Abstract
Full characterization of complex reaction mixtures is necessary to understand mechanisms, optimize yields, and elucidate secondary reaction pathways. Molecular-level information for species in such mixtures can be readily obtained by coupling mass spectrometry imaging (MSI) with thin layer chromatography (TLC) separations. User-guided investigation of imaging data for mixture components with known m/z values is generally straightforward; however, spot detection for unknowns is highly tedious, and limits the applicability of MSI in conjunction with TLC. To accelerate imaging data mining, we developed DetectTLC, an approach that automatically identifies m/z values exhibiting TLC spot-like regions in MS molecular images. Furthermore, DetectTLC can also spatially match m/z values for spots acquired during alternating high and low collision-energy scans, pairing product ions with precursors to enhance structural identification. As an example, DetectTLC is applied to the identification and structural confirmation of unknown, yet significant, products of abiotic pyrazinone and aminopyrazine nucleoside analog synthesis.
Introduction
The origin of life is a yet unsolved problem that continues to fascinate generations of scientists [1]. Unravelling the routes that led the transition from an abiotic scenario composed of complex chemical species to simple living systems is perhaps one of the biggest scientific questions of our time. This transition requires synthesis of the building blocks of life (amino acids, sugars, nucleotides), the conditions leading to polycondensation of these blocks into far-from-equilibrium proto-biopolymers (proto-nucleic acids, proto-peptides, proto-oligosaccharides), the emergence of biochemical function, and the later organization and compartmentalization into supramolecular systems resembling proto-cells. Analytical chemistry thus plays a central role by providing new tools to probe the extremely complex, seemingly intractable mixtures involved in these prebiotic reactions [2].
Prebiotic chemistry experiments investigating these questions typically produce a large and complex inventory of chemical precursors rather than a single product of biological relevance, and only a select few that react further to form larger-order structures. As is common practice in synthetic organic chemistry, preliminary screening of complex prebiotic reaction mixtures is often performed via thin layer chromatography (TLC) because of its simplicity and speed [3]. Visualization of developed TLC plates most commonly uses dyes or fluorescence to identify spots corresponding to the separated components [4].
Standard TLC analysis is rapid, but lacks chemical specificity. For more comprehensive TLC analysis, mass spectrometry (MS) can be employed to read the plates through extraction or direct probing of their surface [5–8]. Direct MS can be performed statically on select spots, or dynamically in microprobe MS imaging mode (MSI), the latter eliminating the need for knowing a priori where the separated compounds are located. Microprobe MSI involves spatially resolved desorption/ionization of analytes from the surface, followed by mass analysis, occasionally in tandem fashion (MS/MS). Experiments are performed by rastering the ion source probe in unidirectional line scans across the sample, recording mass spectra, and translating those into a chemical image [9–11]. MSI of 1D and 2D high performance (HP) TLC plates has been successfully demonstrated with a number of MS techniques, including matrix assisted laser desorption ionization (MALDI) [12], desorption electrospray ionization (DESI) [9, 13–15], and liquid microjunction-surface sampling probes [16], among others [17, 18]. Targeted interpretation of microprobe MSI data from HPTLC plates is straightforward if m/z values of analytes of interest are known: images for these m/z values are simply extracted from the (x,y,m/z) data cube. However, untargeted detection of unknown species can detract from the high-throughput appeal of TLC, as MSI datacubes typically contain thousands of chemical images. Mining useful information from untargeted MSI HPTLC datasets can easily take hours or even days, depending on mixture complexity. An automated workflow to detect spots in MS images from HPTLC plates, even if their m/z values are a priori unknown, would significantly increase the usability of HPTLC-MSI approaches.
Algorithms commonly used for MSI data mining have included principal component analysis (PCA) [19, 20], non-negative matrix factorization (NMF) [21], and clustering [22]. Despite their popularity, these approaches are not intended for individually detecting the many components present in a complex mixture, but rather patterns combining the most salient components. Therefore, we developed a new approach for automatically detecting individual spots on TLC MSI images, named DetectTLC. Its core innovation is that m/z images containing elevated ion abundances in spot-like clusters are distinguished from noninformative images on the basis of differences in their quantitative features, such as compactness, eccentricity, extent, solidity, and entropy.
