandary UL. Important role of the amino acid attached to tRNA in formylation and in initiation of protein synthesis in Escherichia coli. J Biol Chem. 1996;271:1022–1028. doi: 10.1074/jbc.271.2.1022. [DOI] [PubMed] [Google Scholar]
- Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol. 2015;24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA N Y N. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus DD, Paredes JA, Español Y, Ribas de Pouplana L, Moura GR, Santos MAS. Molecular reconstruction of a fungal genetic code alteration. RNA Biol. 2013;10:969–980. doi: 10.4161/rna.24683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenova SR, Stein KR, Bartlett L, Sheppard K. Relaxed tRNA specificity of the Staphylococcus aureus aspartyl-tRNA synthetase enables RNA-dependent asparagine biosynthesis. FEBS Lett. 2014;588:1808–1812. doi: 10.1016/j.febslet.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Moras D, Comarmond MB, Fischer J, Weiss R, Thierry JC, Ebel JP, Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980;288:669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Morgens DW. The protein invasion: a broad review on the origin of the translational system. J Mol Evol. 2013;77:185–196. doi: 10.1007/s00239-013-9592-x. [DOI] [PubMed] [Google Scholar]
- Moura GR, Paredes JA, Santos MAS. Development of the genetic code: insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. doi: 10.1016/j.febslet.2009.11.066. [DOI] [PubMed] [Google Scholar]
- Moutiez M, Seguin J, Fonvielle M, Belin P, Jacques IB, Favry E, et al. Specificity determinants for the two tRNA substrates of the cyclodipeptide synthase AlbC from Streptomyces noursei. Nucleic Acids Res. 2014;42:7247–7258. doi: 10.1093/nar/gku348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Fujimoto H, Nishimura K, Charoensuk K, Nagamitsu H, Raina S, et al. Molecular strategy for survival at a critical high temperature in Eschierichia coli. PloS One. 2011;6:e20063. doi: 10.1371/journal.pone.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Saito N, Takeuchi R, Kadokura H, Nakahigashi K, Wanner BL, Mori H. The tRNA thiolation pathway modulates the intracellular redox state in Escherichia coli. J Bacteriol. 2013;195:2039–2049. doi: 10.1128/JB.02180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton DT, Niemkiewicz M, Lo RY, Mangroo D. Recognition of the initiator tRNA by the Pseudomonas aeruginosa methionyl-tRNA formyltransferase: importance of the base-base mismatch at the end of the acceptor stem. FEMS Microbiol Lett. 1999;178:289–298. doi: 10.1111/j.1574-6968.1999.tb08690.x. [DOI] [PubMed] [Google Scholar]
- O’Brian MR, Thöny-Meyer L. Biochemistry, regulation and genomics of haem biosynthesis in prokaryotes. Adv Microb Physiol. 2002;46:257–318. doi: 10.1016/s0065-2911(02)46006-7. [DOI] [PubMed] [Google Scholar]
- O’Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. The evolutionary history of Cys-tRNACYS formation. Proc Natl Acad Sci U S A. 2005;102:19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- Palioura S, Sherrer RL, Steitz TA, Söll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BC, Ólafsson Ó, Lundgren HK, Hederstedt L, Björk GR. The ms2io6A37 Modification of tRNA in Salmonella typhimurium Regulates Growth on Citric Acid Cycle Intermediates. J Bacteriol. 1998;180:3144–3151. doi: 10.1128/jb.180.12.3144-3151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RajBhandary UL, Söll D. Aminoacyl-tRNAs, the bacterial cell envelope, and antibiotics. Proc Natl Acad Sci U S A. 2008;105:5285–5286. doi: 10.1073/pnas.0801193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Erickson S, Witzky A, Branson OE, Seo J, Gafken PR, et al. Cyclic Rhamnosylated Elongation Factor P Establishes Antibiotic Resistance in Pseudomonas aeruginosa. mBio. 2015;6:e00823. doi: 10.1128/mBio.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias T, Sheppard K, Söll D. The archaeal transamidosome for RNA-dependent glutamine biosynthesis. Nucleic Acids Res. 2010;38:5774–5783. doi: 10.1093/nar/gkq336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Schauer S, Ambrogelly A, Salazar JC, Moser J, Sekine S, et al. tRNA Recognition by Glutamyl-tRNA Reductase. J Biol Chem. 2004;279:34931–34937. doi: 10.1074/jbc.M401529200. [DOI] [PubMed] [Google Scholar]
