Abstract
Glucosinolates, a class of secondary metabolites from cruciferous plants, are derived from amino acids and have diverse biological activities, such as in biotic defense, depending on their side chain modification. The first structural modification step in the synthesis of aliphatic (methionine-derived) glucosinolates—S-oxygenation of methylthioalkyl glucosinolates to methylsulfinylalkyl glucosinolates—was found to be catalyzed by five flavin-containing monooxygenases (FMOs), FMOGS-OX1-5. Here, we report two additional FMOGS-OX enzymes, FMOGS-OX6, and FMOGS-OX7, encoded by At1g12130 and At1g12160, respectively. The overexpression of both FMOGS-OX6 and FMOGS-OX7 decreased the ratio of methylthioalkyl glucosinolates to the sum of methylthioalkyl and methylsulfinylalkyl glucosinolates, suggesting that the introduction of the two genes converted methylthioalkyl glucosinolates into methylsulfinylalkyl glucosinolates. Analysis of expression pattern revealed that the spatial expression of the two genes is quite similar and partially overlapped with the other FMOGS-OX genes, which are primarily expressed in vascular tissue. We further analyzed the responsive expression pattern of all the seven FMOGS-OX genes to exogenous treatment with abscisic acid, 1-aminocyclopropane-1-carboxylic acid (ACC), jasmonic acid (JA), salicylic acid, indole-3-acetic acid (IAA), and low and high temperatures. Although these genes showed same tendency toward the changing stimulus, the sensitivity of each gene was quite different. The variety in spatial expression among the FMOGS-OX genes while responding to environmental stimulus indicated a complex and finely tuned regulation of glucosinolates modifications. Identification of these two novel FMOGS-OX enzymes will enhance the understanding of glucosinolates modifications and the importance of evolution of these duplicated genes.
Keywords: Arabidopsis thaliana, flavin-containing monooxygenase (FMO), glucosinolates, S-oxygenation, expression pattern
Introduction
Glucosinolates (GSLs) are amino acid-derived natural products primarily present in plants belonging to the Brassicaceae family, such as cabbage, broccoli, and the model plant Arabidopsis thaliana. Upon wounding or mastication, GSLs are hydrolyzed by myrosinases (thioglucosidases) and release hydrolysis products, primarily isothiocyanates and nitriles (Halkier and Gershenzon, 2006; Bones and Rossiter, 2006; Zhang et al., 2006). These breakdown products exert diverse biological effects such as induction of direct toxic effects or other defense responses against pathogens and generalist herbivores (Bednarek et al., 2009; Clay et al., 2009; Hopkins et al., 2009; Laluk et al., 2012). In humans, the isothiocyanates derived from some aliphatic (methionine-derived) GSLs are considered to have numerous health benefits including potent anti-cancer property (Fahey et al., 1997, 2002; Rose et al., 2000; Zalcmann and Talalay, 2001). The most well-studied isothiocyanate is sulforaphane, which is derived from 4-methylsulfinylbutyl GSL; it can decrease the risk of various cancers such as breast cancer, prostate cancer, gastric cancer, and skin cancer (Fahey et al., 2002; Rose et al., 2005; Talalay et al., 2007; Atwell et al., 2015; Chang et al., 2015; Fofaria et al., 2015). Aliphatic GSLs are synthesized in three steps—elongation of methionine chain, formation of GSL core structure, and modification of side chain. The diverse biological activities of GSLs are largely dependent on the chemical modifications in their side chain.
The basic aliphatic glucosinolate molecules, methylthioalkyl (MT) GSLs, which contain only the core structure, may undergo a variety of secondary modifications such as oxidation, hydroxylation, or desaturation. The first step in side-chain modification is S-oxygenation, which converts MT GSLs to methylsulfinylalkyl (MS) GSLs. Thus, S-oxygenation is of biological as well as biochemical interest because it influences not only the biosynthesis of MS GSLs, but also the further modification and the resulting activity of their hydrolysis products.