We present DetectTLC by applying it to a case study where DESI MSI experiments were performed on proto-nucleic acid building block reaction mixtures separated onto 2D HPTLC plates. These mixtures contained pyrazinone (PZO) or aminopyrazine (APZ) nucleoside analogs that are potentially capable of complementary self-assembly (hydrogen bonding) required for replication [23]. DetectTLC, however, is flexible enough to be applicable to any other HPTLC-MSI experiment.
Experimental Methods
All DetectTLC development was done in MATLAB (MathWorks, Natick, MA, USA). Full details of the algorithms used and assessment of their function are included in the Supporting Information. DetectTLC can be downloaded from the following address: http://www.bio-miblab.org/software/for_testing.zip with username: miblab, password: br6TrUja. Full details of the synthesis and TLC separation of the PZO and APZ mixtures is included in Supporting Information Section 1.1.
Mass spectrometry images were acquired using combinations of two ion source/imaging stages and mass spectrometers to test the ability of DetectTLC to process datasets from instruments with different spatial and mass spectral resolution. The PZO synthesis mixture was imaged using a 2D OmniSpray automated DESI ion source (Prosolia Inc., Indianapolis, IN, USA) coupled to a Synapt G2 HDMS mass spectrometer (Waters, Milford, MA, USA) in positive-ion mode. The m/z range acquired for each pixel was 50–250. A DESI spray of 1% acetic acid (ACS Reagent grade; Sigma-Aldrich, St. Louis, MO, USA) in acetonitrile (OmniSolv LC-MS grade; EMD Millipore Corporation, Billerica, MA, USA) with a flow rate of 5 μL min−1 and a N2 nebulizing gas (150 psi) was used for maximum sensitivity and minimal impact spot size. Stage motion was programmed such that data was acquired in 200 μm × 200 μm pixels for a total image size of 30 mm2. Mass spectral data was converted using Firefly conversion software (Prosolia, Inc., Indiandapolis, IN, USA) into Analyze 7.5 format and images were visualized using omniSpect (http://omnispect.bme.gatech.edu/) [21] and DetectTLC.
The APZ reaction mixture was imaged using an OmniSpray DESI probe (Prosolia Inc., Indianapolis, IN, USA) spraying acetonitrile at a flow rate of 8 μL min−1 and a N2 nebulizing gas pressure of 130 psi. The sample was rastered using a motorized microscope stage (OptiScan II; Prior Scientific Inc., Rockland, MA, USA) controlled by a Labview VI described elsewhere [21,24], moving at a speed of 160 μm s−1 with a line step of 200 μm. Mass spectra were acquired on an Exactive Plus Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA, USA) in positive-ion mode as centroided data with the following settings remaining constant throughout the experiment; mass resolution (35,000), no. of microscans (2), automatic gain control target (3 × 106), maximum injection time (50 ms), spray voltage (4 kV), capillary temperature (250°C), and S lens rf level (60 V). The imaging data was acquired in a multimodal fashion by alternating each scan between Full MS (no collision-energy applied) and All-Ion Fragmentation (AIF, collision-energy = 60 arb.) modes. There is no precursor ion selection during AIF scans prior to higher-energy collisional dissociation (HCD); therefore, all ions entering the HCD cell were fragmented and detected. In this manner, every other pixel in the MS image represented ions from the alternating modes. The dataset may be visualized as (1) a complete dataset with pixels from both modes included, or (2) as separate Full MS or AIF images after separating the scan modes using Xcalibur software (Thermo Scientific, Inc.). The pixel area corresponding to single scans were 80 × 200 μm for the complete dataset based on stage motion and data acquisition speed. When the dataset is separated into the individual Full MS or AIF datasets, the m/z vs time data is interpolated, resulting in a doubling of effective pixel size (160 × 200 μm). RAW data files were converted to the .mzxml format using `msconvert' for visualization using omniSpect and DetectTLC.
Results and Discussion
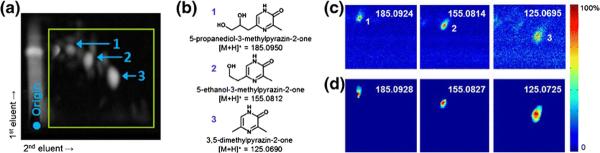
The synthesis of pyrazin-2-one (PZO) and 2-aminopyrazine (APZ) monomers is of interest to prebiotic chemistry [23] (Supplementary Scheme SI 1, 2). These were used as model organic reactions to demonstrate the capabilities of DetectTLC, but the approach is widely applicable to any TLC-separated complex mixture. Reaction products were separated using 2D reversed phase HPTLC. The fluorescence image obtained from HPTLC separation of the PZO mixture using solvent system “A” is shown in Figure 1a. Three major PZO products (1–3) that had been previously identified (chemical structures shown in Figure 1b) were observed in the MSI data from MS images generated manually (Figure 1c).