- Richards J, Sundermeier T, Svetlanov A, Karzai AW. Quality control of bacterial mRNA decoding and decay. Biochim Biophys Acta. 2008;1779:574–582. doi: 10.1016/j.bbagrm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin AS, Szathmáry E, Rodin SN. On origin of genetic code and tRNA before translation. Biol Direct. 2011;6:14. doi: 10.1186/1745-6150-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TE, Ataide SF, Dare K, Katz A, Seveau S, Roy H, Ibba M. A pseudo-tRNA modulates antibiotic resistance in Bacillus cereus. PloS One. 2012;7:e41248. doi: 10.1371/journal.pone.0041248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AH, Gefter ML. An iron-dependent modification of several transfer RNA species in Escherichia coli. J Mol Biol. 1969;46:581–584. doi: 10.1016/0022-2836(69)90197-1. [DOI] [PubMed] [Google Scholar]
- Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, et al. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat Chem Biol. 2011;7:667–669. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JC, Ambrogelly A, Crain PF, McCloskey JA, Söll D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc Natl Acad Sci U S A. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samhita L, Virumäe K, Remme J, Varshney U. Initiation with Elongator tRNAs. J Bacteriol. 2013;195:4202–4209. doi: 10.1128/JB.00637-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Moutiez M, Li Y, Belin P, Seguin J, Du M-H, Le, et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 2011;39:4475–4489. doi: 10.1093/nar/gkr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers D-J, Pinho MG. Bacterial Cell Wall Synthesis: New Insights from Localization Studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P, Giegé R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci U S A. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JO, Schubert W-D, Moser J, Jahn D, Heinz DW. Evolutionary relationship between initial enzymes of tetrapyrrole biosynthesis. J Mol Biol. 2006;358:1212–1220. doi: 10.1016/j.jmb.2006.02.064. [DOI] [PubMed] [Google Scholar]
- Shafritz DA, Anderson WF. Isolation and Partial Characterization of Reticulocyte Factors M1 and M2. J Biol Chem. 1970;245:5553–5559. [PubMed] [Google Scholar]
- Shepherd J, Ibba M. Lipid II-independent trans editing of mischarged tRNAs by the penicillin resistance factor MurM. J Biol Chem. 2013a;288:25915–25923. doi: 10.1074/jbc.M113.479824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J, Ibba M. Direction of aminoacylated transfer RNAs into antibiotic synthesis and peptidoglycan-mediated antibiotic resistance. FEBS Lett. 2013b;587:2895–2904. doi: 10.1016/j.febslet.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepotinovskaya I, Uhlenbeck OC. tRNA residues evolved to promote translational accuracy. RNA N Y N. 2013;19:510–516. doi: 10.1261/rna.036038.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrer RL, Ho JML, Söll D. Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res. 2008;36:1871–1880. doi: 10.1093/nar/gkn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S, Bhattacharyya S, Varshney U. Is the cellular initiation of translation an exclusive property of the initiator tRNAs? RNA Biol. 2015;12:675–680. doi: 10.1080/15476286.2015.1043507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy DC, Fadl AA. tRNA Modification Enzymes GidA and MnmE: Potential Role in Virulence of Bacterial Pathogens. Int J Mol Sci. 2014;15:18267–18280. doi: 10.3390/ijms151018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Björk GR, Ehrenberg M. Over expression of a tRNA(Leu) isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol. 2005;354:16–24. doi: 10.1016/j.jmb.2005.08.076. [DOI] [PubMed] [Google Scholar]