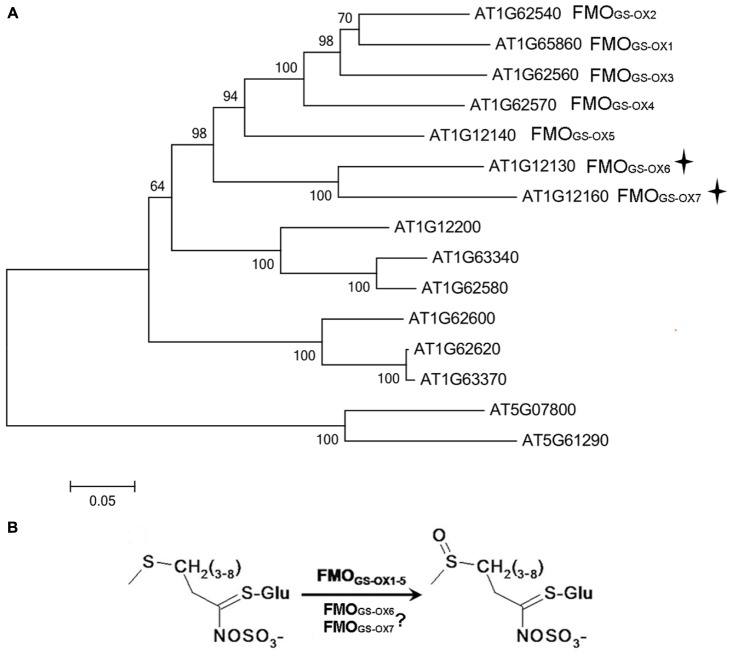
Five flavin-containing monooxygenases (FMOs), FMOGS-OX1-5, have been identified to catalyze S-oxygenation during the conversion of MT GSLs to MS GSLs (Hansen et al., 2007; Li et al., 2008). In phylogenetic analysis of FMOs containing the conserved FMOGS-OX domains in Arabidopsis, At1g12130 and At1g12160 clustered to the same subclade and, together with FMOGS-OX1-5, formed a seven-protein group (Figure 1A). In a previous study, a phylogenetic tree of plant-derived FMOs in rice, Arabidopsis, and poplar was analyzed, and this seven-protein group was involved in the predicted S-oxygenation clade and was present only in the GSL-producing Arabidopsis plant. Thus, these genes were predicted to be involved in the S-oxygenation of GSL (Figure 1B). Further, another FMO-coding gene, At1g12200, was suggested as a candidate FMOGS-OX due to its strong co-expression with other GSL biosynthetic genes in response to Sclerotinia sclerotiorum (Stotz et al., 2011) infection. Therefore, in our study, we investigated At1g12130, At1g12160, and At1g12200 as FMOGS-OX candidate genes. It was discovered that both At1g12130 (named FMOGS-OX6) and At1g12160 (named FMOGS-OX7) are involved in S-oxygenation of aliphatic GSLs, while At1g12200 does not possess the expected activity.
FIGURE 1.
(A) Phylogenetic tree of Arabidopsis flavin-containing monooxygenases (FMOs) possessing the conserved GS-OX domains. (B) FMOGS-OX6 and FMOGS-OX7 are predicted to catalyze S-oxygenation of methylthioalkyl (MT) Glucosinolates (GSLs) to methylsulfinylalkyl (MS) GSLs.
To better understand the coordination of these FMOGS-OXs and the upstream aliphatic GSL biosynthetic genes, responsive expression pattern of these genes toward several hormone treatments and environmental conditions are detected. Integrating the knowledge of catalysis, distribution and responsive pattern of these FMOGS-OXs, we speculate that on one hand, these duplicated genes are redundant and might functionally compensate and increase genetic robustness, whereas on the other hand, they show delicate functional variation and possibly contribute to the complex and finely tuned regulation of GSLs modification.
Materials and Methods
Generation of Phylogenetic Tree
All the FMO protein sequences with conserved GS-OX domains in Arabidopsis were obtained from NCBI (Marchler-Bauer et al., 2015) and the full-length amino acid sequences of these proteins were aligned using ClustalX with default parameters. A phylogenetic tree was constructed using the neighbor-joining method with Mega software (version 5.0). Bootstrap support for the topology was estimated from 1000 replicates, and nodes occurring in less than 50% of the replicates were collapsed.
Plant Material and Growth Conditions
Wild-type (Col-0) and mutants of Arabidopsis seeds were obtained from Arabidopsis Biological Resource Center. Genotype of FMOGS-OX6 T-DNA insertion mutant (SALK_209008) was analyzed by PCR using the following three primers, 5′-CCGCCTAATTCAATCAGCTC-3′, 5′-GAGGCTCAGTGAGATGACC-3′, and the T-DNA-specific primer LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′. Genotype of FMOGS-OX7 T-DNA insertion mutant (SALK_062129) was analyzed by PCR using the following three primers, 5′-AAACCGTGAAGATTGCAATTG-3′, 5′-GACGAGGCTCAGTGAAGTGAC-3′, and the T-DNA-specific primer LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′. Genotype of At1g12200 T-DNA insertion mutant (SALK_136282) was analyzed by PCR using the following three primers, 5′-CCGCCTAATTCAATCAGCTC-3′, 5′-GAGGCTCAGTGAAGTGACC-3′, and the T-DNA-specific primer LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′.
Plants were grown in a growth chamber with 16 h light/8 h dark photoperiod at a photosynthetic photon flux density of 100 μmolm-2 s-1 at 20 and 70% relative humidity, respectively.