Figure 1.
Fluorescence image of a developed HPTLC plate with the area imaged by DESI-MSI outlined in green (a). Identified products of the PZO reaction (Supplementary Scheme SI 1, only 5-isomer is shown), with the numbers indicating the spatial location of each product in the fluorescence and MS images (b). Selected ion images acquired by DESI-MSI of reaction products previously identified (c), and the corresponding spot-like images as they were automatically generated by DetectTLC using the preset Protocol 1 processing pipeline (d)
NMF and PCA were applied to the 2D-HPTLC-MSI datasets to assess the ability of these models to identify individual product spots (Supplementary Figures SI 2, 3). For NMF, the data was separated into five components. Three of the component images showed a combination of 1–3 spot-like features, at two unique spatial distributions. The two other components resulted in images and spectra corresponding to chemical background. For PCA, the first five components accounted for 73% of the variation within the dataset. PC 1 and PC 2 showed spot-like features, each with unique spatial distributions. PC 3–5 largely represented chemical noise within the dataset. These results are to be expected considering that both the NMF and PCA models aim to identify predominant patterns within the dataset, not to extract a complete set of spot-like features. The two spot-like features identified by NMF and PCA are the areas with the highest ion intensities, indicating a notable pattern within the dataset. However, additional product spots may have high chemical significance, but are not identified by NMF or PCA, making these methods unsuitable for reliable TLC plate analysis. These results highlight the need for the DetectTLC tool, which can automatically characterize the chemical diversity within a 2D TLC separation,
Fully automated analysis of the MSI data using DetectTLC generated 50+ images containing individual spots that corresponded to various analytes on the TLC plate. Included in those were the known products, as shown in Figure 1d.
Experimental m/z values for these products were consistent with theoretical values, with an average error of 1 mmu. Interestingly, a systematic difference of approximately 5 mmu on average was observed between the raw ion image m/z values and the DetectTLC images, which was attributed to the centroiding process used. Mass spectra were acquired in profile mode, but the m/z peaks observed are not truly Gaussian; therefore, the local m/z maximum value observed differed slightly from the centroided m/z value used by DetectTLC [25]. Differences in image contrast between the manually-obtained and DetectTLC images were also observed and attributed to the median filtering step that removes background noise pixels in the DetectTLC image processing pipeline, discussed below.
One key advantage of DetectTLC is its ability to identify reaction products that may not be anticipated. When the PZO mixture was separated by a second solvent system “B”, a new, intense fluorescence spot, different from 1–3, was detected (Supplementary Figure SI 1). DetectTLC was applied to the MSI dataset generated with the new solvent system, and the results included an ion image at m/z 167.0353 co-localized with the unknown fluorescent spot, yielding a chemical formula of C8H11N2O2 (Δm = 0.8 mmu). Taking into consideration the reaction chemistry, this species was tentatively identified as 3,5(6)-dimethyl-4-oxoethyl-2-oxopyrazin-4-ium, a plausible reaction side-product (Supplementary Schemes SI 1, 4).
The m/z 167.0353 image ranked 13th in the DetectTLC results and was displayed on the first page of 24 images in the main graphical user interface (GUI) window. This exemplifies the ability of DetectTLC to detect chemical species in an untargeted, automatic manner, revealing species that may have gone undetected with a more targeted approach.