- Subramaniam AR, Deloughery A, Bradshaw N, Chen Y, O’Shea E, Losick R, Chai Y. A serine sensor for multicellularity in a bacterium. eLife. 2013;2:e01501. doi: 10.7554/eLife.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Pan T, Cluzel P. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc Natl Acad Sci U S A. 2013;110:2419–2424. doi: 10.1073/pnas.1211077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O’Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F-J, Caetano-Anollés G. The origin and evolution of tRNA inferred from phylogenetic analysis of structure. J Mol Evol. 2008;66:21–35. doi: 10.1007/s00239-007-9050-8. [DOI] [PubMed] [Google Scholar]
- Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 2006;25:5942–5950. doi: 10.1038/sj.emboj.7601433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Vial L, Panvert M, Schmitt E, Watanabe K, Mechulam Y, Blanquet S. Recognition of tRNAs by Methionyl-tRNA transformylase from mammalian mitochondria. J Biol Chem. 2001;276:20064–20068. doi: 10.1074/jbc.M101007200. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci U S A. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U, Lee CP, Seong BL, RajBhandary UL. Mutants of initiator tRNA that function both as initiators and elongators. J Biol Chem. 1991;266:18018–18024. [PubMed] [Google Scholar]
- Vial L, Gomez P, Panvert M, Schmitt E, Blanquet S, Mechulam Y. Mitochondrial methionyl-tRNAfMet formyltransferase from Saccharomyces cerevisiae: gene disruption and tRNA substrate specificity. Biochemistry (Mosc) 2003;42:932–939. doi: 10.1021/bi026901x. [DOI] [PubMed] [Google Scholar]
- Villet R, Fonvielle M, Busca P, Chemama M, Maillard AP, Hugonnet J-E, et al. Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis. Nucleic Acids Res. 2007;35:6870–6883. doi: 10.1093/nar/gkm778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar HP de. Amino acid fermentation at the origin of the genetic code. Biol Direct. 2012;7:6. doi: 10.1186/1745-6150-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Fegley MW, Warner JB, Grindley CLJ, Marotta NP, Petersson EJ. N-terminal protein modification using simple aminoacyl transferase substrates. J Am Chem Soc. 2011;133:15139–15147. doi: 10.1021/ja2055098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell. 2014;56:5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007;449:867–871. doi: 10.1038/nature06167. [DOI] [PubMed] [Google Scholar]
- Watanabe Y-I, Suematsu T, Ohtsuki T. Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front Genet. 2014;5:109. doi: 10.3389/fgene.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F-Y, Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr J. 2011;58:819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- Weisblum B. Back to Camelot: defining the specific role of tRNA in protein synthesis. Trends Biochem Sci. 1999;24:247–250. doi: 10.1016/s0968-0004(99)01396-1. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- Wettstein FO, Stent GS. Physiologically induced changes in the property of phenylalanine tRNA in Escherichia coli. J Mol Biol. 1968;38:25–40. doi: 10.1016/0022-2836(68)90126-5. [DOI] [PubMed] [Google Scholar]
- Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev MMBR. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Sheppard K, Söll D. Amino acid modifications on tRNA. Acta Biochim Biophys Sin. 2008;40:539–553. doi: 10.1111/j.1745-7270.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-M, Liu C, Slater S, Hou Y-M. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys) Nat Struct Mol Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ntai I, Kelleher NL, Walsh CT. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci U S A. 2011;108:12249–12253. doi: 10.1073/pnas.1109539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Korostelev A, Lancaster L, Noller HF. Crystal structures of 70S ribosomes bound to release factors RF1, RF2 and RF3. Curr Opin Struct Biol. 2012;22:733–742. doi: 10.1016/j.sbi.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]