Plasmid Construction and Plant Transformation
To prepare the overexpression constructs 35S::FMOGS-OX6, 35S::FMOGS-OX7, and 35S::At1g12200, coding sequences (CDS) of FMOGS-OX6, FMOGS-OX7 and At1g12200 were amplified through reverse transcription-PCR (RT-PCR) method. The total RNA from plant was isolated with TRIzol reagent according to the manufacturer’s instructions. First-strand cDNA was synthesized using the SupermoIII RT kit (Bioteke). The CDS of FMOGS-OX6, FMOGS-OX7 and At1g12200 were amplified with Pfu Turbo Cx Hotstart DNA polymerase (Stratagene) from the first-strand products using the following primers, respectively: 5′-GGCTTAAUATGACACCACCGCCTAATTC-3′, 5′-GGTTTAAUTTATGTAGACCAAACTTTGCTCG-3′ and 5′-GGCTTAAUATGACCAGCGTAATCACCTC-3′, 5′-GGTTTAAUCTAAGAAGAACAATCTTGGTTCG-3′, and 5′-ATGGCAACGAGTCATCCTGA-3′, 5′-TTAGGTTTTGAGCATCGGCAAAAG-3′. The PCR products were then cloned into the expression vector pCAMBIA230035Su using the USER Cloning method as described by Nour-Eldin et al. (2006).
To identify the spatial expression pattern of FMOGS-OX6 and FMOGS-OX7, two constructs FMOGS-OX6Pro::GUS (GUS was driven by the promoter of FMOGS-OX6) and FMOGS-OX7Pro::GUS (GUS was driven by the promoter of FMOGS-OX7), respectively, were prepared. For FMOGS-OX6Pro::GUS, a 999bp DNA fragment containing the FMOGS-OX6 promoter was amplified with Pfu Turbo Cx Hotstart DNA polymerase (Stratagene) from genomic DNA using the PCR primers 5′-GGCTTAAUGAGTGGTTAATGTGCAACATCAGC-3′ and 5′-GGTTTAAUAGTATCAGTCAAAGTATTTGTTTCCTCG-3′. For FMOGS-OX7Pro::GUS, a 417 bp DNA fragment containing the FMOGS-OX7 promoter was amplified with Pfu Turbo Cx Hotstart DNA polymerase (Stratagene) from genomic DNA using the PCR primers 5′-GGCTTAAUAACGGGATTTTTGATTGGTT-3′ and 5′-GGTTTAAUTGTCAAGTCAATTAATGTCAATCTATCAG-3′. The PCR products were then cloned into the expression vector pCAMBIA3300 NLS-GUS by using the USER method as described by Nour-Eldin et al. (2006). pCAMBIA3300 NLS-GUS is generated from the original vector pCAMBIA3300 by adding the NLS (nuclear localization signal) and CDS of the GUS with User cloning sites (Nour-Eldin et al., 2006).
Using Agrobacterium-mediated transformation, all the constructs were transformed into Arabidopsis using the floral-dip method (Clough and Bent, 1998). T0 transgenic plants were selected on 1/2 Murashige and Skoog (MS) medium containing 50 μg⋅mL-1 kanamycin. For each construct, three independent confirmed T2 transgenic lines were used in the following analysis.
GSL Extraction and Analysis
35S::FMOGS-OX6, 35S::FMOGS-OX7, wild-type, FMOGS-OX6 knockout mutant, and FMOGS-OX7 knockout mutant plants were grown simultaneously for 24 days. Individual leaves from each plant were harvested for GSL measurement. 50 to 100 mg leaves were used for the extraction. GSL extraction was performed as previously described (Hansen et al., 2007). High performance liquid chromatography (HPLC) analysis was performed by using the method described by Pang et al. (2009).
Spatial Expression Analysis
The histochemical detection of GUS expression was performed as previously described (Jefferson et al., 1987). Plant materials were cut and incubated in substrate solution at 37°C for 12 h followed by removal of chlorophyll by submerging the samples in 96% ethanol. The samples were examined and photographed using a stereo microscope (Nikon SMZ1270) or a research microscope (Olympus CX21).
For analysis of the responsive expression pattern, seeds of FMOGS-OX6Pro::GUS, FMOGS-OX7Pro::GUS, and wild-type plants were vernalized at 4°C for 72 h and allowed to germinate in 1/2 MS medium for 5 days. The plants were then transferred to 1/2 MS medium containing 50 μM methyl jasmonate (MeJA), 200 μM salicylic acid (SA), and 10 μM abscisic acid (ABA), respectively, and were treated for 48 h. The whole seedling was used for GUS detection using the abovementioned method.
Responsive Gene Expression Analysis
Wild-type seeds were vernalized at 4°C for 72 h and allowed to germinate in 1/2 MS medium for 5 days. The seedlings were then transferred to 1/2 MS medium containing 100 μM indole acetic acid (IAA), 200 μM SA, 50 μM MeJA, 20 mM 1-aminocyclopropane-1-carboxylic acid (ACC), and 10 μM ABA, respectively. For the cold stress and heat stress treatments, seedlings were transferred to 4 and 30°C growing chamber, respectively. All the treatments were performed for 24 h.
Total RNA of the treated and control plants were isolated using TRIzol reagent (Invitrogen) and first-strand cDNA was synthesized using the SupermoIII RT kit (Bioteke). The primer pairs used in qRT-PCR for the detection of genes are listed in Supplementary Table S1. qRT-PCR was performed using SYBR Green Master Mix on an ABI 7500 sequence detection system. Relative transcript levels were normalized using ACTIN II as a control.