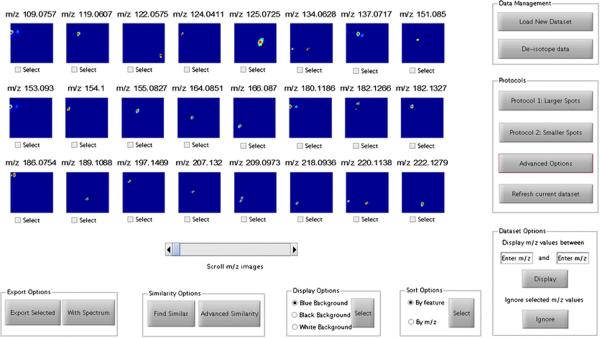
The DetectTLC workflow is shown in Figure 2. An outline of the operations involved is presented here and additional details are given in the Supporting Information. First, 2D median filtering and pixel count-based filtering are applied to each m/z image in order to remove spatially-disperse, low-intensity noise and to reject sparse or high-intensity noisy images, respectively. The images that pass this step are in the second step thresholded to binary images in order to identify spot-like regions regardless of signal intensity. This facilitates the detection of low-abundance species that are separated by TLC, and leaves the decision of significance to the user. Third, morphologic opening is applied to smooth discontinuous pixel clusters into spot-like regions, thus correcting for signal variability and enhancing the quantitative image feature-based detection method. Fourth, quantitative image features are used to score each m/z image on the basis that it contains a spot. Eight image features are currently available in DetectTLC, seven of which are shape-based (area, compactness, convex area, eccentricity, extent, number of connected regions, and solidity) and the eighth is texture-based (entropy). Each of these steps is automatically performed during routine analysis using two preset protocols, one tailored to identify larger TLC spots, and one for smaller spots, both with no user input required. Finally, m/z images are ranked in terms of their feature values and visualized in the DetectTLC GUI (Figure 3). Through this GUI, the user may save the average mass spectrum across a TLC spot together with the corresponding m/z image for further analysis. Selected ion images and a list of m/z values that produce images with spots may also be batch-exported to a target folder (Supplementary Figure SI 4). Secondary windows, along with a full description of all data processing and visualization possibilities available—including advanced options facilitating manual data filtering and processing by the user—are discussed in the Supporting Information (Supplementary Figures SI 5, 9, Supplementary Table SI 1).
Figure 2.
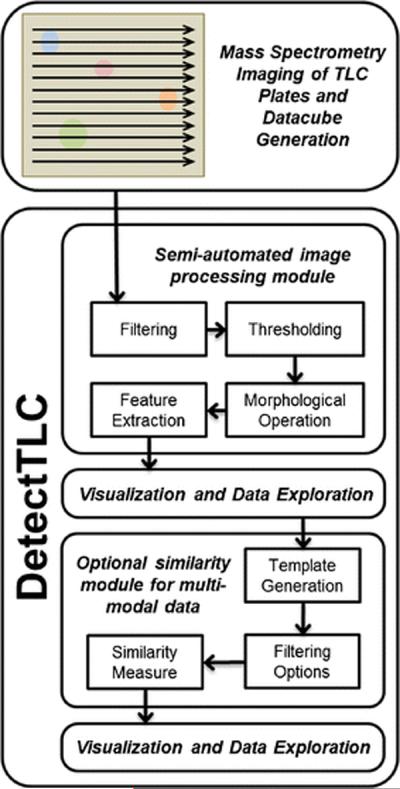
An overview of the workflow used in the DetectTLC software tool
Figure 3.
The main graphical user interface with the top 24 images containing spot-like regions are displayed. Data is first uploaded and (optionally) de-isotoped, following which the users may select from Protocol 1 or Protocol 2 for feature selection, or design their own processing pipeline through the Advanced Options tab. Spots with similar spatial distributions may be identified using the Similarity Options. Selected images and/or spectra may be exported using the Export Options
Various processing pipelines within the DetectTLC framework were assessed for their ability to identify TLC spots. Each possible image processing combination, 128 pipelines in all, were evaluated based on the number of true spots identified in the top 40 images generated when applied to the PZO synthesis dataset. A full assessment of the results for all pipelines is included in Supporting Information (Tables SI 2, 3), along with the top 40 images found by the area, compactness, and entropy features (Supplementary SI Figures 6–8). It was observed that for thresholding purposes, Otsu's method and selected manual thresholds yielded comparable results across all other processing variables. The key difference between these two approaches is the user input required for manual thresholding versus the user-independent Otsu's method. In almost every combination of processing methods, morphologic opening yielded a higher number of spot-containing images than dilation (on average twice as many true spots for all combinations). The other processing step that significantly affected the number of true spots identified was median filtering. For this dataset, the 7 × 7 median filter was found to perform notably better than no filter. The user can input different filter sizes through the Advanced Options menu and select the most appropriate size for the dataset under analysis.
Ultimately, the analysis pipeline of (1) removal of images with <5 and >1500 non-zero pixels, (2) 7 × 7 median filtering, (3) Otsu's threshold, and (4) morphologic opening, provided the best results for the PZO dataset. Combining this pipeline with entropy as the image feature for ranking, two of the known products in the PZO reaction mixture were included in the top five images output by DetectTLC, and all three predicted products were within the top 40 ranked images. For other chemical systems, DetectTLC settings could be tailored to provide optimum results and to obtain a complete list of m/z images containing spots.
To confidently assign analyte structure by mass spectrometry, MS fragmentation data must be obtained. Modern mass spectrometers allow for data to be acquired in multiple modes sequentially, in real time. For example, alternating between low and high collision-energy scans enables data corresponding to precursor and product ions to be acquired in a single acquisition cycle. In true tandem mass spectrometers, the precursor ion is first selected for activation, but when many analytes are simultaneously ionized as in MSI, it becomes challenging to mass-select all individual precursor ions while maintaining high imaging throughput. Matching of precursor to product ions in alternating scan experiments can, for example, be performed based on 1D chromatographic profile similarity [26].
Similarly, DetectTLC includes algorithms capable of automatically pairing precursors and product ions from multimodal imaging datasets using their 2D spatial distributions on the TLC plate. This kind of similarity matching is performed in DetectTLC by first allowing the user to select the precursor ion spot of interest from a low collision-energy image (Supplementary Figure SI 9). This region of interest is then used to generate a template against which to measure similarity of the high collision-energy images using either Pearson correlation or the hypergeometric similarity measure [27, 28]. The results are displayed as a set of ion images, but the user also has the option of generating an all-fragment mass spectrum displaying peaks for m/z values with similar spatial distribution as the precursor. Further details of the similarity algorithms implemented in DetectTLC are provided under Supporting Information.
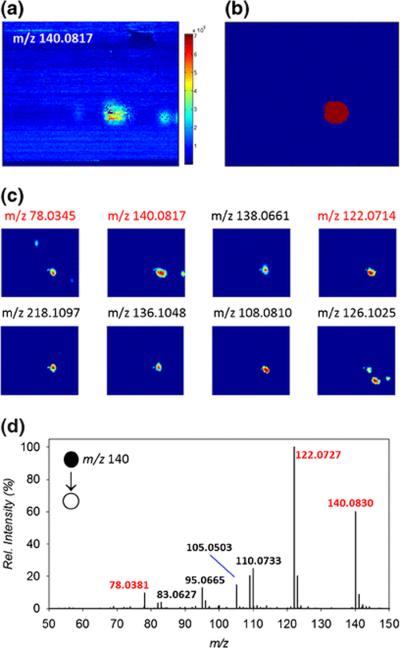
To demonstrate this capability, a TLC plate with the separated APZ synthesis reaction mixture (Supplemental Scheme SI 2) was imaged using multi-modal MSI, alternating high and low collision-energy scans. The spot corresponding to the intact protonated precursor ion of 5(6)-ethanol-2-aminopyrazine (m/z 140.0817, Schemes SI 2, 2), a predicted side-product of the APZ reaction, was first tentatively identified by DetectTLC from the low collision-energy dataset, and was selected to create a template of the spot's location (Figure 4). Pearson correlation and the hypergeometric similarity measure were applied to pair the precursor ion with product ions for structural confirmation (Supplementary Figures SI 10, 11). The top eight m/z images matched using the hypergeometric similarity measure from the high collision-energy dataset are shown in Figure 4c. The same top spot-like m/z images were obtained using the Pearson correlation, but ranked in a different order. Upon further investigation, two of the product ions (m/z 122.0714 and 78.0345) were assigned to H2O and CH6N2O losses from the precursor ion, respectively. This observed fragmentation pattern supports the structural assignment of 5(2-hydroxyethyl)-2-aminopyrazine to the precursor at m/z 140.0817. To validate these DetectTLC matches, liquid chromatography-tandem MS analysis was performed on the m/z 140.0817 precursor ion (see Supporting Information). Two of expected product ions were detected at m/z 122.0727 and 78.0381. Additional products at m/z 83.0627, 95.0665, 105.0503, and 110.0733 were also detected in UPLC-MS/MS experiments (Figure 4d), probably due to sensitivity and ion activation efficiency differences in the instrumentation used.