Results
Phylogenetic Analysis of FMOGS-OX Genes in Arabidopsis
Flavin-containing monooxygenases protein sequences containing the conserved FMOGS-OX domains were obtained from TAIR1 and were aligned using ClustalX. It was found that At1g12130 and At1g12160 were located in the same subclade, and together with the annotated FMOGS-OX1-5 formed a seven-protein group (Figure 1A), while At1g12200 was located in a subclade adjacent to the seven-protein group (Figure 1A).
Because both the overexpressor and knockout mutant of At1g12200 did not show the expected phenotype (Supplementary Tables S2 and S3), the possibility of At1g12200 to be a FMOGS-OX could be excluded. Thus, in the following study, we focused on At1g12130 (named FMOGS-OX6) and At1g12160 (named FMOGS-OX7) (Figure 1B).
Overexpression of FMOGS-OX6 and FMOGS-OX7 Altered MT GSLs and MS GSLs Profile
To confirm whether FMOGS-OX6 and FMOGS-OX7 possess S-oxygenation activity, transgenic plants 35S::FMOGS-OX6 and 35S::FMOGS-OX7 were generated and three independent lines of each FMOGS-OX6 and FMOGS-OX7 overexpressors were, respectively, used to analysis GSLs profile. Data of the line with the most significant phenotype was shown in Tables 1 and 2. These transgenic plants were confirmed to have much higher expression level than that in wild-type plants by RT-PCR analysis (Supplementary Figure S1). For each plant, the content of GSLs was detected in leaves and seeds of segregating progeny obtained from a heterozygous transgenic parent, thus the possible maternal effects could be minimized. Since the conversion of MT GSLs to MS GSLs rather than the absolute content of MS GSLs could better represent the S-oxygenation activity, MT: (MT+MS) was calculated from the HPLC data. Aliphatic GSLs with side chains of different lengths, including C3 (propyl), C4 (butyl), C5 (pentyl), C6 (hexyl), C7 (heptyl), and C8 (octyl), in the leaf and seed tissues of 35S::FMOGS-OX6, 35S::FMOGS-OX7, and wild-type are shown in Tables 1 and 2. In the seed, MT:(MT+MS) of all the detectable GSLs except propyl, were significantly lower in both 35S::FMOGS-OX6 and 35S::FMOGS-OX7 than in the wild-type. In the leaf, MT:(MT+MS) was lower in 35S::FMOGS-OX7 than in the wild-type, while it was not significantly changed in 35S::FMOGS-OX6. The significantly decreased MT:(MT+MS) in the seed tissue of the two overexpressors suggested that the FMOGS-OX6 and FMOGS-OX7 possess S-oxygenation activity for both short-chain and long-chain GSLs. The weak phenotype of overexpressors in the leaf tissue had been previously observed for other FMOGS-OX enzymes (Hansen et al., 2007; Li et al., 2008); this is possibly because of the low content of the substrate (MT GSLs) and the high content of the product (MS GSL) in the leaf, which showed impaired MT converting reaction.
Table 1.
Glucosinolates profile in 35S::FMOGS-OX6.
| MT:(MS+MT) | Leaf tissue |
Seed tissue |
||||
|---|---|---|---|---|---|---|
| WT | 35S:: FMOGS-OX6 | P-value | WT | 35S:: FMOGS-OX6 | P-value | |
| Propyl GSL (C3) | 0.11 ± 0.009 | 0.10 ± 0.005 | NS | 0.02 ± 0.001 | 0.01 ± 0.000 | NS |
| Butyl GSL (C4) | 0.28 ± 0.021 | 0.27 ± 0.022 | NS | 0.90 ± 0.010 | 0.57 ± 0.021 | <0.05 |
| Pentyl GSL (C5) | 0.33 ± 0.013 | 0.31 ± 0.011 | NS | 0.75 ± 0.039 | 0.41 ± 0.024 | <0.05 |
| Hexyl GSL (C6) | ND | ND | ND | ND | ||
| Heptyl GSL (C7) | 0.30 ± 0.012 | 0.31 ± 0.012 | NS | 0.77 ± 0.025 | 0.40 ± 0.019 | <0.05 |
| Octyl GSL (C8) | 0.17 ± 0.011 | 0.17 ± 0.012 | NS | 0.31 ± 0.013 | 0.14 ± 0.011 | <0.05 |
Data presented are mean values of MT:(MS+MT) ± Standard Error for at least three replicates per sample. P-value for MT:(MS+MT) differences between the two genotypes were determined by Student’s t-test. ND means given GSL was not detectable; therefore, no statistical analyses were conducted. NS means non-significant P-value (P > 0.05).
Table 2.