Figure 4.
A selected ion image of the precursor ion at m/z 140.1 (a), and the template used to apply the similarity measure (b). The top eight ion images matched with the precursor ion using hypergeometric similarity measure (c). Product ion spectrum obtained by UPLC-MS/MS used to validate the DetectTLC output (d). The fragment ions detected in both UPLC-MS/MS and DetectTLC datasets are indicated with red labels
Conclusions
Through automated analysis of MSI data, DetectTLC is able to pinpoint spots on 2D HPTLC plates corresponding to both expected and unexpected reaction products, including those undetectable via fluorescence, or those that co-migrated, preventing selective optical visualization. This type of processing ensures that the user gains information describing all chemical species present on the plate regardless of their optical or chromatographic properties. Structural identification of reaction products displaying spot-like regions can be accomplished when multimodal imaging is performed, detecting intact and fragment ions in a single acquisition. Although DetectTLC was designed for processing TLC-MSI datasets, it could also be more broadly applied to MSI datasets where other types of spot-like features are expected.
Supplementary Material
Acknowledgments
The authors acknowledge support for his research by the Center for Chemical Evolution, jointly supported by NSF and the NASA Astrobiology Program (NSF CHE-1004570). Initial work was supported by a seed grant from the BioImaging Mass Spectrometry Initiative at Georgia Tech to M.D.W. and F.M.F. M.D.W. also acknowledges support from The Parker H. Petit Institute for Bioengineering and Bioscience (IBB), Johnson & Johnson, National Institutes of Health (R01CA108468, U54CA119338), Georgia Cancer Coalition (Distinguished Cancer Scholar Award), and Microsoft Research. C.D.K. acknowledges support from an NSF GRF and a P.E.O. International Scholar Award.
References
- 1.Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic systems chemistry: new perspectives for the origins of life. Chem. Rev. 2014;114:285–366. doi: 10.1021/cr2004844. CrossRef. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz AW. Intractable mixtures and the origin of life. Chem. Biodivers. 2007;4:656–664. doi: 10.1002/cbdv.200790056. CrossRef. [DOI] [PubMed] [Google Scholar]
- 3.Poole F, C., Poole SK. Instrumental thin-layer chromatography. Anal. Chem. 1994;66:27A–37A. CrossRef. [Google Scholar]
- 4.Sherma J. Planar chromatography. Anal. Chem. 1998;70:7r–26r. CrossRef. [Google Scholar]
- 5.Busch KL. Mass-spectrometric detection for thin-layer chromatographic separations. TrAC, Trends Anal. Chem. 1992;11:314–324. CrossRef. [Google Scholar]
- 6.Unger SE, Vincze A, Cooks RG, Chrisman R, Rothman LD. Identification of quaternary alkaloids in mushroom by chromatography-secondary ion mass-spectrometry. Anal. Chem. 1981;53:976–981. CrossRef. [Google Scholar]
- 7.Cheng SC, Huang MZ, Shiea J. Thin layer chromatography/mass spectrometry. J. Chromatogr. A. 2011;1218:2700–2711. doi: 10.1016/j.chroma.2011.01.077. CrossRef. [DOI] [PubMed] [Google Scholar]
- 8.Morlock G, Schwack W. Coupling of planar chromatography to mass spectrometry. TrAC, Trends Anal. Chem. 2010;29:1157–1171. CrossRef. [Google Scholar]
- 9.Van Berkel GJ, Kertesz V. Automated sampling and imaging of analytes separated on thin-layer chromatography plates using desorption electrospray ionization mass spectrometry. Anal. Chem. 2006;78:4938–4944. doi: 10.1021/ac060690a. CrossRef. [DOI] [PubMed] [Google Scholar]
- 10.Wiseman JM, Ifa DR, Song QY, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. CrossRef. [DOI] [PubMed] [Google Scholar]
- 11.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. CrossRef. [DOI] [PubMed] [Google Scholar]
- 12.Gusev AI, Vasseur OJ, Proctor A, Sharkey AG, Hercules DM. Imaging of thin-layer chromatograms using matrix/assisted laser desorption/ionization mass-spectrometry. Anal. Chem. 1995;67:4565–4570. CrossRef. [Google Scholar]