Glucosinolates profile in 35S::FMOGS-OX7.
| MT:(MS+MT) | Leaf tissue |
Seed tissue |
||||
|---|---|---|---|---|---|---|
| WT | 35S:: FMOGS-OX7 | P-value | WT | 35S:: FMOGS-OX7 | P-value | |
| Propyl GSL (C3) | 0.09 ± 0.004 | 0.06 ± 0.003 | <0.05 | 0.03 ± 0.007 | 0.04 ± 0.010 | NS |
| Butyl GSL (C4) | 0.39 ± 0.014 | 0.11 ± 0.003 | <0.05 | 0.83 ± 0.024 | 0.59 ± 0.011 | <0.05 |
| Pentyl GSL (C5) | 0.43 ± 0.009 | 0.30 ± 0.010 | <0.05 | 0.74 ± 0.021 | 0.38 ± 0.012 | <0.05 |
| Hexyl GSL (C6) | ND | ND | ND | ND | ||
| Heptyl GSL (C7) | 0.35 ± 0.011 | 0.20 ± 0.009 | <0.05 | 0.66 ± 0.012 | 0.31 ± 0.012 | <0.05 |
| Octyl GSL (C8) | 0.13 ± 0.008 | 0.06 ± 0.005 | <0.05 | 0.32 ± 0.014 | 0.13 ± 0.008 | <0.05 |
Data presented are mean values of MT:(MS+MT) ± Standard Error for at least three replicates per sample. P-value for MT:(MS+MT) differences between the two genotypes were determined by Student’s t-test. ND means given GSL was not detectable; therefore, no statistical analyses were conducted. NS means non-significant P-value (P > 0.05).
T-DNA knockout mutants FMOGS-OX6 and FMOGS-OX7 were, respectively, analyzed but no significant changes in the GSL profiles were detected (Supplementary Tables S4 and S5). Considering that there are five other identified FMOGS-OX enzymes with the same catalytic activity (Hansen et al., 2007; Li et al., 2008), the absence of observable phenotypic effects in the T-DNA knockout mutants could be attributable to the redundancy in gene function.
Expression Pattern of FMOGS-OX6 and FMOGS-OX7
To investigate the spatial expression patterns of FMOGS-OX6 and FMOGS-OX7, we generated the transgenic plants FMOGS-OX6Pro::GUS and FMOGS-OX7Pro::GUS, respectively, expressing the GUS under the control of FMOGS-OX6 and FMOGS-OX7 promoters. Promoter activities were detected by GUS staining, a histochemical technique. As shown in Figure 2, the two genes presented similar expression patterns. In seedlings, the GUS signal was detected in the vascular tissue throughout the radicle, hypocotyls, cotyledon, foliage leaf, and root in both FMOGS-OX6Pro::GUS and FMOGS-OX7Pro::GUS transgenic plants (Figures 2A–D,A’–D’). This expression pattern partially overlaps with that of other FMOGS-OX (Li et al., 2011). Similarly, FMOGS-OX5, FMOGS-OX6, and FMOGS-OX7 were expressed in the vascular tissue of the foliage, including the mid-vein and almost all side-veins, while the other four FMOGS-OX genes were expressed either only in the mid-vein and some major big veins or only in the side-veins (Li et al., 2011). Both the genes were expressed in the vascular tissue throughout the whole root (Figures 2D,D’), which is again similar to the expression pattern of FMOGS-OX5 and different from that of the other FMOGS-OX genes, which are expressed only in the connections of the taproot and lateral roots (Li et al., 2011).
FIGURE 2.
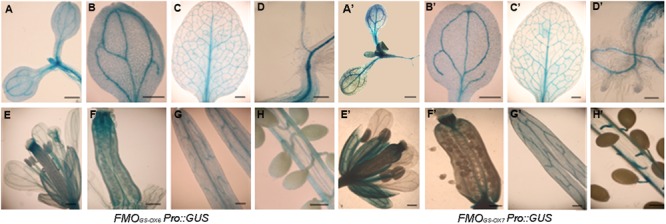
Expression patterns of FMOGS-OX6 and FMOGS-OX7 detected by promoter-driven GUS expression. (A,B,A’,B’) Cotyledons; (C,C’) foliage leaf; (D,D’) root and hypocotyl; (E,E’) flower; (F,F’) pistil; (G,G’) silique wall; (H,H’) Pseudoseptum and seeds; Bar = 0.3 mm.
For both FMOGS-OX6Pro::GUS and FMOGS-OX7Pro::GUS, in flowers, GUS signals were detected in the vascular bundles of calyces, carpels, and stamen filaments, but very little signal was detected in petals (Figures 2E–G,E’–G’). Dense GUS signal was clearly observed at the top and bottom of the ovary and seed funicles (Figures 2F,H,F’,H’). This expression pattern is quite similar to that of other FMOGS-OX genes (Li et al., 2011). In general, the spatial expression of FMOGS-OX6 and FMOGS-OX7 overlapped with each other to a large extent and showed a similar pattern in the vascular tissue to that of other GSL biosynthetic genes (Mikkelsen et al., 2000; Reintanz et al., 2001; Tantikanjana et al., 2001; Grubb et al., 2004; Levy et al., 2005; Skirycz et al., 2006; Li et al., 2011).
We further detected the expression of the two genes in response to exogenous ABA, MeJA, and SA. Both FMOGS-OX6 and FMOGS-OX7 were strongly inhibited by MeJA and induced by SA, whereas ABA promoted the expression of FMOGS-OX6 significantly but did not affect FMOGS-OX7 considerably (Figure 3).