- 13.Van Berkel GJ, Ford MJ, Deibel MA. Thin-layer chromatography and mass spectrometry coupled using desorption electrospray ionization. Anal. Chem. 2005;77:1207–1215. doi: 10.1021/ac048217p. CrossRef. [DOI] [PubMed] [Google Scholar]
- 14.Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. Using HPTLC/DESI-MS for peptide identification in 1D separations of tryptic protein digests. Anal. Bioanal. Chem. 2008;391:317–324. doi: 10.1007/s00216-008-1874-6. CrossRef. [DOI] [PubMed] [Google Scholar]
- 15.Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. HPTLC/DESI-MS imaging of tryptic protein digests separated in two dimensions. J. Mass Spectrom. 2008;43:1627–1635. doi: 10.1002/jms.1431. CrossRef. [DOI] [PubMed] [Google Scholar]
- 16.Van Berkel GJ, Sanchez AD, Quirke JME. Thin-layer chromatography and electrospray mass spectrometry coupled using a surface sampling probe. Anal. Chem. 2002;74:6216–6223. doi: 10.1021/ac020540+. CrossRef. [DOI] [PubMed] [Google Scholar]
- 17.Haddad R, Milagre HM, Catharino RR, Eberlin MN. Easy ambient sonic-spray ionization mass spectrometry combined with thin-layer chromatography. Anal. Chem. 2008;80:2744–2750. doi: 10.1021/ac702216q. CrossRef. [DOI] [PubMed] [Google Scholar]
- 18.Stanley MS, Busch KL, Vincze A. Imaging analysis of thin layer chromatograms by secondary ion mass spectrometry: analysis of neostigmine and pyridostigmine bromides. J. Planar Chromatogr.–Mod. 1988;TLC 1:76–78. [Google Scholar]
- 19.Sjovall P, Lausmaa J, Johansson B. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 2004;76:4271–4278. doi: 10.1021/ac049389p. CrossRef. [DOI] [PubMed] [Google Scholar]
- 20.Eijkel GB, Kaletas BK, van der Wiel IM, Kros JM, Luider TM, Heeren RMA. Correlating MALDI and SIMS imaging mass spectrometric datasets of biological tissue surfaces. Surf. Interface Anal. 2009;41:675–685. CrossRef. [Google Scholar]
- 21.Parry RM, Galhena AS, Gamage CM, Bennett RV, Wang MD, Fernandez FM. omniSpect: an open MATLAB-based tool for visualization and analysis of matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry images. J. Am. Soc. Mass Spectrom. 2013;24:646–649. doi: 10.1007/s13361-012-0572-y. CrossRef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov T, Chernyavsky I, Becker M, von Eggeling F, Nikolenko S. Analysis and interpretation of imaging mass spectrometry data by clustering mass-to-charge images according to their spatial similarity. Anal. Chem. 2013;85:11189–11195. doi: 10.1021/ac401420z. CrossRef. [DOI] [PubMed] [Google Scholar]
- 23.Weber AL. Sugar-driven prebiotic synthesis of 3,5(6)-dimethylpyrazin-2-one: a possible nucleobase of a primitive replication process. Orig. Life Evol. Biosph. 2008;38:279–292. doi: 10.1007/s11084-008-9141-6. CrossRef. [DOI] [PubMed] [Google Scholar]
- 24.Bennett RV, Gamage CM, Galhena AS, Fernandez FM. Contrast-enhanced differential mobility-desorption electrospray ionization-mass spectrometry imaging of biological tissues. Anal. Chem. 2014;86:3756–3763. doi: 10.1021/ac5007816. CrossRef. [DOI] [PubMed] [Google Scholar]
- 25.Urban J, Afseth NK, Stys D. Fundamental definitions and confusions in mass spectrometry about mass assignment, centroiding and resolution. TrAC, Trends Anal. Chem. 2014;53:126–136. CrossRef. [Google Scholar]
- 26.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MS(E): a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 2006;20:1989–1994. doi: 10.1002/rcm.2550. CrossRef. [DOI] [PubMed] [Google Scholar]
- 27.Kaddi C, Parry RM, Wang MD. Hypergeometric similarity measure for spatial analysis in tissue imaging mass spectrometry. Proceedings of the 2011 I.E. International Conference on Bioinformatics and Biomedicine; 2011. pp. 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaddi CD, Parry RM, Wang MD. Multivariate hypergeometric similarity measure. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013 doi: 10.1109/TCBB.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.