FIGURE 3.
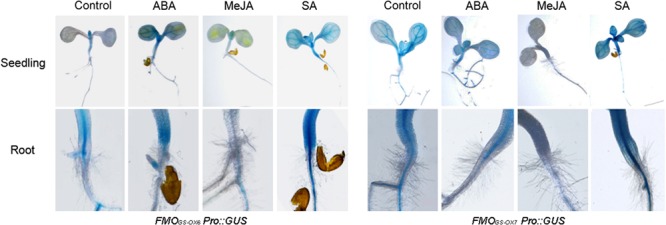
Expression of FMOGS-OX6 and FMOGS-OX7 in response to exogenous abscisic acid (ABA), methyl jasmonate (MeJA), and salicylic acid (SA).
Responsive Expression of FMOGS-OX Genes to Different Stimuli
The S-oxygenation of MT GSLs to MS GSLs is highly important for plant defense because of the bioactivities of MS GSLs against both biotic and abiotic stresses (Stotz et al., 2011; Martínez-Ballesta et al., 2013, 2015). To date, including FMOGS-OX6 and FMOGS-OX7, seven FMOGS-OX enzymes with redundant functions and overlapping spatial distribution have been identified. Therefore, we investigated whether the response of these seven FMOGS-OX genes to environmental stimuli was co-ordinated or varied. The promoter sequences of the seven FMOGS-OX genes were obtained from NCBI2 and analyzed using the web tool Plantcare 3. The elements in each of the promoter are listed in Supplementary Table S6.
Some common elements were found frequently among these promoters and the function of these elements is responsiveness to MeJA, ABA, auxin, ethylene, SA, cold, and heat stresses. To investigate the responsive expression of FMOGS-OX and other aliphatic GSL biosynthetic genes toward these hormone and environmental signals, 5-day old seedlings were treated with ABA, ACC, MeJA, SA, IAA, low temperature (4°C), and high temperature (30°C). Further, the expression level of the seven FMOGS-OX genes, together with that of MYB28 (encodes the main transcription factor that regulates aliphatic GSL biosynthetic genes) and CYP83A1 (an aliphatic GSL biosynthetic gene), were determined by qRT-PCR. Except FMOGS-OX4, which maintained relatively stable expression under different conditions, all the genes responded to the treatments (Figure 4). In general, the FMOGS-OX genes showed responsive expression pattern similar to that of the upstream genes (MYB28 and CYP83A1) in the aliphatic GSL biosynthesis pathway. For example, almost all genes were upregulated by ABA and ACC. However, to the same stimulus, different genes presented different level of sensitivity. FMOGS-OX1, FMOGS-OX2, FMOGS-OX6, and MYB28 showed upregulation (fivefolds), i.e., high sensitivity, toward ABA; FMOGS-OX5, FMOGS-OX6, FMOGS-OX7, and MYB28, toward ACC; FMOGS-OX1 and MYB28, toward MeJA; FMOGS-OX7 and MYB28, toward SA; and FMOGS-OX2, FMOGS-OX5, and MYB28, toward IAA. None of the aliphatic biosynthetic genes were very sensitive to cold and heat stresses.
FIGURE 4.

Responsive expression patterns of FMOGS-OX genes to different exogenous stimuli. The mRNA abundance of nine genes was analyzed by qRT-PCR. Samples for qRT-PCR were run in three biological replicates with three technical replicates and the data were represented as the Mean ± SD (n = 3). The relative gene expression was calculated using the ΔΔCt algorithm. The expression data were normalized using the invariant expression of ACTIN II. The leaves from mock control seedlings were used as reference sample, which was set to 1. Significant differences according to Student’s t-test are indicated: ∗P < 0.05, ∗∗P < 0.01.
Discussion
Aliphatic GSLs display high structural diversity due to variation in chain length and secondary modification, and the bioactivity of these compounds is largely dependent on the structure of the side chain. The chemical modification catalyzed by the different FMOGS-OX enzymes is highly important because MT GSLs and MS GSLs account for a large proportion of total GSLs, and production of MS GSLs is the basis of further modification of aliphatic GSLs.
The five annotated FMOGS-OX enzymes were identified by Hansen (Hansen et al., 2007) and Li (Li et al., 2008). In the study conducted by Hansen, a phylogenetic tree of plant FMOs from rice, Arabidopsis, and poplar was analyzed. Within the clade considered to be involved in S-oxygenation, the presence of a subclade of seven proteins, including the annotated FMOGS-OX1-5, FMOGS-OX6, and FMOGS-OX7, only in the GSL-producing Arabidopsis plant suggests that these genes may be responsible for S-oxygenation of GSLs. Our result verified Hansen’s prediction. Compared with the known FMOGS-OX enzymes, FMOGS-OX6 and FMOGS-OX7 was quite similar to FMOGS-OX1-4, which contribute to the conversion of both short and long-chain MT GSLs, and was different from FMOGS-OX5, which specifically recognizes long-chain substrates.
FMOGS-OX6 and FMOGS-OX7 showed very similar spatial expression patterns and it overlapped with that of most other GSL biosynthetic genes, which are frequently found in the vascular bundle. However, some small variations can be observed between these FMOGS-OX genes. Li et al. (2011) reported that in the leaf, FMOGS-OX1-4 were distributed partially in the veins and FMOGS-OX5 was expressed in the mid-vein and all the branched veins, whereas in the root, FMOGS-OX1-4 was limited to the connections between the taproot and lateral roots and FMOGS-OX5 was present throughout the vascular tissue in the whole root. Interestingly, FMOGS-OX6 and FMOGS-OX7, like FMOGS-OX1-4, catalyze S-oxygenation of GSLs without substrate specificity and exhibit less specific spatial expression like FMOGS-OX5. Based on the phylogenetic analysis of the FMOGS-OX genes in Arabidopsis (Figure 1A), our results suggest an FMOGS-OX ancestor with a broad range of non-specific substrates for catalysis and broad spatial expression. The gene encoding this ancestor could probably have been duplicated into two groups: one (comprising FMOGS-OX1-5) might have become more specific either in spatial expression (FMOGS-OX1-4) or in substrate recognition (FMOGS-OX5) and the other group (comprising FMOGS-OX6 and FMOGS-OX7) remained non-specific in both aspects.
Some common elements were detected in the promoter sequences of these FMOGS-OX genes. Consistently, the FMOGS-OX genes presented similar responsive expression tendency under several hormone treatments and temperature stresses. However, the sensitivity of each gene differed in response to the same treatment.
Integrating the knowledge of the various FMOGS-OX genes, we observed that they are quite redundant in their catalytic activity, distribution pattern, and response to exogenous stimuli. It is considered that duplicate genes play a more important role for the compensation of secondary metabolites than that of primary metabolites because there are fewer alternative pathways in secondary metabolite synthesis (Hanada et al., 2011). This redundancy of FMOGS-OX genes might play a significant role in functional compensation and increasing genetic robustness. Additionally, the variations found among these FMOGS-OX genes could possibly result in delicate functional difference and contribute to the finely tuned modification of aliphatic GSLs.
Aliphatic GSLs are primarily considered to contribute to plant resistance toward pests (Beekwilder et al., 2008). Recent studies discovered multiple functions of these compounds; they are involved in toxicity against pathogens and have been suggested to contribute to maintaining water balance under salt stress (Tierens et al., 2001; Stotz et al., 2011; Martínez-Ballesta et al., 2015). The versatile bioactivity of aliphatic GSLs is largely dependent on the length and modification of their side chain (Stotz et al., 2011). However, the contribution of specific metabolites to the biotic or abiotic stress defenses is still poorly understood. Identification of new FMOGS-OX enzymes provides a way to investigate the specific bioactivity of aliphatic GSLs with different side chain structures. As an application for humans, these FMOGS-OX genes can potentially be used in breeding Brassica vegetables with improved anti-cancer properties conferred by the MS GSLs.
Author Contributions
JL and WK carried out experiments and analyzed experiment results. QY, WC, and RX carried out partial of the experiments. JL and WK wrote the manuscript. WJ designed the experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) 31370334, NSFC 31170368, and Science and Technology Research Project of Education Department of Heilongjiang Province, 2531003.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01292
Expression levels of FMOGS-OX6 and FMOGS-OX7 in the overexpression mutants.
References
- Atwell L. L., Beaver L. M., Shannon J., Williams D. E., Dashwood R. H., Ho E. (2015). Epigenetic regulation by sulforaphane: opportunities for breast and prostate cancer chemoprevention. Curr. Pharmacol. Rep. 2 102–111. 10.1007/s40495-014-0002-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. 10.1126/science.1163732 [DOI] [PubMed] [Google Scholar]
- Beekwilder J., Leeuwen W. V., Dam N. M. V., Bertossi M., Grandi V., Mizzi L., et al. (2008). The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3:e2068 10.1371/journal.pone.0002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bones A. M., Rossiter J. T. (2006). The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67 1053–1067. 10.1016/j.phytochem.2006.02.024 [DOI] [PubMed] [Google Scholar]
- Chang Y. W., Jang J. Y., Kim Y. H., Kim J. W., Shim J. J. (2015). The effects of broccoli sprout extract containing sulforaphane on lipid peroxidation and Helicobacter pyloriinfection in the gastric mucosa. Gut Liver. 9 486–493. 10.5009/gnl14040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Fahey J. W., Haristoy X., Dolan P. M., Kensler T. W., Scholtus I., Stephenson K. K., et al. (2002). Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[α] pyreneinduced stomach tumors. Proc. Natl. Acad. Sci. U.S.A. 99 7610–7615. 10.1073/pnas.112203099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey J. W., Zhang Y., Talalay P. (1997). Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against?chemical?carcinogens. Proc. Natl. Acad. Sci. U.S.A. 94 10367–10372. 10.1073/pnas.94.19.10367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofaria N. M., Ranjan A., Kim S. H., Srivastava S. K. (2015). Mechanisms of the anticancer effects of isothiocyanates. Enzymes 37 111–137. 10.1016/bs.enz.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Grubb C. D., Zipp B. J., Ludwig-Muller J., Masuno M. N., Molinski T. F., Abel S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. 10.1111/j.1365-313X.2004.02261.x [DOI] [PubMed] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hanada K., Sawada Y., Kuromori T., Klausnitzer R., Saito K., Toyoda T., et al. (2011). Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol. Biol. Evol. 1 377–382. 10.1093/molbev/msq204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B. G., Kliebenstein D. J., Halkier B. A. (2007). Identification of a flavinmonooxygenaseas the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50 902–910. 10.1111/j.1365-313X.2007.03101.x [DOI] [PubMed] [Google Scholar]
- Hopkins R. J., Dam N. M. V., Loon J. J. A. V. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interaction. Annu. Rev. Entomol. 54 57–83. 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Jefferson R., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: ß-glucuronidase as a sensitive and versatile gene fusion marker inhigher plants. EMBO J. 16 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K., Prasad K. V., Savchenko T., Celesnik H., Dehesh K., Levy M., et al. (2012). The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 53 2008–2015. 10.1093/pcp/pcs143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Wang Q., Kaspi R., Parrella M. P., Abel S. (2005). Arabidopsis IQD1 a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43 79–96. 10.1111/j.1365-313X.2005.02435.x [DOI] [PubMed] [Google Scholar]
- Li J., Hansen B. G., Ober J. A., Kliebenstein D. J., Halkier B. A. (2008). Subclade of flavinmonooxygenasesinvolved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148 1721–1733. 10.1104/pp.108.125757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kristiansen K. A., Hansen B. G., Halkier B. A. (2011). Cellular andsubcellular localization of flavin-monooxygenases involved in glucosinolatebiosynthesis. J. Exp. Bot. 62 1337–1346. 10.1093/jxb/erq369 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43 222–226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ballesta M. C., Moreno-Fernández D. A., Carvajal M. (2013). The Physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int. J. Mol. Sci. 14 11607–11625. 10.3390/ijms140611607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ballesta M. C., Moreno-Fernández D. A., Castejón D., Ochando C., Morandini P. A., Carvajal M. (2015). The impact of the absence of aliphatic glucosinolates on water transport under salt stress in Arabidopsis thaliana. Front. Plant Sci. 6:524 10.3389/fpls.2015.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M. D., Hansen C. H., Wittstock U., Halkier B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. 10.1074/jbc.M001667200 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A. (2006). Advancing uracil-excision based cloning towards an ideal techniquefor cloning PCR fragments. Nucleic Acids Res. 34:e122 10.1093/nar/gkl635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Chen S., Li L., Yan X. (2009). Characterizationof glucosinolate-myrosinase system in developing salt cress Thellungiella halophila. Physiol. Plant 136 1–9. 10.1111/j.1399-3054.2009.01211.x [DOI] [PubMed] [Google Scholar]
- Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., et al. (2001). Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367. 10.2307/3871281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P., Faulkner K., Williamson G., Mithen R. (2000). 7-Methylsulfinylheptyl and 8-methylsulfinyloctyl isothiocyanates from watercress are potent inducers of phase II enzymes. Carcinogenesis 21 1983–1988. 10.1093/carcin/21.11.1983 [DOI] [PubMed] [Google Scholar]
- Rose P., Huang Q., Ong C. N., Whiteman M. (2005). Broccoli and watercress suppress matrix metalloproteinase-9 activity and invasiveness of human MDA-MB-231 breast cancer cells. Toxicol. Appl. Pharmacol. 209 105–113. 10.1016/j.taap.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47 10–24. 10.1111/j.1365-313X.2006.02767.x [DOI] [PubMed] [Google Scholar]
- Stotz H. U., Sawada Y., Shimada Y., Hirai M. Y., Sasaki E., Krischke M., et al. (2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 67 81–93. 10.1111/j.1365-313X.2011.04578.x [DOI] [PubMed] [Google Scholar]
- Talalay P., Fahey J. W., Healy Z. R., Wehage S. L., Benedict A. L., Min C., et al. (2007). Sulforaphane mobilizes cellular defenses that protect skin against damage by UV radiation. Proc. Natl. Acad. Sci. U.S.A. 104 17500–17505. 10.1073/pnas.0708710104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T., Yong J. W., Letham D. S., Griffith M., Hussain M., Ljung K., et al. (2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Gene Dev. 15 1577–1588. 10.1101/gad.887301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierens K. J., Thomma B., Brower M., Schmidt J., Kistner K., Porzel A., et al. (2001). Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125 1688–1699. 10.1104/pp.125.4.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalcmann A. T., Talalay P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ober J. A., Kliebenstein D. J. (2006). The gene controlling thequantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolatehydrolysis and insect resistance in Arabidopsis. Plant Cell 18 1524–1536. 10.1105/tpc.105.039602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of FMOGS-OX6 and FMOGS-OX7 in the overexpression mutants.